Dynamics of allosteric regulation of the phospholipase C-γ isozymes upon recruitment to membranes
Figures
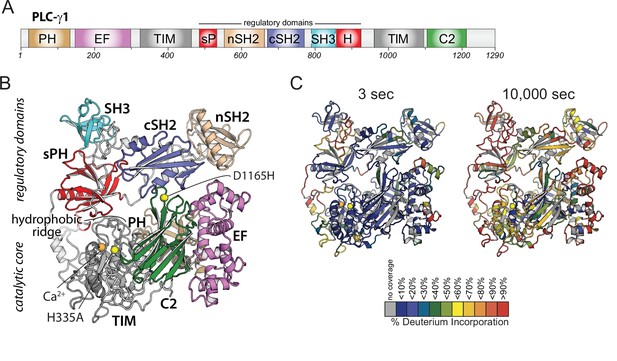
Domain architecture of phospholipase C-γ1 (PLC-γ1) and initial hydrogen-deuterium exchange mass spectrometry time course.
(A) Domain schematic of PLC-γ1. Full-length PLC-γ1 possesses a set of regulatory domains inserted within its catalytic core. (B) Structure of PLC-γ1. In the basally autoinhibited state shown here (PDB ID: 6PBC), the regulatory domains prevent access to membranes by the catalytic core demarked by a Ca2+ cofactor (orange). His335 is also within the catalytic core and its substitution (H335A) renders the isozyme catalytically inactive (yellow). Asp1165 substitution (D1165H) disrupts the interface between the core and the regulatory array leading to a constitutively active lipase (also yellow). (C) Relative levels of deuterium incorporation are colored on the structure of PLC-γ1 (residue 21–1215) according to the legend at either 3 or 10,000 s of D2O exposure.
-
Figure 1—source data 1
Raw mass spectral data of deuterium incorporation per peptide.
- https://cdn.elifesciences.org/articles/77809/elife-77809-fig1-data1-v1.xlsx
-
Figure 1—source data 2
Unprocessed images.
Raw images of gels used to produce final figures.
- https://cdn.elifesciences.org/articles/77809/elife-77809-fig1-data2-v1.zip
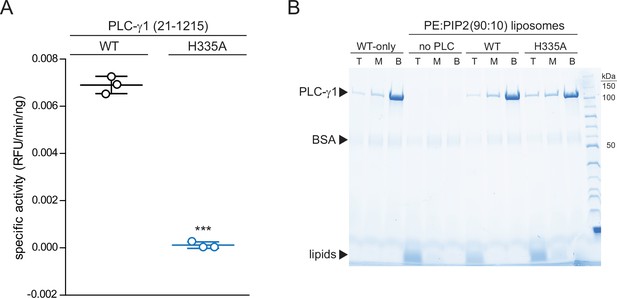
Phospholipase C-γ1 (PLC-γ1) (H335A) is catalytically inactive while retaining capacity to bind liposomes.
(A) Specific activities measured with the membrane-embedded substrate XY-69. XY-69 (0.5 μM) was incorporated into liposomes containing phosphatidylethanolamine:phosphatidylinositol 4,5-bisphosphate (PE:PIP2) (80:20) prior to addition of 10 nM of wild-type (WT) PLC-γ1 or PLC-γ1 (H335A). Phospholipase activities were determined by quantifying XY-69 hydrolysis in real time and presented as the mean ± SD of three independent experiments, each with three or more technical replicates. Statistical significance was determined with a t-test represented as *** for a p<0.05. (B) Membrane flotation assay. Indicated proteins were incubated with liposomes of PE:PIP2:nitro-2–1,3-benzoxadiazol-4-yl-PE (89.8:10:0.2) and centrifuged in a sucrose gradient (0–30%). After centrifugation, top (T), middle (M), and bottom (B) fractions were collected followed by SDS-PAGE and staining with Coomassie Brilliant Blue.
