Effects of an IgE receptor polymorphism acting on immunity, susceptibility to infection, and reproduction in a wild rodent
Figures
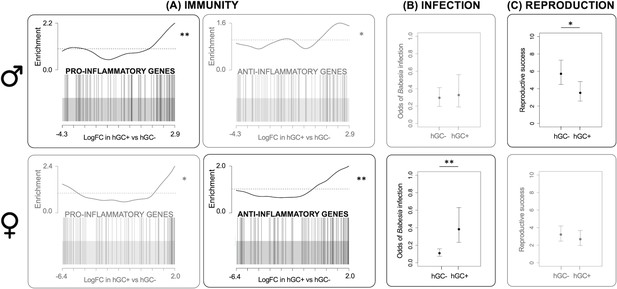
Effects of GC haplotype (hGC).
Upper panel: males. Lower panel: females: (A) Unstimulated immune gene expression: Barcode plots showing enrichment of the GO terms GO:0050729 (pro-inflammatory genes) and GO:0050728 (anti-inflammatory genes) in unstimulated splenocytes taken from individuals with (hGC+) vs. without (hGC-) the haplotype, showing that males with the haplotype have a pro-inflammatory bias, whereas females have an anti-inflammatory bias. In each plot, x-axis shows log fold change (logFC) in hGC+ vs. hGC-, black bars represent genes annotated with the GO terms and the worm shows relative enrichment. (B) Susceptibility to infection: association between hGC and the odds of infection with Babesia microti, showing that females with the haplotype have an increased susceptibility to infection (from a GLMM). (C) Reproduction: association between hGC and reproductive success, showing that males with the haplotype have lower reproductive success (from a GLM) (error bars represent ± standard error; *p<0.05; **p<0.01; see Table 1 for sample sizes). GLM = generalized linear model; GLMM = generalized linear mixed effects model.
Tables
Model specifications including, for each main model, covariates included in the full model, datasets used, and sample sizes (F = included as a fixed effect; R = included as a random effect).
| DGE analysis | Haplotype association analyses | ||||
|---|---|---|---|---|---|
| Response variable | |||||
| Immune gene expression | Parasite infection | Reproductive success | SOD1activity | ||
| Covariates Snout-vent length | F | F | |||
| Eye lens weight | F | F | |||
| Reproductive status | F | F | F | ||
| Body condition | F | F | F | ||
| Birth month | F | ||||
| Culled or not | F | ||||
| Site | F; R (Il17a LMM) | F (Macro); R (Micro) | F | F | |
| Year | F | F | F | F | |
| Season | F; R (Il17a LMM) | F (Macro); R (Micro) | F | ||
| Individual ID | R (Micro) | ||||
| Assay plate | R (Il17a LMM) | ||||
| Dataset | C | C | C (Macro) L (Micro) | C+L | C |
| Sample size | ♀31 ♂53 | Il17a LMM: ♀73 ♂220 | Macro: ♀82 ♂235 B. microti: ♀1075 ♂1247 Bartonella spp.: ♀1283 ♂1104 | ♀419 ♂232 | ♀81 ♂227 |
-
C = cross-sectional; L = longitudinal; DGE = differential gene expression; LMM = linear mixed effects model.
Position of single-nucleotide polymorphisms (SNPs) and other key features in the Fcer1a gene.
Features lie in scaffold CADCXT010006977 within assembly GCA_902806775.
| Feature | Start | End |
|---|---|---|
| Fcer1a gene | 10745528 | 10745528 |
| Exon 5 | 10745528 | 10745881 |
| Exon 4 | 10746818 | 10747076 |
| Exon 3 | 10748609 | 10748864 |
| Exon 2 | 10750044 | 10750065 |
| Exon 1 | 10750465 | 10750642 |
| 5’ UTR | 10750520 | 10750642 |
| CDS | 10750465 | 10750520 |
| CDS | 10750044 | 10750065 |
| CDS | 10748609 | 10748864 |
| CDS | 10746818 | 10747076 |
| CDS | 10745729 | 10745881 |
| 3’ UTR | 10745528 | 10745729 |
| SNP 2 | 10748718 | 10748719 |
| SNP 1 | 10746846 | 10746847 |
Top 10 annotated genes that were differentially expressed between males with vs. without the GC haplotype, including associated log fold changes (logFC), p-values, and q-values (false discovery rate [FDR]-corrected p-values).
