Genome-wide detection of imprinted differentially methylated regions using nanopore sequencing
Figures
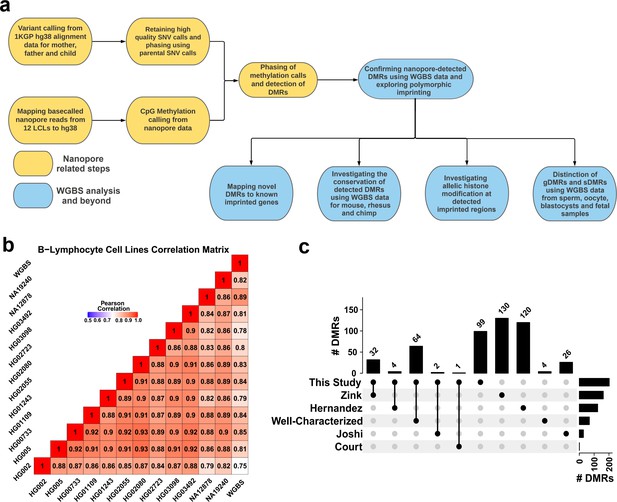
Detection of allelic methylation using nanopore sequencing.
(a) Flowchart of the study representing all the analysis steps. (b) Pearson correlation matrix of the nanopore CpG methylation frequencies for the 12 cell lines and NA12878 whole-genome bisulfite sequencing (WGBS) from ENCODE (ENCFF835NTC). (c) Upset plot of the number of differentially methylated regions (DMRs) detected in our study and previous studies, including overlaps.
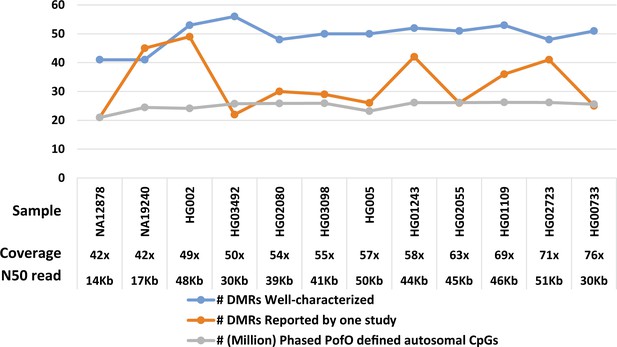
Number of allelic differentially methylated regions (DMRs) overlapped to the reported DMRs and parent-of-origin (PofO)-defined phased CpG methylation in each sample examined for differential methylation analysis by DSS R package.
To calculate coverage, the genome length is considered as 3.2G.
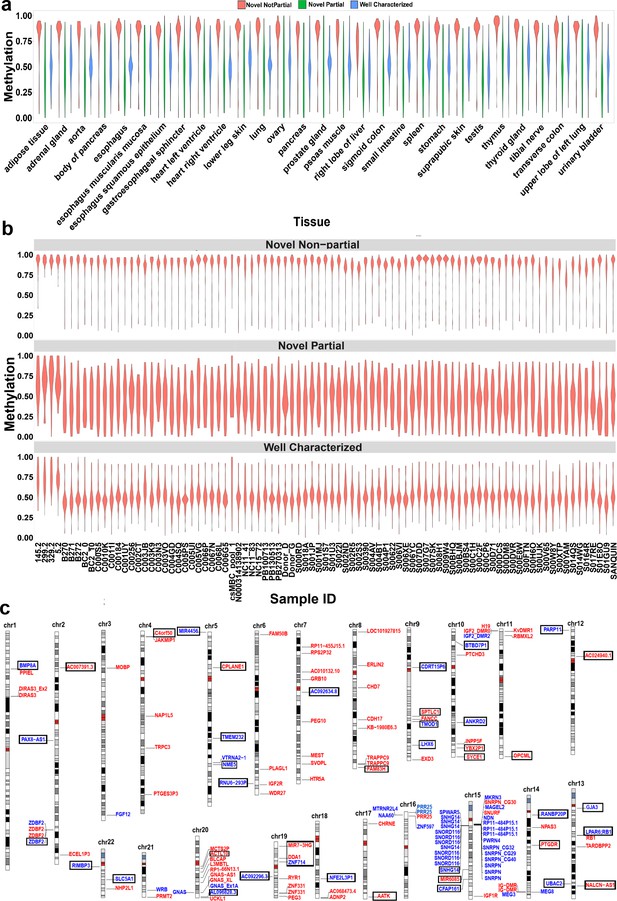
Confirmation of nanopore-detected differentially methylated regions (DMRs) using whole-genome bisulfite sequencing (WGBS) data.
(a) and (b) Violin plots representing the average methylation of each DMR in WGBS tissue and blood samples. (c) Idiogram of the 101 DMRs overlapping to reported intervals and 42 novel DMRs which were confirmed by WGBS. Paternally methylated DMRs are labelled on the left side of each chromosome while maternally methylated DMRs are on the right. Red labels represent germline DMRs while blue labels represent somatic DMRs. Novel DMRs are boxed and named based on their nearest gene (Ensembl Gene 103 GRCh38.p13).
-
Figure 2—source data 1
The average methylation of each DMR in human WGBS tissue samples.
- https://cdn.elifesciences.org/articles/77898/elife-77898-fig2-data1-v1.txt
-
Figure 2—source data 2
The average methylation of each DMR in human WGBS blood samples.
- https://cdn.elifesciences.org/articles/77898/elife-77898-fig2-data2-v1.txt
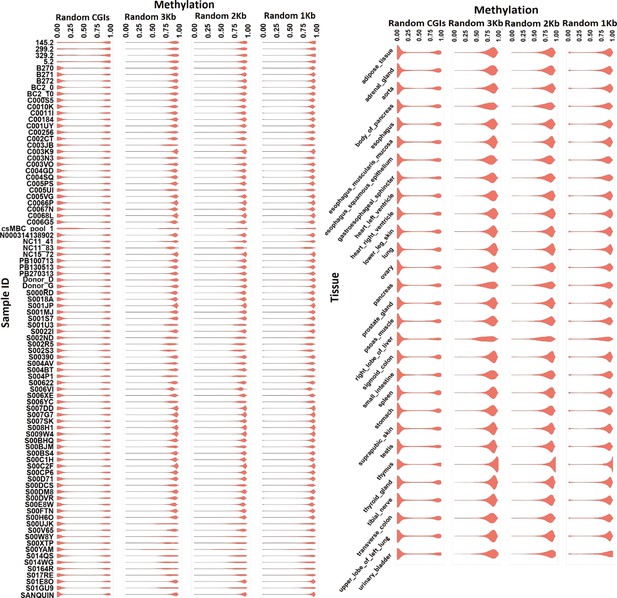
Violin plots representing methylation in whole-genome bisulfite sequencing (WGBS) blood (left) and tissue (right) samples at randomly selected CpG islands (CGIs), 1, 2, and 3 kb intervals.
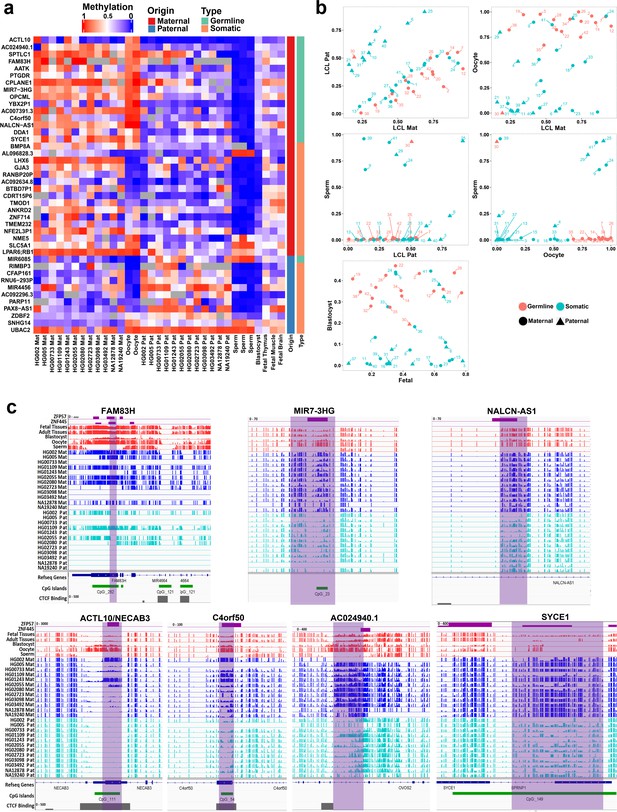
Detection of novel germline and somatic differentially methylated regions (DMRs).
(a) Heatmap displaying average methylation of the 42 nanopore-detected DMRs confirmed by whole-genome bisulfite sequencing (WGBS). DMRs are named based on their nearest gene (Ensembl Gene 103 GRCh38.p13). (b) Dot plots representing the methylation of novel germline and somatic DMRs in each sample with respect to other samples. (c) IGV screenshots from six novel germline DMRs overlapping with ZNF445 and/or ZFP57 chromatin immunoprecipitation sequencing (ChIP-seq) peaks. The range for all methylation tracks is 0–1.