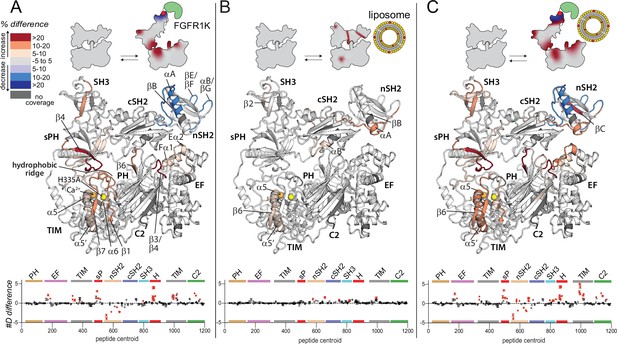
Widespread changes in deuterium exchange of phospholipase C-γ1 (PLC-γ1) upon binding kinase domain of fibroblast growth factor receptor (FGFR1K).
Significant differences in deuterium incorporation are mapped on the structure of PLC-γ1 (H335A) according to the legend for the following three states: alone versus bound to phosphorylated kinase domain of fibroblast growth factor receptor (FGFR1K) (A), alone versus in the presence of liposomes containing phosphatidylethanolamine:phosphatidylinositol 4,5-bisphosphate (90:10) (B), or alone versus bound to both pFGFR1K and liposomes (C). In addition, Figure 2—figure supplement 4 shows differences in exchange of PLC-γ1 (H335A) bound to either liposomes or pFGFR1K relative to a final state with PLC-γ1 (H335A) bound to both liposomes and pFGFR1K. Significant differences in any peptide required three specific conditions: greater than both a 5% and 0.4 Da difference in exchange at any time point (3, 30, 300, 3000, and 10,000 s), and a two-tailed, unpaired t-test of p<0.01. The #D difference for each condition is graphed below, which shows the total difference in deuterium incorporation over the entire hydrogen-deuterium exchange time course, with each point indicating a single peptide (error shown as SD [n=3]). Each circle represents the central residue of a corresponding peptide with the full deuterium exchange information for all peptides available in the source data. Individual peptides with a significant difference as defined above are colored red.
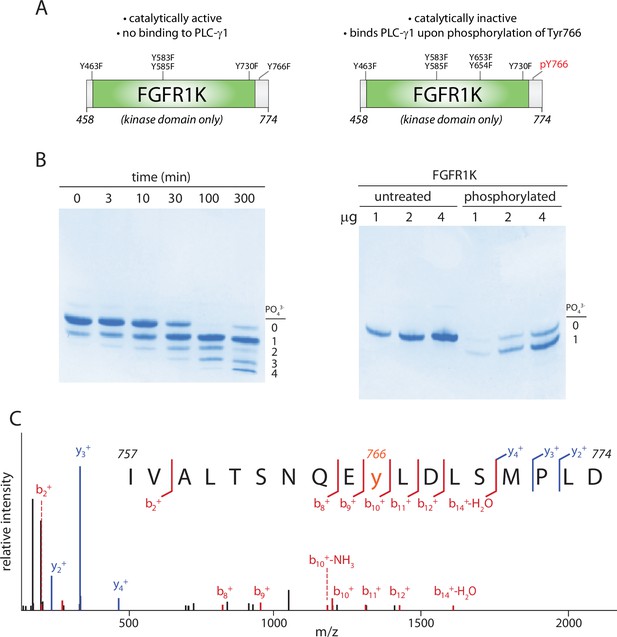
Kinase domain of fibroblast growth factor receptor (FGFR1K) specifically phosphorylated on Tyr766.
(A) Schematic representation of the kinase domain (residues 458–774) of FGFR1K. The six tyrosines mutated to phenylalanine to prevent phosphorylation and render the kinase resistant to phosphorylation-dependent activation are listed. Phosphorylation of Tyr766 is shown in red. (B) Time course of phosphorylation (left) and preparative phosphorylation (right) of catalytically inactive FGFR1K (100 μM) using equimolar concentrations of an equivalent, catalytically active fragment. Reactions were terminated by separating the two forms of FGFR1K using affinity chromatography; native PAGE was subsequently used to separate phosphorylated forms of the catalytically inactive version followed by staining with Coomassie Brilliant Blue. Preparative phosphorylation was carried out for 100 min. (C) Catalytically inactive FGFR1K was phosphorylated specifically at Tyr766 as determined by LC-MS/MS.