| Gene | Protein | LogFC | p-value | q-value |
|---|---|---|---|---|
| Snai3 | Snail family zinc finger 3 | 1.698 | 1.220 × 10–9 | 1.574 × 10–5 |
| Pla2g4c | Phospholipase A2, group IVC (cytosolic, calcium-independent) | 2.904 | 6.386 × 10–9 | 4.120 × 10–5 |
| Il33 | Interleukin 33 | 2.756 | 1.474 × 10–7 | 4.755 × 10–4 |
| Mmp13 | Matrix metallopeptidase 13 | 2.571 | 4.427 × 10–7 | 1.142 × 10–3 |
| Uba7 | Ubiquitin-like modifier activating enzyme 7 | 0.771 | 5.979 × 10–7 | 1.286 × 10–3 |
| Robo4 | Roundabout guidance receptor 4 | 0.892 | 3.738 × 10–6 | 4.385 × 10–3 |
| Ttn | Titin | 1.665 | 8.995 × 10–6 | 7.254 × 10–3 |
| Flnc | Filamin C, gamma | 1.554 | 1.245 × 10–5 | 9.447 × 10–3 |
| Crb2 | Crumbs family member 2 | 1.327 | 1.567 × 10–5 | 1.063 × 10–2 |
| Muc16 | Mucin 16 | 1.937 | 1.971 × 10–5 | 1.156 × 10–2 |
Annotated genes that were differentially expressed (q ≤ 0.1) between females with vs. without the GC haplotype, including associated log fold changes (logFC), p-values, and q-values (false discovery rate [FDR]-corrected p-values).
| Gene | Protein | LogFC | p-value | q-value |
|---|---|---|---|---|
| Tldc1 | TBC/LysM associated domain containing 1 | 1.489 | 2.502 × 10–8 | 3.228 × 10–4 |
| Peg3 | Paternally expressed 3 | 1.440 | 5.369 × 10–5 | 0.049 |
| Socs3 | Suppressor of cytokine signaling 3 | 1.071 | 6.061 × 10–5 | 0.052 |
Top 10 annotated genes that were differentially expressed between males with vs. without the GC haplotype when controlling for cestode burden, including associated log fold changes (logFC), p-values, and q-values (false discovery rate [FDR]-corrected p-values).
| Gene | Protein | LogFC | p-value | q-value |
|---|---|---|---|---|
| Il33 | Interleukin 33 | 3.091 | 2.473 × 10–10 | 3.191 × 10–6 |
| Snai3 | Snail family zinc finger 3 | 1.672 | 5.259 × 10–9 | 3.393 × 10–5 |
| Pla2g4c | Phospholipase A2, group IVC (cytosolic, calcium-independent) | 2.883 | 1.562 × 10–8 | 6.721 × 10–5 |
| Uba7 | Ubiquitin-like modifier activating enzyme 7 | 0.809 | 2.477 × 10–8 | 7.990 × 10–5 |
| Mmp13 | Matrix metallopeptidase 13 | 2.606 | 4.090 × 10–7 | 8.796 × 10–4 |
| Robo4 | Roundabout guidance receptor 4 | 0.892 | 8.160 × 10–6 | 9.572 × 10–3 |
| Ttn | Titin | 1.599 | 2.532 × 10–5 | 1.951 × 10–2 |
| Flnc | Filamin C, gamma | 1.493 | 2.721 × 10–5 | 1.951 × 10–2 |
| Crb2 | Crumbs family member 2 | 1.261 | 3.512 × 10–5 | 2.151 × 10–2 |
Annotated genes that were differentially expressed (q ≤ 0.1) between females with vs. without the GC haplotype when controlling for cestode burden, including associated log fold changes (logFC), p-values, and q-values (false discovery rate [FDR]-corrected p-values).
| Gene | Protein | LogFC | p-value | q-value |
|---|---|---|---|---|
| Tldc1 | TBC/LysM associated domain containing 1 | 1.489 | 2.890 × 10–8 | 3.730 × 10–4 |
| Socs3 | Suppressor of cytokine signaling 3 | 1.083 | 4.133 × 10–5 | 0.041 |
| Peg3 | Paternally expressed 3 | 1.411 | 6.375 × 10–5 | 0.055 |
| Ppp1r3c | Protein phosphatase 1, regulatory (inhibitor) subunit 3C | 1.662 | 8.798 × 10–5 | 0.071 |
Significance values from hapassoc models for expression of 18 genes (assayed by Q-PCR) in splenocytes.