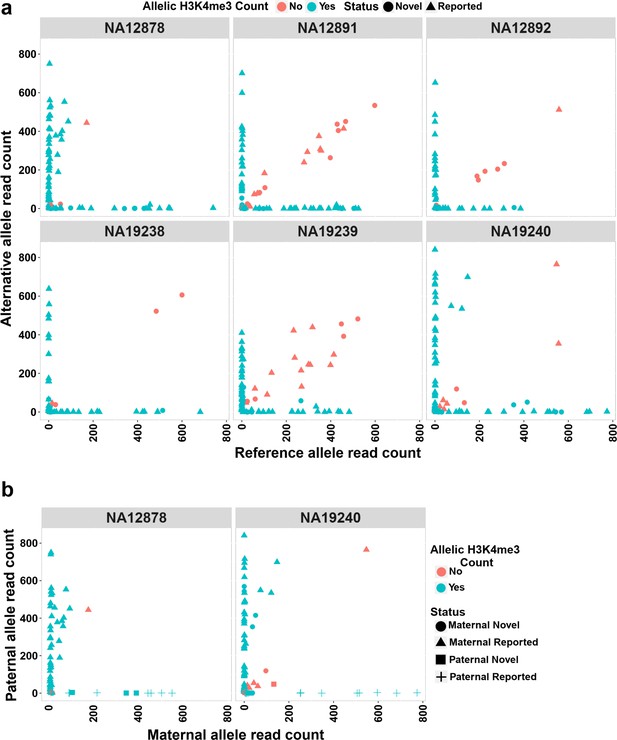
Allelic H3K4me3 histone mark at detected imprinted differentially methylated regions (DMRs).
(a) The plots representing reference and alternative alleles H3K4me3 read counts for the heterozygous single-nucleotide variants (SNVs) mapped to the detected DMRs for the six examined samples. Each point represents an SNV. Blue color displays SNVs with Fisher’s combined p-value binomial <0.05 and at least 80% of the reads on one allele and red color represent those SNVs that did not satisfy either of these thresholds. (b) The plots representing paternal and maternal H3K4me3 read counts for the heterozygous SNVs at DMRs in NA12878 and NA19240. Each point represents an SNV. The ‘Status’ indicates the methylation origin of the DMR and if the DMR is novel or reported.
-
Figure 4—source data 1
H3K4me3 allelic read counts for the heterozygous single-nucleotide variants (SNVs) mapped to the detected DMRs.
- https://cdn.elifesciences.org/articles/77898/elife-77898-fig4-data1-v1.zip
-
Figure 4—source data 2
H3K4me3 allelic read counts for the paternal and maternal heterozygous single-nucleotide variants (SNVs) mapped to the detected DMRs.
- https://cdn.elifesciences.org/articles/77898/elife-77898-fig4-data2-v1.zip
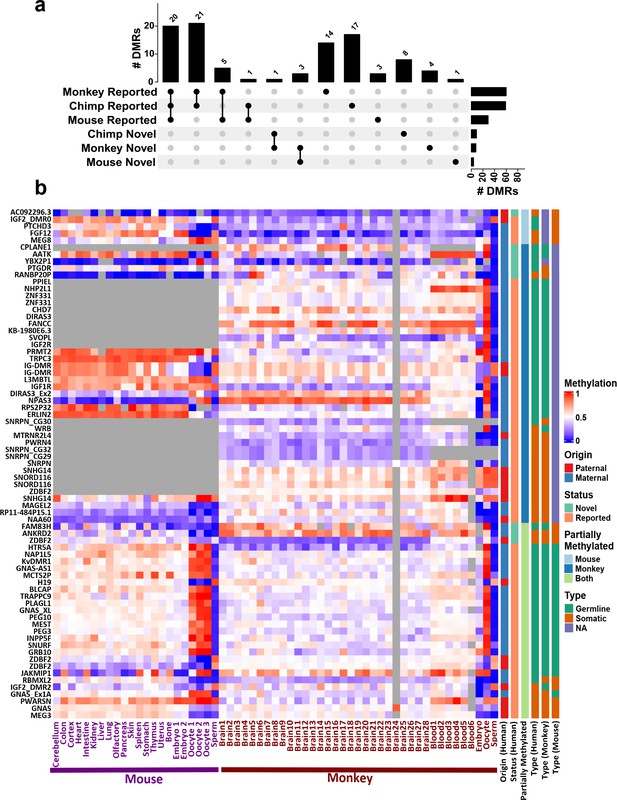
Conservation of detected differentially methylated regions (DMRs).
(a) Upset plot representing the number of previously reported and novel DMRs with evidence of conservation (partial methylation) in each of the mammals. (b) Heatmap representing human DMRs (DMR names on the left) and average methylation of their orthologous intervals in mouse and macaque in different tissues and also in sperm, oocyte, and embryonic samples. Gray regions represent NA values that either did not have an ortholog or enough CpG in the whole-genome bisulfite sequencing (WGBS) data.
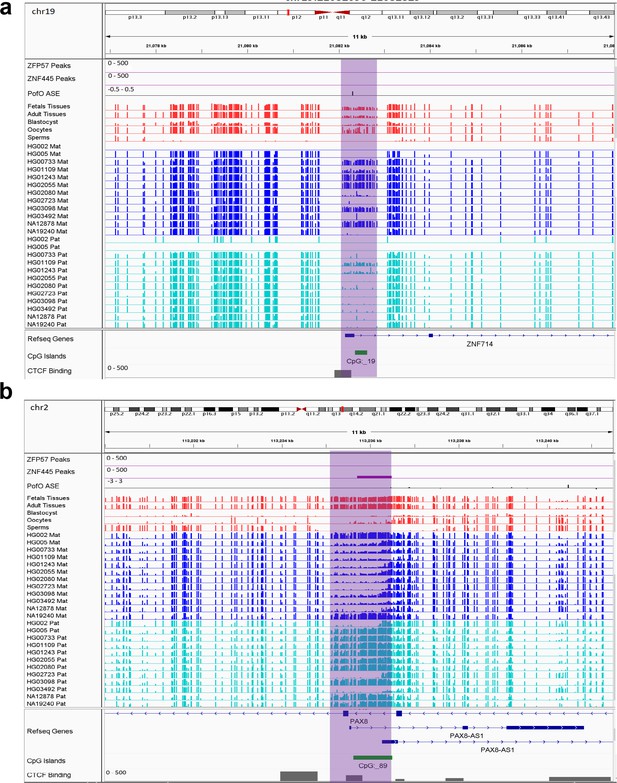
IGV screenshots of two novel somatic differentially methylated regions (DMRs).
(a) Novel maternally methylated somatic DMR overlapping with the promoter of paternally expressed ZNF714 gene. (b) Novel paternally methylated somatic DMR overlapping with the promoter of maternally expressed PAX8-AS1gene. Highlighted box regions represent the DMRs. Parent-of-origin (PofO) allele-specific expression (ASE) track is created using publicly available ASE data from Zink et al., 2018 (see Materials and methods). Positive vertical bars (upward) represent paternal expression and negative bars (downward) represent maternal expression. The range for all methylation tracks is 0–1.
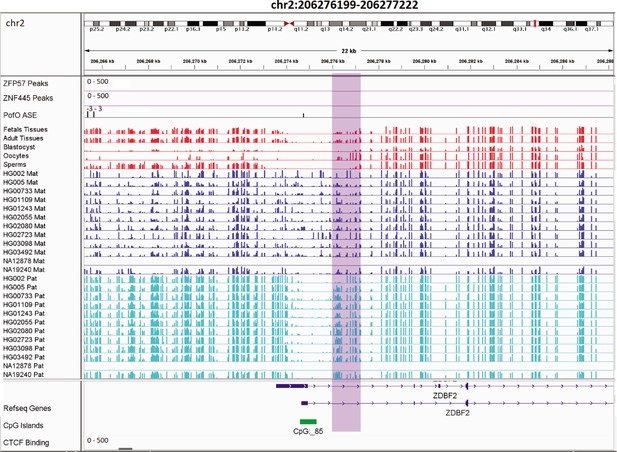
Novel somatic paternally methylated differentially methylated region (DMR) in paternally expressed ZDBF2 gene.
The range for all methylation tracks is 0–1. In parent-of-origin (PofO) ASE track, positive or upward bars represent paternal expression bias and negative or downward bars represent maternal expression bias.