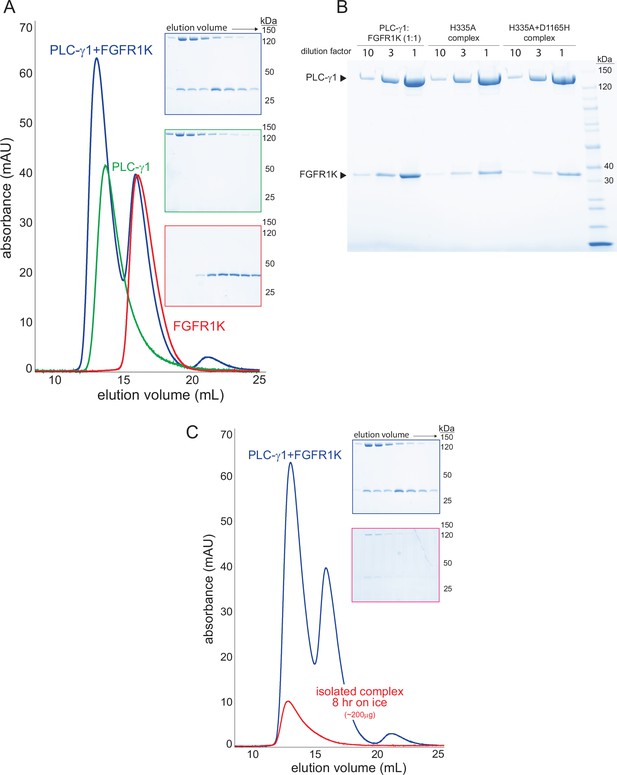
Stable complexes of phospholipase C-γ1 (PLC-γ1) and phosphorylated kinase domain of fibroblast growth factor receptor (FGFR1K).
(A) PLC-γ1 (H335A) or PLC-γ1 (H335A+D1165H) were mixed with a two-fold molar excess of phosphorylated FGFR1K and isolated by size exclusion chromatography; representative chromatograms are shown with fractions visualized after SDS-PAGE and staining with Coomassie Brilliant Blue (insets). (B) Final complexes were visualized similarly. For reference, a 1:1 mixture of PLC-γ1 and FGFR1K was also visualized. (C) Stability of the complex of PLC-γ1 (H335A) and FGFR1K was assessed by size exclusion chromatography after 8 hr on ice (red curve). Blue curve is duplicated from panel A and shown for reference.

Potential mechanism of priming of phospholipase C-γ1 (PLC-γ1) by kinase domain of fibroblast growth factor receptor (FGFR1K).
Structural superimposition of autoinhibited PLC-γ1 (PDB code: 6PBC) and the tandem SH2 domains of PLC-γ1 (light pink) bound to FGFR1K (green; PDB code: 3GQI). Structures superimposed using the nSH2 domains only. Autoinhibited PLC-γ1 is colored gray with regions of differential exchange upon binding FGFR1K colored as in Figure 2A. AMPPCP is a non-hydrolyzable analog of ATP. Red arrows highlight changes in the position of loops within the tandem SH2 domains.
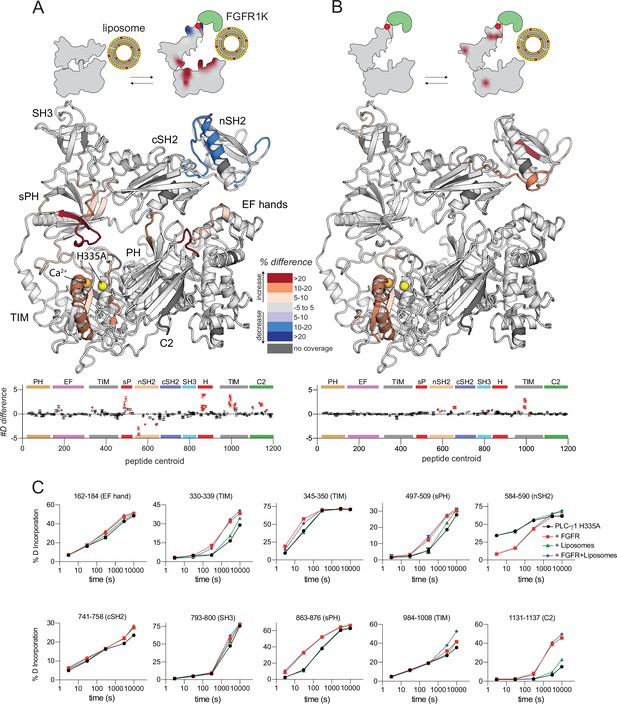
Kinase domain of fibroblast growth factor receptor (FGFR1K) and liposomes act essentially independently to affect sites of exchange within phospholipase C-γ1 (PLC-γ1).