Splenocytes were stimulated with anti-CD3 and anti-CD28 antibodies in order to promote the proliferation of T-cells. q-values (false discovery rate [FDR]-corrected p-values) are reported alongside original p-values for the genotype by sex interaction.
| Gene | p-value | q-value |
|---|---|---|
| Cd4 | 0.124 | 0.822 |
| Cd8a | 0.744 | 0.866 |
| Foxp3 | 0.499 | 0.845 |
| Gata3 | 0.563 | 0.845 |
| Il10 | 0.866 | 0.866 |
| Mpo | 0.173 | 0.822 |
| Tbx21 | 0.650 | 0.866 |
| Tgfb1 | 0.271 | 0.822 |
| Ifng | 0.399 | 0.822 |
| Il17a | 0.002 | 0.037 |
| Il1b | 0.749 | 0.866 |
| Il6 | 0.282 | 0.822 |
| Ms4a1 | 0.281 | 0.822 |
| Orai1 | 0.353 | 0.822 |
| Tnfa | 0.858 | 0.866 |
| Il2 | 0.411 | 0.822 |
| Apobr | 0.857 | 0.866 |
| Arg1 | 0.552 | 0.845 |
Estimates, standard errors, and z-statistics from best LMM for Yeo-Johnson-transformed Il17a expression levels.
| Estimate | SE | z | |
|---|---|---|---|
| (Intercept) | 0.107 | 0.023 | 4.61 |
| Genotype | –0.115 | 0.037 | –3.10 |
| Sex male | –0.055 | 0.019 | –2.89 |
| Year 2016 | 0.084 | 0.018 | 4.60 |
| Genotype × sex male | 0.143 | 0.044 | 3.25 |
Effect sizes, standard errors, z-statistics, and associated significance from Gaussian hapassoc model for SOD1 activity.
| Estimate | SE | z | p-value | |
|---|---|---|---|---|
| (Intercept) | 0.614 | 1.514 | 0.406 | 0.685 |
| hAT | –0.453 | 0.223 | –2.032 | 0.042 |
| Reproductive status active | –0.112 | 0.266 | –0.422 | 0.673 |
| Sex male | 0.031 | 0.250 | 0.124 | 0.901 |
| SVL | 0.016 | 0.017 | 0.945 | 0.345 |
| Lens weight 2 | 0.568 | 0.531 | 1.070 | 0.284 |
| Lens weight 3 | 0.440 | 0.549 | 0.801 | 0.423 |
| Lens weight 4 | 0.445 | 0.587 | 0.758 | 0.448 |
| Lens weight 5 | 0.415 | 0.653 | 0.636 | 0.525 |
| Lens weight 6 | 0.301 | 0.752 | 0.400 | 0.689 |
| Lens weight 7 | –0.307 | 1.095 | –0.280 | 0.779 |
| Body condition | 0.020 | 0.030 | 0.677 | 0.498 |
| Site CHE | 1.429 | 0.431 | 3.316 | 0.001 |
| Site COL | 2.675 | 1.099 | 2.434 | 0.015 |
| Site GRD | 1.476 | 0.437 | 3.379 | 0.001 |
| Site RAV | 0.904 | 0.918 | 0.984 | 0.325 |
| Site SCP | 1.107 | 0.419 | 2.643 | 0.008 |
| Year 2016 | 0.593 | 0.242 | 2.457 | 0.014 |
| Season early summer | –0.392 | 0.389 | –1.007 | 0.314 |
| Season late summer | –0.098 | 0.266 | –0.369 | 0.712 |
| Season spring | –0.969 | 0.383 | –2.529 | 0.011 |
Effect sizes, standard errors, and z-statistics from best binomial GLMM for probability of infection with Babesia microti.
| Estimate | SE | z | |
|---|---|---|---|
| (Intercept) | –2.214 | 0.377 | –5.87 |
| Genotype | 1.254 | 0.416 | 3.02 |
| Sex male | 0.957 | 0.229 | 4.19 |
| Year 2016 | 1.260 | 0.277 | 4.55 |
| Year 2017 | 1.047 | 0.302 | 3.46 |
| Reproductive status active | 0.754 | 0.144 | 5.24 |
| Body condition | 0.044 | 0.016 | 2.76 |
| Genotype × sex male | –1.116 | 0.615 | –1.82 |
Effect sizes, standard errors, and z-statistics from best binomial GLMM for probability of infection with Bartonella spp.
| Estimate | SE | z | |
|---|---|---|---|
| (Intercept) | 0.236 | 0.369 | 0.64 |
| Year 2016 | 0.740 | 0.135 | 5.49 |
| Year 2017 | 2.280 | 0.175 | 13.01 |
Effect sizes, standard errors, z-statistics, and associated significance from Gaussian hapassoc model for macroparasite infection summarized by a single principal component.