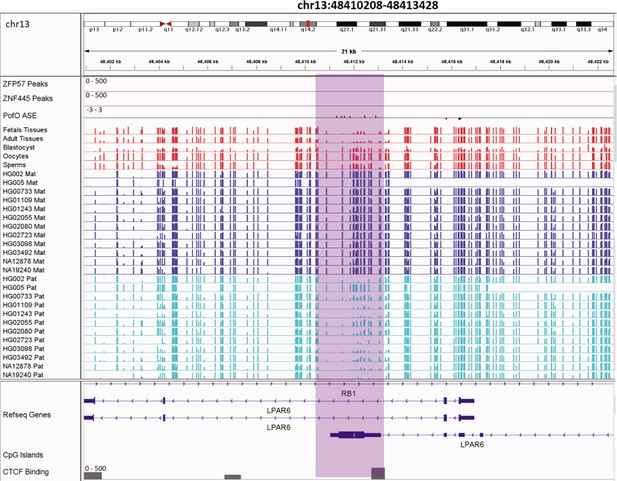
Novel somatic maternally methylated differentially methylated region (DMR) in maternally expressed RB1 gene and isoform dependent (or in some studies paternally) expressed LPAR6 gene.
The range for all methylation tracks is 0–1. In parent-of-origin (PofO) ASE track, positive or upward bars represent paternal expression bias and negative or downward bars represent maternal expression bias.
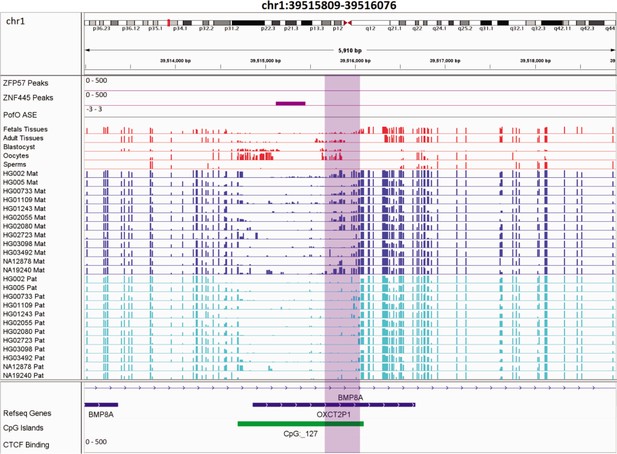
Novel somatic maternally methylated differentially methylated region (DMR) in paternally expressed BMP8A gene.
The range for all methylation tracks is 0–1. In parent-of-origin (PofO) ASE track, positive or upward bars represent paternal expression bias and negative or downward bars represent maternal expression bias.
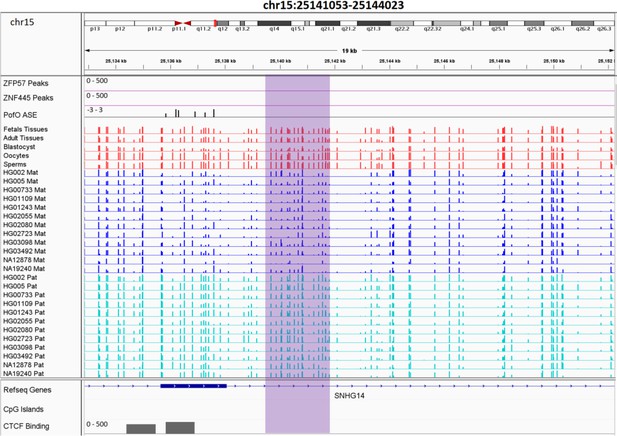
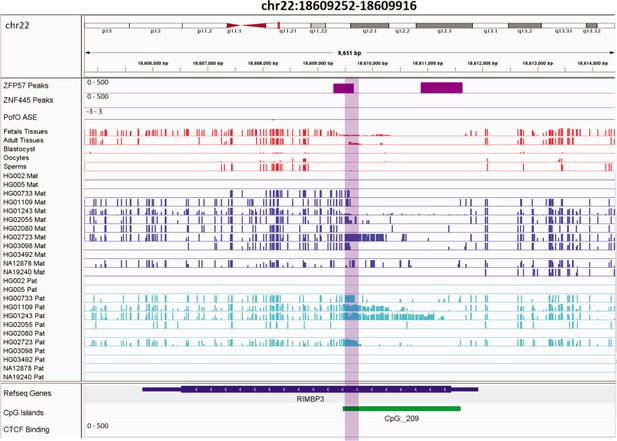
Novel somatic paternally methylated differentially methylated region (DMR) 3 kb away from paternally expressed PWAR1 gene.
The range for all methylation tracks is 0–1. In parent-of-origin (PofO) ASE track, positive or upward bars represent paternal expression bias and negative or downward bars represent maternal expression bias.
Novel somatic paternally methylated differentially methylated region (DMR) 90 kb away from maternally expressed LINC00665 gene.
The range for all methylation tracks is 0–1. In parent-of-origin (PofO) ASE track, positive or upward bars represent paternal expression bias and negative or downward bars represent maternal expression bias.
Novel somatic paternally methylated differentially methylated region (DMR) 296 kb away from randomly/maternally expressed DGCR6 gene.
The range for all methylation tracks is 0–1. In parent-of-origin (PofO) ASE track, positive or upward bars represent paternal expression bias and negative or downward bars represent maternal expression bias.
Novel somatic paternally methylated differentially methylated region (DMR) 1.03 Mb away from maternally expressed IGF2R.
The range for all methylation tracks is 0–1. In parent-of-origin (PofO) ASE track, positive or upward bars represent paternal expression bias and negative or downward bars represent maternal expression bias.
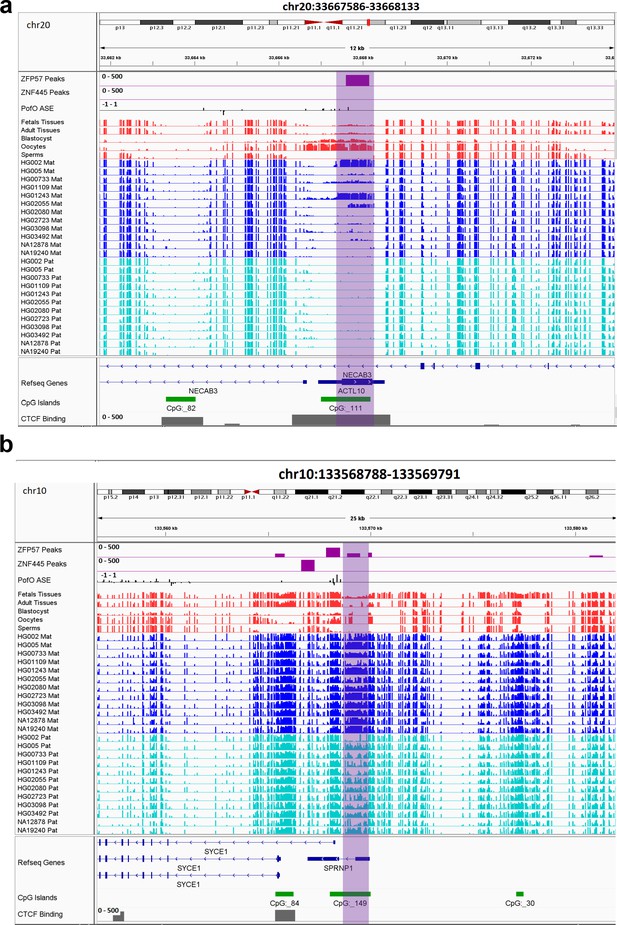
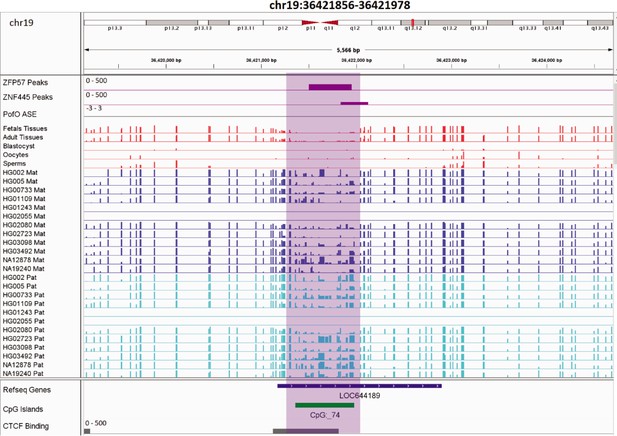
IGV screenshots of two novel maternal germline differentially methylated regions (DMRs).
(a) Novel maternally methylated germline DMR overlapping with the promoter of the paternally expressed ACTL10 gene. (b) Novel maternally methylated germline DMR overlapping with the promoter of the SYCE1 gene, which demonstrates paternal expression bias from parent-of-origin (PofO) allele-specific expression (ASE) track. Highlighted box regions represent the DMRs. PofO ASE track is created using publicly available ASE data from Zink et al., 2018 (see Materials and methods). Positive vertical bars (upward) represent paternal expression and negative bars (downward) represent maternal expression. The range for all methylation tracks is 0–1.