Differences in deuterium incorporation was measured for PLC-γ1 (H335A) bound to FGFR1K and in the presence of liposomes containing phosphatidylethanolamine:phosphatidylinositol 4,5-bisphosphate (PE:PIP2; 90:10). Differences in deuterium incorporation were calculated relative to PLC-γ1 (H335A) in the presence of PE:PIP2 (90:10) liposomes (A) or PLC-γ1 (H335A) bound to pFGFR1K (B). Peptides that showed significant difference in deuterium incorporation (>5% and >0.4 Da difference in exchange at any time point, with a two-tailed, unpaired t-test of p<0.01) are mapped onto the structure. For each comparison, the difference in the number of incorporated deuterons was averaged over all time points and shown below the structures as the mean ± SD (n=3). Each circle represents the central residue of a corresponding peptide; red circles indicate peptides with a significant difference between conditions. (C) Percent deuterium incorporation for select peptides spanning PLC-γ1 (H335A). Percent deuterium incorporation is shown for PLC-γ1 (H335A) alone (Apo; black line), bound to FGFR1K (red line), in the presence of PE:PIP2 (90:10) liposomes (green line), and with both kinase and liposomes (blue line). Percent deuterium incorporation is the mean ± SD (n=3), with most SD smaller than the size of the point. The full deuterium incorporation data for all peptides are shown in the source data.
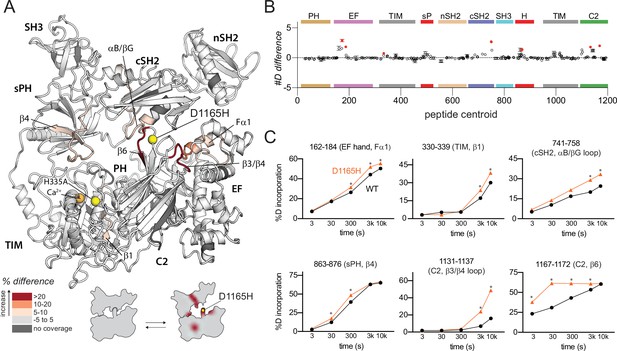
Oncogenic substitution of phospholipase C-γ1 (PLC-γ1) mimics kinase engagement.
(A) Significant differences in deuterium incorporation for PLC-γ1 (H335A) versus the oncogenic mutant of PLC-γ1 (H335A+D1165H) were determined and mapped on the structure of PLC-γ1. (B) The #D difference upon mutation shows the total difference in deuterium incorporation over the entire hydrogen-deuterium exchange time course, with each point indicating a single peptide (error shown as SD [n=3]). Individual peptides with a significant difference between conditions (defined as greater than both a 5% and 0.4 Da difference in exchange at any time point, and a two-tailed, unpaired t-test of p<0.01) are colored red. (C) A selection of peptides showing significant differential exchange at any time point (data shown as mean ± SD [n=3]), asterisks indicate significant time points for peptides as defined above. Full deuterium exchange data is available in the source data.
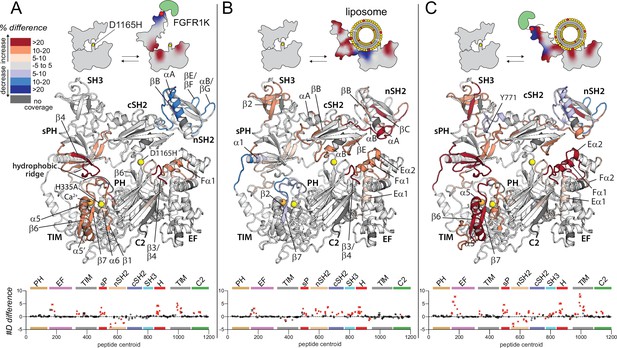
Oncogenic substitution uncovers functional cooperativity within phospholipase C-γ1 (PLC-γ1).