| Estimate | SE | z | p-value | |
|---|---|---|---|---|
| (Intercept) | 0.050 | 0.331 | 0.152 | 0.879 |
| hAC | –0.102 | 0.130 | –0.786 | 0.432 |
| hAT | –0.108 | 0.158 | –0.682 | 0.495 |
| Sex male | –0.271 | 0.126 | –2.146 | 0.032 |
| Season early summer | –0.103 | 0.182 | –0.567 | 0.571 |
| Season late summer | 0.012 | 0.132 | 0.090 | 0.929 |
| Season spring | 0.327 | 0.176 | 1.854 | 0.064 |
| Reproductive status active | –0.222 | 0.119 | –1.868 | 0.062 |
| Body condition | –0.019 | 0.014 | –1.328 | 0.184 |
| Year 2016 | –0.473 | 0.124 | –3.810 | 0.000 |
| Site CHE | 0.714 | 0.226 | 3.157 | 0.002 |
| Site COL | 0.762 | 0.575 | 1.327 | 0.185 |
| Site GRD | 0.707 | 0.229 | 3.090 | 0.002 |
| Site RAV | 1.100 | 0.481 | 2.288 | 0.022 |
| Site SCP | 0.747 | 0.220 | 3.389 | 0.001 |
Effect sizes, standard errors, z-statistics, and associated significance from best quasi-Poisson GLM for reproductive success in males.
| Estimate | SE | z | p-value | |
|---|---|---|---|---|
| (Intercept) | 1.743 | 0.243 | 7.171 | 1.05 × 10–11 |
| Genotype | –0.484 | 0.237 | –2.042 | 0.042 |
| Year 2016 | 0.592 | 0.185 | 3.193 | 0.002 |
| Year 2017 | –0.437 | 0.199 | –2.193 | 0.029 |
| Birth month | –0.358 | 0.048 | –7.449 | 1.97 × 10–12 |
| Culled yes | 0.422 | 0.150 | 2.809 | 0.005 |
Effect sizes, standard errors, z-statistics, and associated significance from best quasi-Poisson GLM for reproductive success in females.
| Estimate | SE | z | p-value | |
|---|---|---|---|---|
| (Intercept) | 1.137 | 0.264 | 4.298 | 2.15 × 10–5 |
| Year 2016 | 0.520 | 0.211 | 2.466 | 0.014 |
| Year 2017 | –0.118 | 0.215 | –0.549 | 0.584 |
| Birth month | –0.298 | 0.047 | –6.391 | 4.42 × 10–10 |
Effect sizes, standard errors, z-statistics, and associated significance from best quasi-Poisson GLM for reproductive success in males when controlling for Babesia microti and Bartonella spp. infection.
| Estimate | SE | z | p-value | |
|---|---|---|---|---|
| (Intercept) | 1.467 | 0.418 | 3.513 | 7.64 × 10–4 |
| Genotype | –1.081 | 0.425 | –2.544 | 0.013 |
| Year 2016 | 0.238 | 0.279 | 0.854 | 0.396 |
| Year 2017 | –0.912 | 0.386 | –2.338 | 0.022 |
| Birth month | –0.464 | 0.091 | –5.090 | 2.69 × 10–6 |
| Proportion of samples Babesia-positive | 0.849 | 0.276 | 3.076 | 0.003 |
| Proportion of samples Bartonella-positive | 0.997 | 0.440 | 2.268 | 0.026 |
Effect sizes, standard errors, z-statistics, and associated significance from best quasi-Poisson GLM for reproductive success in females when controlling for Babesia microti and Bartonella spp. infection.
| Estimate | SE | z | p-value | |
|---|---|---|---|---|
| (Intercept) | 1.483 | 0.305 | 4.867 | 3.20 × 10–6 |
| Birth month | –0.310 | 0.073 | –4.229 | 4.38 × 10–5 |
| Proportion of samples Babesia-positive | 0.569 | 0.247 | 2.308 | 0.0226 |
Panel of 18 genes for which expression levels in splenocytes stimulated with anti-CD3 and anti-CD28 antibodies were measured using two-step reverse transcription quantitative PCR (Q-PCR).
| Gene | Protein |
|---|---|
| Cd4 | T-cell surface glycoprotein CD4 |
| Cd8a | T-cell surface glycoprotein CD8 alpha chain |
| Foxp3 | Forkhead box protein P3 |
| Gata3 | GATA binding protein 3 |
| Il10 | Interleukin-10 |
| Mpo | Myeloperoxidase |
| Tbx21 | T-box transcription factor TBX21 |
| Tgfb1 | Transforming growth factor beta 1 |
| Ifng | Interferon gamma |
| Il17a | Interleukin-17a |
| Il1b | Interleukin-1 beta |
| Il6 | Interleukin-6 |
| Ms4a1 | B-lymphocyte antigen CD20 |
| Orai1 | Calcium release-activated calcium channel protein 1 |
| Tnfa | Tumor necrosis factor alpha |
| Il2 | Interleukin-2 |
| Apobr | Apolipoprotein B receptor |
| Arg1 | Arginase-1 |
Loadings from principal component analysis summarizing infection by macroparasites (ticks, fleas, and cestodes).
| Macroparasite | PC1 |
|---|---|
| Cestodes | –0.481 |
| Ticks | –0.592 |
| Fleas | –0.646 |