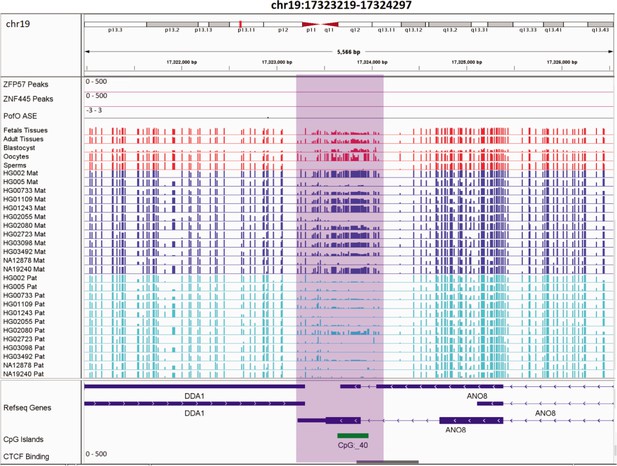
Novel germline maternally methylated differentially methylated region (DMR) in maternally expressed DDA1 gene.
The range for all methylation tracks is 0–1. In parent-of-origin (PofO) ASE track, positive or upward bars represent paternal expression bias and negative or downward bars represent maternal expression bias.
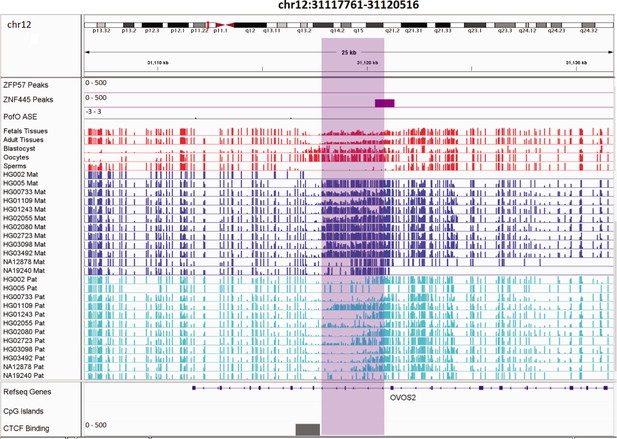
Novel germline maternally methylated differentially methylated region (DMR) in paternally expressed AC024940.1 (OVOS2) gene.
The range for all methylation tracks is 0–1. In parent-of-origin (PofO) ASE track, positive or upward bars represent paternal expression bias and negative or downward bars represent maternal expression bias.
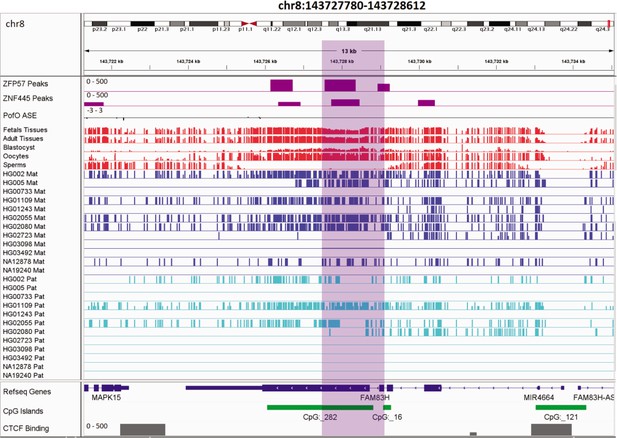
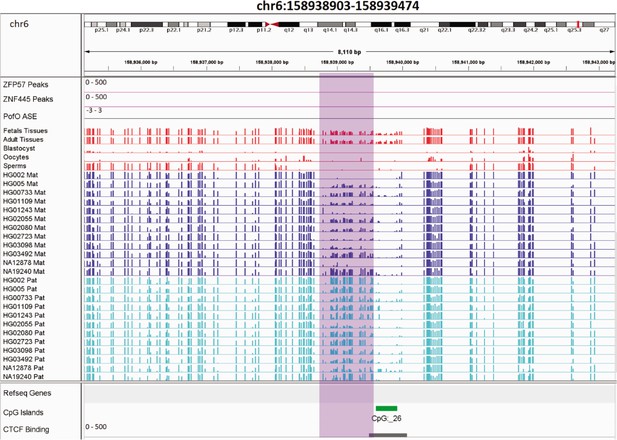
Novel germline maternally methylated differentially methylated region (DMR) 149 kb away from isoform dependent expressed NAPRT gene.
The range for all methylation tracks is 0–1. In parent-of-origin (PofO) ASE track, positive or upward bars represent paternal expression bias and negative or downward bars represent maternal expression bias.
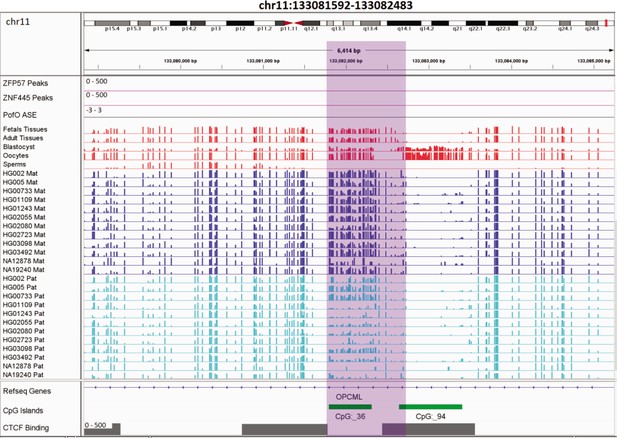
Novel germline maternally methylated differentially methylated region (DMR) 745 kb away from maternally expressed NTM gene.
The range for all methylation tracks is 0–1. In parent-of-origin (PofO) ASE track, positive or upward bars represent paternal expression bias and negative or downward bars represent maternal expression bias.
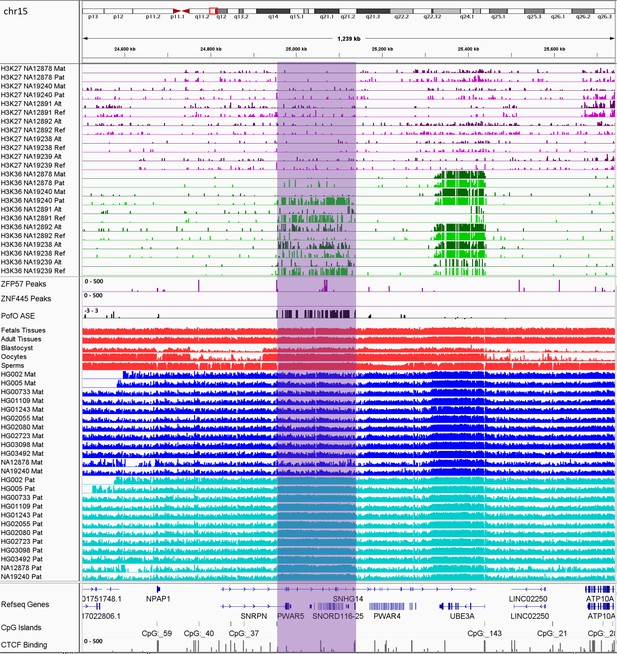
IGV screenshot of 200 kb paternally methylated biased methylation block in the PWS/AS imprinted cluster.
The range for all methylation tracks is 0–1. The histone mark tracks represent allelic read counts for H3K36me3 and H3K27me3 modifications. The range for all histone mark tracks is 0–20. In H3K27 and H3K36 tracks, for NA12878 and NA19240 the parent-of-origin could be determined and specified by maternal (Mat) and paternal (Pat) alleles. While the other samples are specified by reference (Ref) and alternative (Alt) alleles.
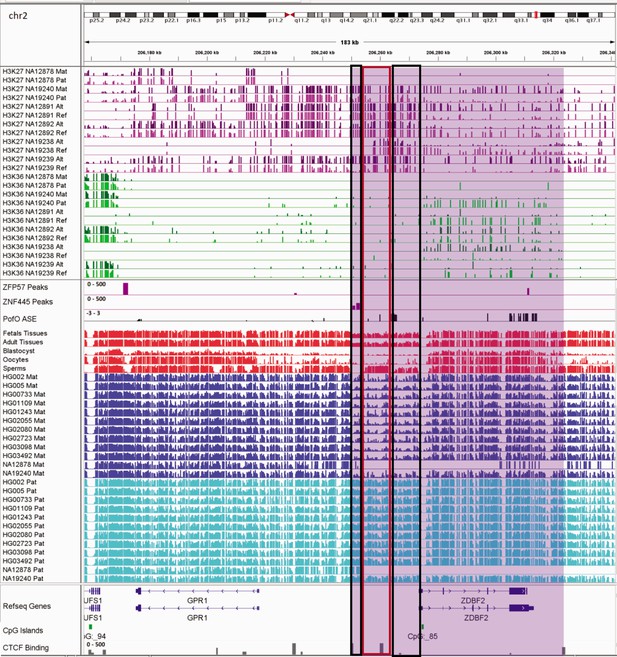
IGV screenshot of ~65 kb paternally methylated biased methylation block in GPR1-AS/ZDBF2 imprinted cluster.