Differences in deuterium incorporation were measured for PLC-γ1 (H335A+D1165H) in three states: alone versus bound to phosphorylated kinase domain of fibroblast growth factor receptor (pFGFR1K) (A), alone versus in the presence of phosphatidylethanolamine:phosphatidylinositol 4,5-bisphosphate (90:10) liposomes (B), or alone versus with both pFGFR1K and liposomes present (C). In addition, Figure 4—figure supplement 1 shows differences in exchange of PLC-γ1 (H335A+D1165H) bound to either liposomes or pFGFR1K relative to a final state with PLC-γ1 (H335A+D1165H) bound to both liposomes and pFGFR1K. Differences in deuterium exchange for a series of time points (3, 30, 300, 3000, and 10,000 s) were calculated relative to PLC-γ1 (H335A+D1165H) alone and peptides with significant differential exchange at any time point (both a 5% and 0.4 Da difference in exchange at any time point, and a two-tailed, unpaired t-test of p<0.01) mapped onto the structure. The #D difference for each condition is graphed below, which shows the total difference in deuterium incorporation over the entire hydrogen-deuterium exchange time course, with each point indicating a single peptide (error shown as SD [n=3]); peptides with significant differences between conditions as defined above are red.
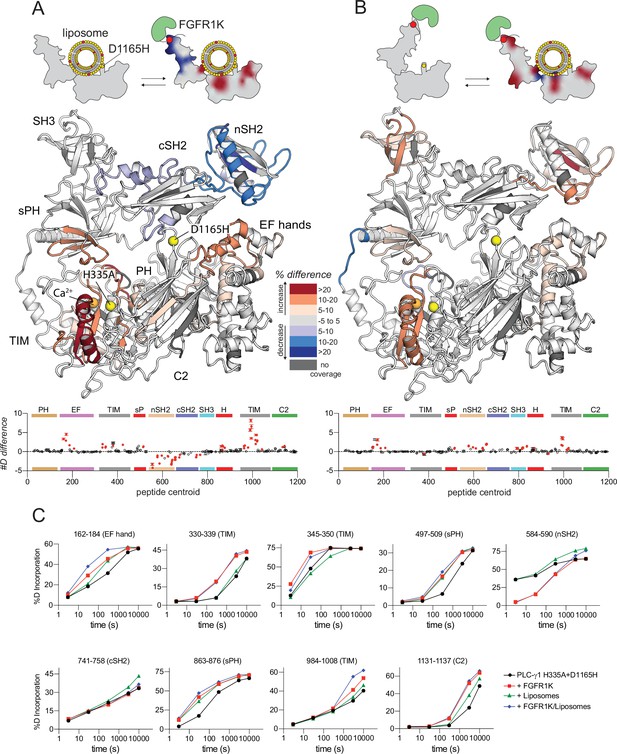
Kinase domain of fibroblast growth factor receptor (FGFR1K) and liposomes cooperate to affect the deuterium exchange of phospholipase C-γ1 (PLC-γ1) (D1165H).
Deuterium incorporation was measured for PLC-γ1 (H335A+D1165H) bound to FGFR1K in the presence of phosphatidylethanolamine:phosphatidylinositol 4,5-bisphosphate (PE:PIP2; 90:10) liposomes. Differences in deuterium incorporation were calculated relative to PLC-γ1 (H335A+D1165H) in the presence of PE:PIP2 liposomes (A) or bound to pFGFR1K (B). Peptides that showed significant H/D exchange differences (by the following criteria: >5% and >0.4 Da difference in exchange at any time point [3, 30, 300, 3000, and 10,000 s], with a two-tailed, unpaired t-test of p<0.01) are mapped onto the structure. For each comparison, the difference in the number of incorporated deuterons was averaged over all time points and shown below the structures as the mean ± SD (n=3). Each circle represents the central residue of a corresponding peptide; red circles indicate peptides with a significant difference between conditions. (C) Percent deuterium incorporation for select peptides of PLC-γ1 (H335A+D1165H) plotted as the mean ± SD (n=3) for the indicated conditions. Most error bars are smaller than the size of the corresponding point. The full deuterium incorporation data for all peptides are shown in the source data.
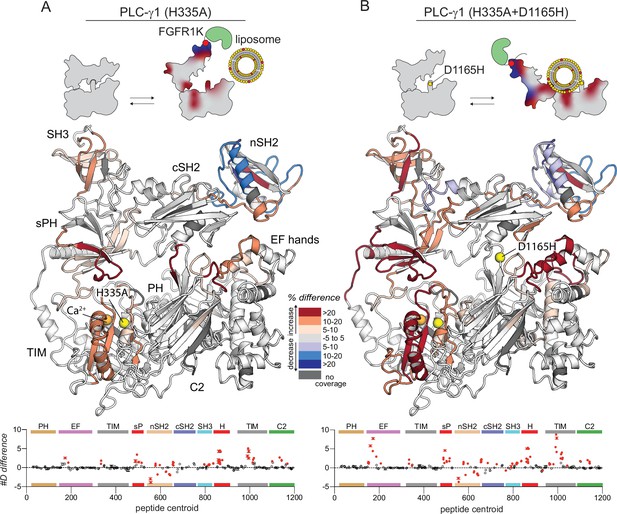
Kinase domain of fibroblast growth factor receptor (FGFR1K) and liposomes have enhanced capacity to affect the deuterium exchange of the oncogenic mutant of phospholipase C-γ1 (PLC-γ1) compared to the wild type (WT).
Differences in deuterium incorporation was measured for PLC-γ1 (H335A) or PLC-γ1 (H335A+D1165H) with both kinase and liposomes. Differences in deuterium incorporation were calculated relative to PLC-γ1 (H335A) alone (A) or to PLC-γ1 (H335A+D1165H) alone (B). Peptides that showed significant H/D exchange differences (by the following criteria: >5% and >0.4 Da difference in exchange at any time point [3, 30, 300, 3000, and 10,000 s], with a two-tailed, unpaired t-test of p<0.01) are mapped onto the structure. For each comparison, the difference in the number of incorporated deuterons was averaged over all time points and shown below the structures as the mean ± SD (n=3). Each circle represents the central residue of a corresponding peptide; red circles indicate peptides with a significant difference between conditions. Figure duplicates portions of Figures 2 and 4 to provide a direct comparison of the effects of FGFR1 and liposomes on the exchange profiles of PLC-γ1 (H335A) and (H335A+D1165H).
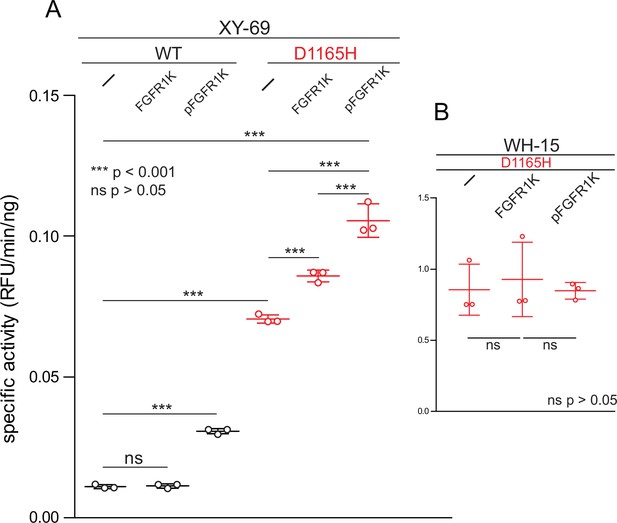
Phosphorylated kinase domain of fibroblast growth factor receptor (FGFR1K) increases phospholipase C-γ1 (PLC-γ1) specific activity.
(A) Specific activities measured with the membrane-embedded substrate XY-69. XY-69 (0.5 μM) was incorporated into liposomes comprised of phosphatidylethanolamine:phosphatidylinositol 4,5-bisphosphate (80:20) prior to addition of indicated variants of PLC-γ1 (10 nM WT; 0.3 nM D1165H). Specific activities were calculated for PLC-γ1 alone and in the presence of either unphosphorylated FGFR1K or its phosphorylated counterpart (pFGFR1K). (B) Specific activities of PLC-γ1 (D1165H) measured with water-soluble WH-15 (5 μM). In all cases, specific activities are presented as the mean ± SD of three independent experiments (n=3), each with three or more technical replicates. Statistical significance was determined with a two-tailed, unpaired t-test.
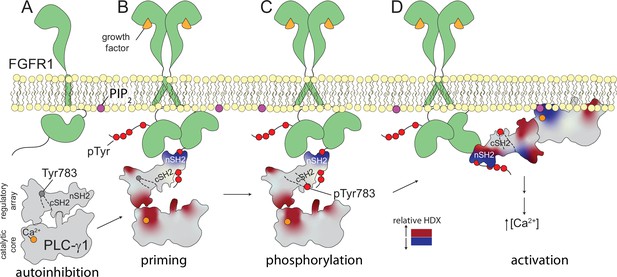
Multi-step activation of phospholipase C-γ1 (PLC-γ1).