The histone mark tracks are representing allelic read counts for H3K36me3 and H3K27me3 modifications. The range for all histone mark tracks is 0–20. Purple highlight demonstrates the whole block. In H3K27 and H3K36 tracks, for NA12878 and NA19240 the parent-of-origin (PofO) could be determined and specified by maternal (Mat) and paternal (Pat) alleles. While the other samples are specified by reference (Ref) and alternative (Alt) alleles. Black box represents known somatic differentially methylated region (DMR) in the block while red box shows known germline DMR in the block. The range for all methylation tracks is 0–1. In PofO ASE track, positive or upward bars represent paternal expression bias and negative or downward bars represent maternal expression bias.
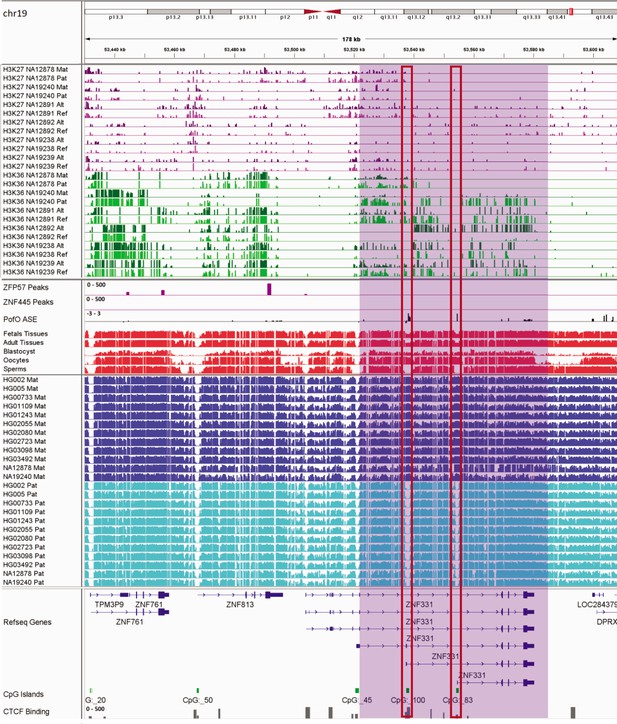
IGV screenshot of ~58 kb paternally methylated biased methylation block in ZNF331/ZNF813 imprinted cluster.
The histone mark tracks are representing allelic read counts for H3K36me3 and H3K27me3 modifications. The range for all histone mark tracks is 0–20. In H3K27 and H3K36 tracks, for NA12878 and NA19240 the parent-of-origin (PofO) could be determined and specified by maternal (Mat) and paternal (Pat) alleles. While the other samples are specified by reference (Ref) and alternative (Alt) alleles. Purple highlight demonstrates the whole block. Red box shows known germline differentially methylated region (DMR) in the block. The range for all methylation tracks is 0–1. In PofO ASE track, positive or upward bars represent paternal expression bias and negative or downward bars represent maternal expression bias.
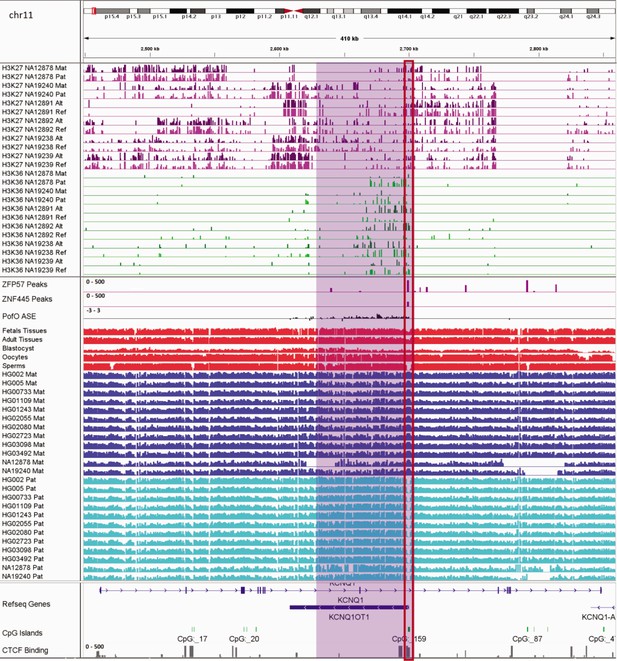
IGV screenshot of ~56 kb paternally methylated biased methylation block in KCNQ1/KCNQ1OT1 imprinted cluster.
The histone mark tracks are representing allelic read counts for H3K36me3 and H3K27me3 modifications. The range for all histone mark tracks is 0–20. In H3K27 and H3K36 tracks, for NA12878 and NA19240 the parent-of-origin (PofO) could be determined and specified by maternal (Mat) and paternal (Pat) alleles. While the other samples are specified by reference (Ref) and alternative (Alt) alleles. Purple highlight demonstrates the whole block. Red box shows known germline differentially methylated region (DMR) in the block. The range for all methylation tracks is 0–1. In PofO ASE track, positive or upward bars represent paternal expression bias and negative or downward bars represent maternal expression bias.
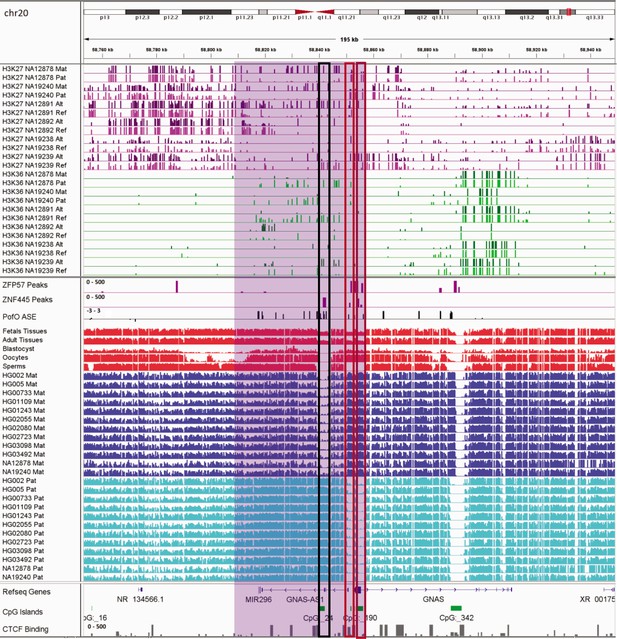
IGV screenshot of ~44 kb paternally methylated biased methylation block in GNAS/GNAS-AS1 imprinted cluster.
The histone mark tracks are representing allelic read counts for H3K36me3 and H3K27me3 modifications. The range for all histone mark tracks is 0–20. In H3K27 and H3K36 tracks, for NA12878 and NA19240 the parent-of-origin (PofO) could be determined and specified by maternal (Mat) and paternal (Pat) alleles. While the other samples are specified by reference (Ref) and alternative (Alt) alleles. Purple highlight demonstrates the whole block. Black box represents known somatic differentially methylated region (DMR) in the block while red box shows known germline DMR in the block. The range for all methylation tracks is 0–1. In PofO ASE track, positive or upward bars represent paternal expression bias and negative or downward bars represent maternal expression bias.
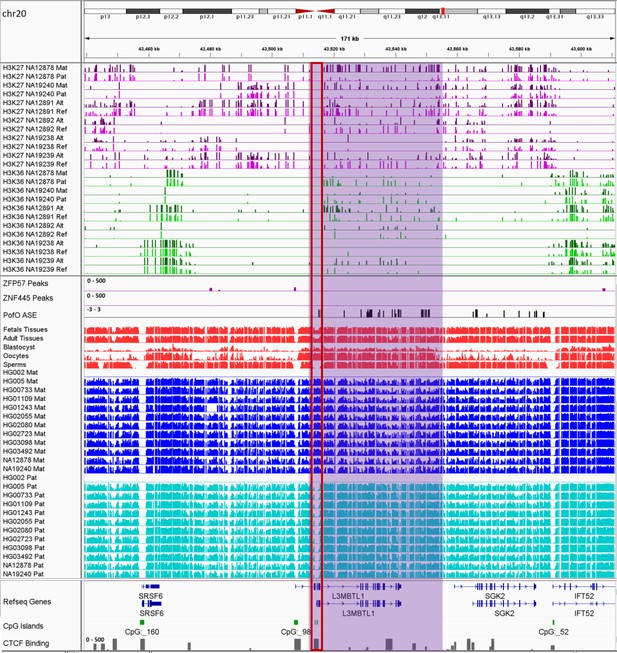
IGV screenshot of ~42 kb paternally methylated biased methylation block in L3MBTL1/SGK2 imprinted cluster.