Initially, fibroblast growth factor receptor (FGFR1) is monomeric and inactive while PLC-γ1 is cytosolic and basally autoinhibited (A). Growth factor-promoted dimerization of FGFR1 leads to its tyrosine phosphorylation and the recruitment of PLC-γ1 through interactions mediated by the nSH2 domain of PLC-γ1 (B). Complex formation reduces deuterium exchange within the nSH2 domain while simultaneously increasing deuterium exchange throughout the interface between the catalytic core and regulatory domains of PLC-γ1. Increased deuterium exchange is consistent with a loosening of the autoinhibited form of PLC-γ1 shown schematically. Active FGFR1 subsequently phosphorylates Tyr783 of PLC-γ1 to favor the interaction of pTyr783 and the cSH2 domain of PLC-γ1 (C). This step reinforces the disruption of the interface between regulatory and catalytic domains to favor a fully open form of PLC-γ1 capable of productively engaging membranes and manifesting as a decrease in the deuterium exchange of the catalytic TIM barrel (D).
Tables
| Reagent type(species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | NEB 5-alpha | New England BioLabs | Cat# C2987I | Chemically competent cells |
| Strain, strain background (E. coli) | DH10Bac | Thermo Fisher Scientific | Cat# 10361012 | Chemically competent cells |
| Cell line (Trichoplusia ni) | HighFive | Invitrogen | Cat# B85502 RRID:CVCL_C190 | |
| Recombinant DNA reagent | pFB-LIC2-PLC-γ1 (21–1215) (plasmid) | PMID:31889510 | Vector is a modified version of pFastBacHT1 containing His6 tag and TEV cleavage sequence | |
| Recombinant DNA reagent | pFB-LIC2-PLC-γ1(21–1215) H335A (plasmid) | This paper | Vector is a modified version of pFastBacHT1 containing His6 tag and TEV cleavage sequence | |
| Recombinant DNA reagent | pFB-LIC2-PLC-γ1(21–1215) H335A, D1165H (plasmid) | This paper | Vector is a modified version of pFastBacHT1 containing His6 tag and TEV cleavage sequence | |
| Recombinant DNA reagent | pUC57-Amp-FGFR1K(458-774) Y463F, Y583F, Y585F, Y730F (plasmid) | This paper | Cloning vector synthesized by Genewiz | |
| Recombinant DNA reagent | pFB-LIC2-FGFR1K(458-774) Y463F, Y583F, Y585F, Y730F (plasmid) | This paper | Vector is a modified version of pFastBacHT1 containing His6 tag and TEV cleavage sequence | |
| Recombinant DNA reagent | pFB-LIC2-FGFR1K(458-774) Y463F, Y583F, Y585F, Y730F, Y653F, Y654F (plasmid) | This paper | Vector is a modified version of pFastBacHT1 containing His6 tag and TEV cleavage sequence | |
| Recombinant DNA reagent | pFB-LIC2-FGFR1K(458-774) Y463F, Y583F, Y585F, Y730F, Y766F (plasmid) | This paper | Vector is a modified version of pFastBacHT1 containing His6 tag and TEV cleavage sequence | |
| Chemical compound, drug | L-α-phosphatidylethanolamine (Liver, Bovine) | Avanti Polar Lipids | Cat# 840,026 | |
| Chemical compound, drug | L-α-phosphatidylinositol-4,5-bisphosphate (Brain, Porcine) (ammonium salt) | Avanti Polar Lipids | Cat# 840,046 | |
| Software, algorithm | PEAKS | Bioinformatics Solutions Inc (BSI) | Version 7 (PEAKS7) | |
| Software, algorithm | HDExaminer | Sierra Analytics | ||
| Software, algorithm | PyMOL | PyMOL by Schrödinger | RRID:SCR_000305 | Version 2.5.2 |
| Other | WH-15 | PMID:21158426 | Fluorescent PIP2 analogue, soluble | |
| Other | XY-69 | PMID:29263090 | Fluorescent PIP2 analogue, membrane-associated |
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/77809/elife-77809-transrepform1-v1.docx
-
Supplementary file 1
Experimental parameters associated with hydrogen-deuterium exchange experiments.
- https://cdn.elifesciences.org/articles/77809/elife-77809-supp1-v1.docx