The histone mark tracks are representing allelic read counts for H3K36me3 and H3K27me3 modifications. The range for all histone mark tracks is 0–20. In H3K27 and H3K36 tracks, for NA12878 and NA19240 the parent-of-origin (PofO) could be determined and specified by maternal (Mat) and paternal (Pat) alleles. While the other samples are specified by reference (Ref) and alternative (Alt) alleles. Purple highlight demonstrates the whole block. Red box shows known germline differentially methylated region (DMR) in the block. The range for all methylation tracks is 0–1. In PofO ASE track, positive or upward bars represent paternal expression bias and negative or downward bars represent maternal expression bias.
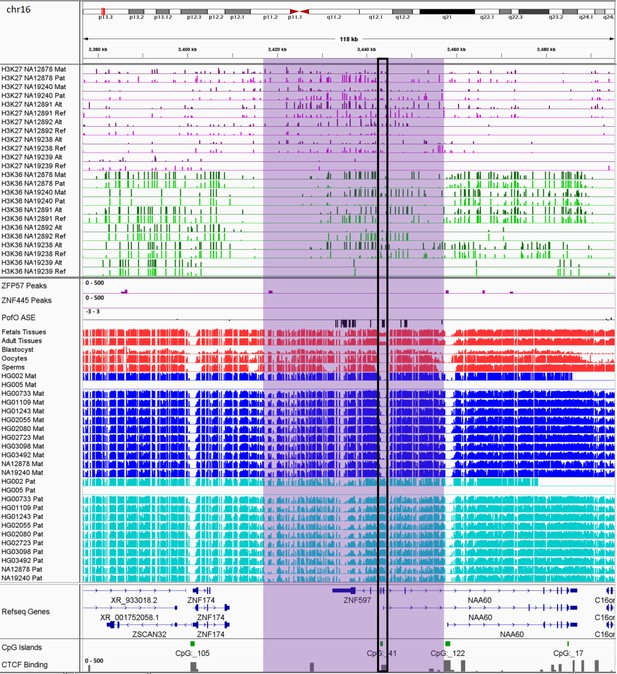
IGV screenshot of ~35 kb maternally methylated biased methylation block in ZNF597/NAA60 imprinted cluster.
The histone mark tracks are representing allelic read counts for H3K36me3 and H3K27me3 modifications. The range for all histone mark tracks is 0–20. In H3K27 and H3K36 tracks, for NA12878 and NA19240 the parent-of-origin (PofO) could be determined and specified by maternal (Mat) and paternal (Pat) alleles. While the other samples are specified by reference (Ref) and alternative (Alt) alleles. Purple highlight demonstrates the whole block. Black box represents known somatic differentially methylated region (DMR) in the block. The range for all methylation tracks is 0–1. In PofO ASE track, positive or upward bars represent paternal expression bias and negative or downward bars represent maternal expression bias.
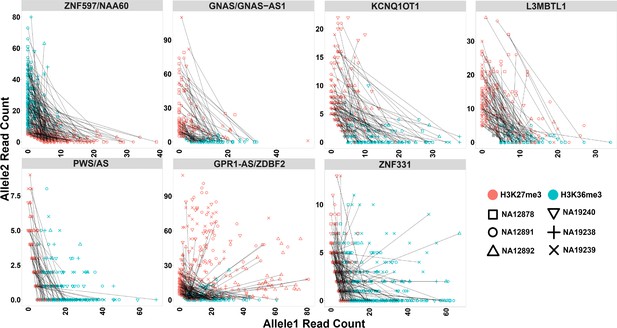
Mutual exclusive allelic H3K36me3 and H3K27me3 histone marks at seven parent-of-origin (PofO) methylation-biased blocks.
All blocks demonstrate allelic H3K36me3 on hypermethylated allele and H3K27me3 on hypomethylated allele. For NA12878 and NA19240, allele1 is the paternal and allele2 is maternal. For sake of visualization in other four cell lines without parental information, allele1 for H3K36me3 mark demonstrates the allele with more mapped reads at all blocks except ZNF597/NAA60. Therefore, for H3K36me3 we swapped the reference allele read count with the alternative allele read count if the reference allele count was less than the alternative allele count. At ZNF597/NAA60, we swapped the reference allele read count with the alternative allele read count if the reference had higher read count. We also swapped the reference and the alternative allele counts for the same SNVs for H3K27me3. Each point represents a heterozygous SNV. Lines are connecting SNVs that have mapped reads for both histone modifications.
-
Figure 9—source data 1
H3K36me3 and H3K27me3 allelic read counts for the heterozygous single-nucleotide variants (SNVs) mapped to the detected PofO-biased methylation blocks.
- https://cdn.elifesciences.org/articles/77898/elife-77898-fig9-data1-v1.zip
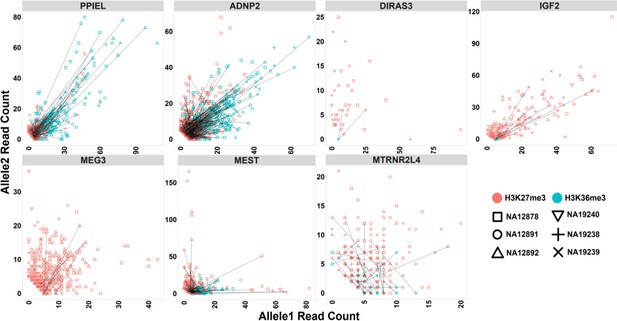
Allelic H3K36me3 and H3K27me3 histone marks read count at seven test blocks.
Test blocks do not display allelic bias for H3K36me3 or H3K27me3 histone modifications. For NA12878 and NA19240, allele1 is the paternal and allele2 is maternal. For sake of visualization in other four cell lines without parental information, allele1 for H3K36me3 mark demonstrates the allele with more mapped reads. Therefore, for H3K36me3 we swapped reference allele read count with alternative allele read count if reference allele count was less than alternative allele count. We also swapped reference and alternative allele counts for the same SNVs for H3K27me3. Each point represents a heterozygous SNV. Lines are connecting SNVs that have mapped reads for both histone modifications.
-
Figure 9—figure supplement 1—source data 1
H3K36me3 and H3K27me3 allelic read counts for the heterozygous single-nucleotide variants (SNVs) mapped to the test blocks.
- https://cdn.elifesciences.org/articles/77898/elife-77898-fig9-figsupp1-data1-v1.zip
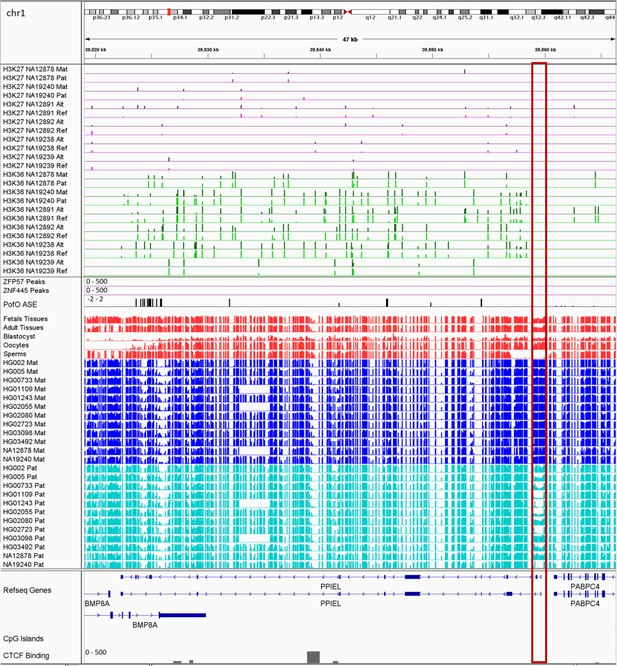
IGV screenshot of the PPIEL imprinted cluster.
The histone mark tracks are representing allelic read counts for H3K36me3 and H3K27me3 modifications. The range for all histone mark tracks is 0–20. In H3K27 and H3K36 tracks, for NA12878 and NA19240 the parent-of-origin (PofO) could be determined and specified by maternal (Mat) and paternal (Pat) alleles. While the other samples are specified by reference (Ref) and alternative (Alt) alleles. Red box shows known germline DMR. The range for all methylation tracks is 0–1. In PofO ASE track, positive or upward bars represent paternal expression bias and negative or downward bars represent maternal expression bias.
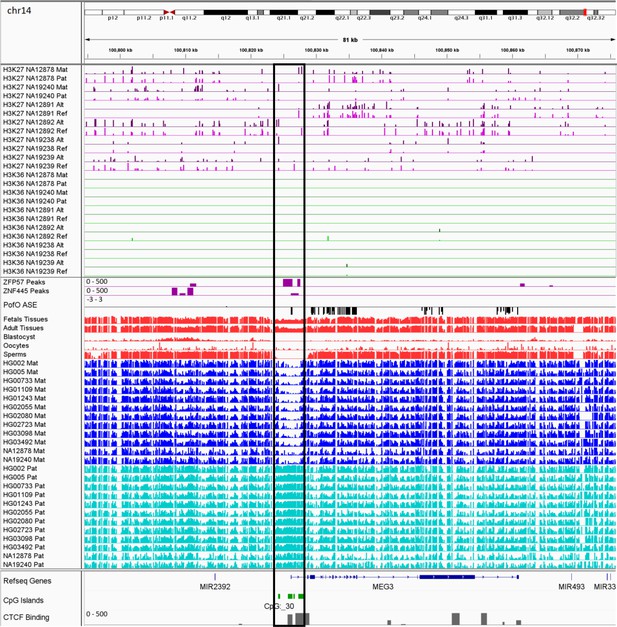
IGV screenshot of the MEG3 imprinted cluster.
The histone mark tracks are representing allelic read counts for H3K36me3 and H3K27me3 modifications. The range for all histone mark tracks is 0–20. In H3K27 and H3K36 tracks, for NA12878 and NA19240 the parent-of-origin (PofO) could be determined and specified by maternal (Mat) and paternal (Pat) alleles. While the other samples are specified by reference (Ref) and alternative (Alt) alleles. Black box shows known somatic differentially methylated region (DMR). The range for all methylation tracks is 0–1. In PofO ASE track, positive or upward bars represent paternal expression bias and negative or downward bars represent maternal expression bias.
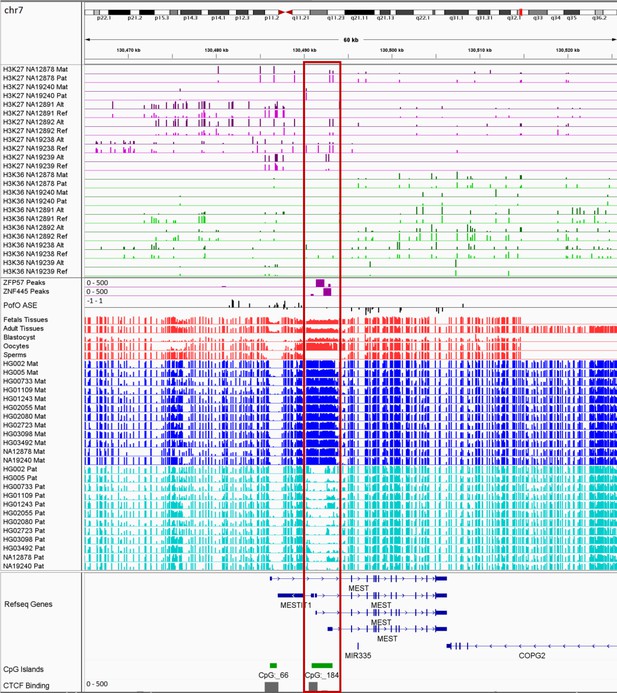
IGV screenshot of the MEST imprinted cluster.
The histone mark tracks are representing allelic read counts for H3K36me3 and H3K27me3 modifications. The range for all histone mark tracks is 0–20. In H3K27 and H3K36 tracks, for NA12878 and NA19240 the parent-of-origin (PofO) could be determined and specified by maternal (Mat) and paternal (Pat) alleles. While the other samples are specified by reference (Ref) and alternative (Alt) alleles. Red box shows known germline differentially methylated region (DMR). The range for all methylation tracks is 0–1. In PofO ASE track, positive or upward bars represent paternal expression bias and negative or downward bars represent maternal expression bias.
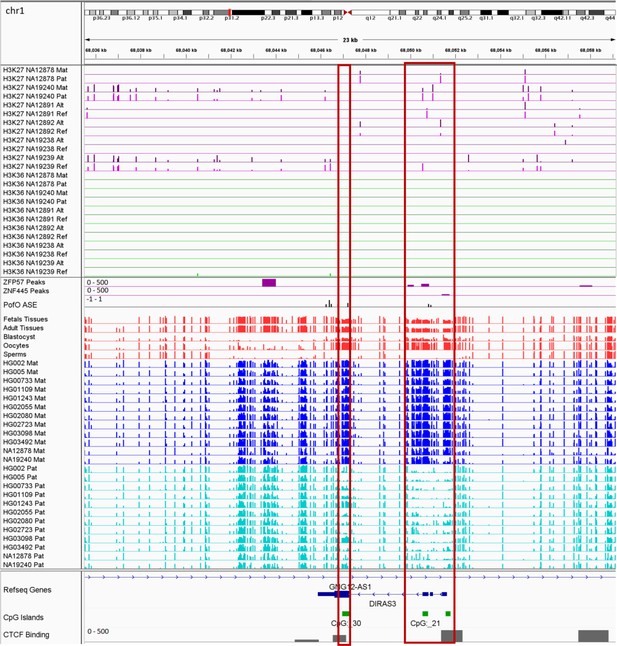
IGV screenshot of the DIRAS3 imprinted cluster.
The histone mark tracks are representing allelic read counts for H3K36me3 and H3K27me3 modifications. The range for all histone mark tracks is 0–20. In H3K27 and H3K36 tracks, for NA12878 and NA19240 the parent-of-origin (PofO) could be determined and specified by maternal (Mat) and paternal (Pat) alleles. While the other samples are specified by reference (Ref) and alternative (Alt) alleles. Red box shows known germline differentially methylated region (DMR). The range for all methylation tracks is 0–1. In PofO ASE track, positive or upward bars represent paternal expression bias and negative or downward bars represent maternal expression bias.
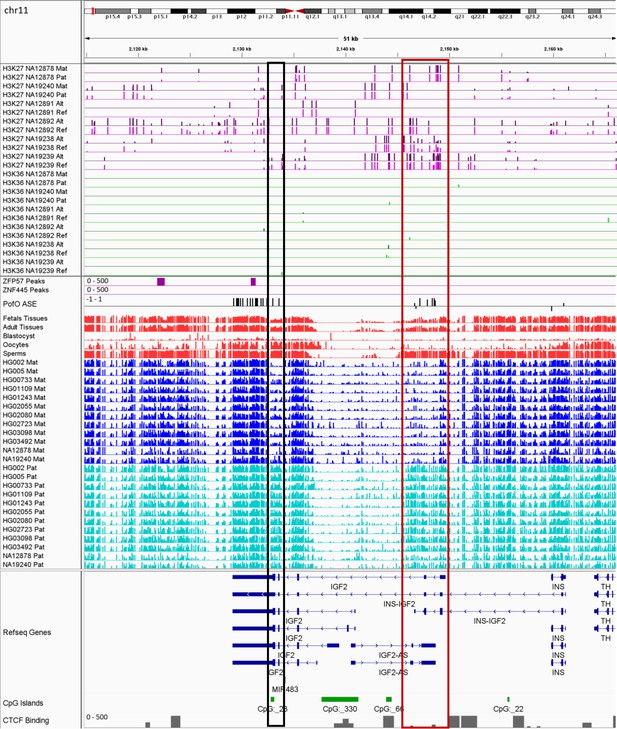
IGV screenshot of the IGF2 imprinted cluster.
The histone mark tracks are representing allelic read counts for H3K36me3 and H3K27me3 modifications. The range for all histone mark tracks is 0–20. In H3K27 and H3K36 tracks, for NA12878 and NA19240 the parent-of-origin (PofO) could be determined and specified by maternal (Mat) and paternal (Pat) alleles. While the other samples are specified by reference (Ref) and alternative (Alt) alleles. Black box represents known somatic differentially methylated region (DMR) and red box shows known germline DMR. The range for all methylation tracks is 0–1. In PofO ASE track, positive or upward bars represent paternal expression bias and negative or downward bars represent maternal expression bias.
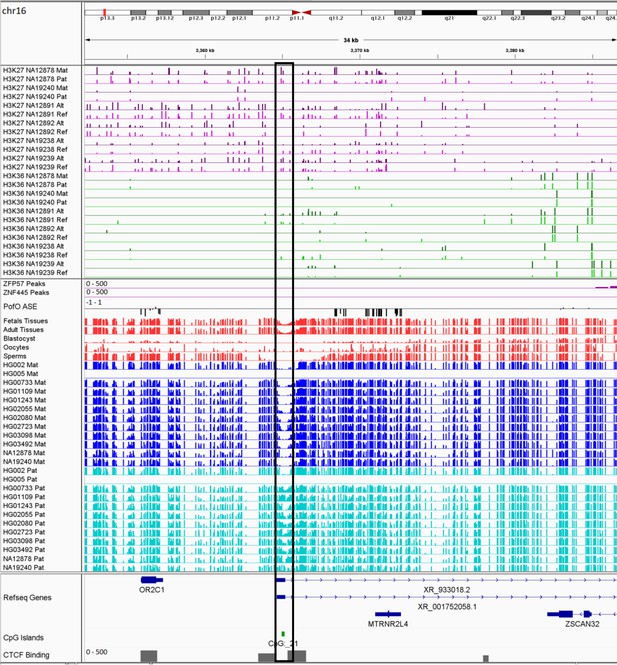
IGV screenshot of the MTRNR2L4 imprinted cluster.
The histone mark tracks are representing allelic read counts for H3K36me3 and H3K27me3 modifications. The range for all histone mark tracks is 0–20. In H3K27 and H3K36 tracks, for NA12878 and NA19240 the parent-of-origin (PofO) could be determined and specified by maternal (Mat) and paternal (Pat) alleles. While the other samples are specified by reference (Ref) and alternative (Alt) alleles. Black box represents known somatic differentially methylated region (DMR). The range for all methylation tracks is 0–1. In PofO ASE track, positive or upward bars represent paternal expression bias and negative or downward bars represent maternal expression bias.
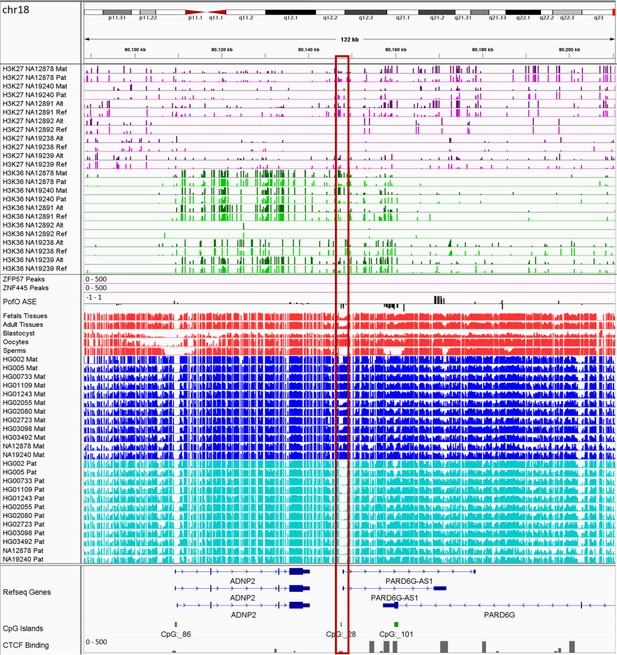
IGV screenshot of the ADNP2 imprinted cluster.
The histone mark tracks are representing allelic read counts for H3K36me3 and H3K27me3 modifications. The range for all histone mark tracks is 0–20. In H3K27 and H3K36 tracks, for NA12878 and NA19240 the parent-of-origin (PofO) could be determined and specified by maternal (Mat) and paternal (Pat) alleles. While the other samples are specified by reference (Ref) and alternative (Alt) alleles. Red box shows known germline differentially methylated region (DMR). The range for all methylation tracks is 0–1. In PofO ASE track, positive or upward bars represent paternal expression bias and negative or downward bars represent maternal expression bias.
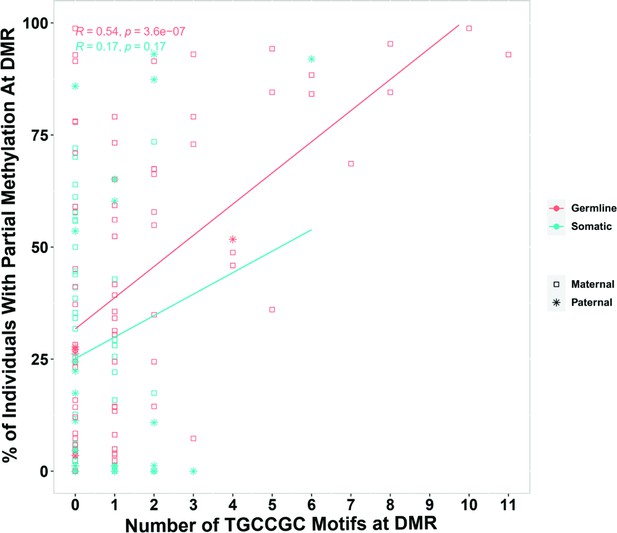
Pearson correlation for the number of ZFP57-binding motif (TGCCGC) at differentially methylated regions (DMRs) and percent of individuals that demonstrated partial methylation in their whole-genome bisulfite sequencing data at the DMRs.
-
Appendix 1—figure 1—source data 1
Number of ZFP57-binding motif (TGCCGC) at differentially methylated regions (DMRs).
- https://cdn.elifesciences.org/articles/77898/elife-77898-app1-fig1-data1-v1.zip
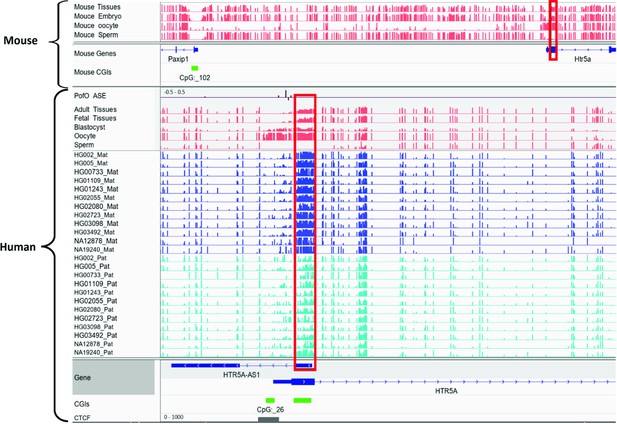
Conserved differentially methylated region (DMR) at HTR5A in human and mouse.
The known DMR at HTR5A reported to be not conserved in mouse by Court et al. (PMID: 24402520; see supplementary figure S6 from Court et al., 2014), however we detected it as conserved due to a different orthologous examined in our study. Court et al. examined CGI 102 which is also not imprinted in our analysis, however the ortholog we examined spans beginning of the HTR5A and is partially methylated which suggests the region is imprinted. Red boxes are showing germline DMRs. The range for all methylation tracks is 0–1. In parent-of-origin (PofO) ASE track, positive or upward bars represent paternal expression bias and negative or downward bars represent maternal expression bias.
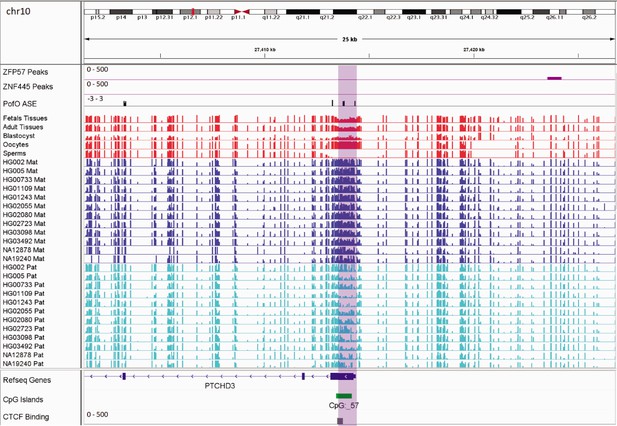
Germline maternally methylated differentially methylated region (DMR) in the promoter of paternally expressed PTCHD3 gene.
The range for all methylation tracks is 0–1. In parent-of-origin (PofO) ASE track, positive or upward bars represent paternal expression bias and negative or downward bars represent maternal expression bias.
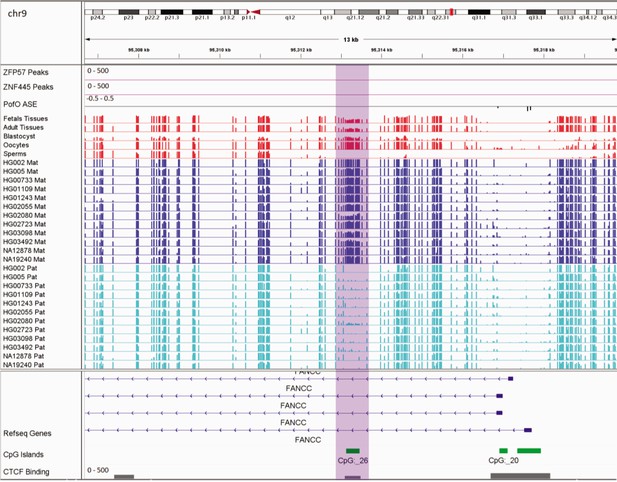
Germline maternally methylated differentially methylated region (DMR) in the intron 1 of maternally expressed FANCC gene.
The range for all methylation tracks is 0–1. In parent-of-origin (PofO) ASE track, positive or upward bars represent paternal expression bias and negative or downward bars represent maternal expression bias.
Tables
Forty-two detected imprinted differentially methylated regions (DMRs) from nanopore data and confirmed using whole-genome bisulfite sequencing (WGBS) data.
DMRs are named after the nearest gene (EnsemblGene 103 GRCh38.p13).
| ID | DMR name | Origin | Type | Distance to nearest imprinted gene | % Individuals with partial methylation | % Tissues with partial methylation |
|---|---|---|---|---|---|---|
| 22 | AC024940.1 | Maternal | Germline | 0 | 15.9 | 3.8 |
| 35 | DDA1 | Maternal | Germline | 0 | 7.3 | 15.4 |
| 38 | ACTL10;NECAB3 | Maternal | Germline | 0 | 3.7 | 8.7 |
| 42 | SYCE1 | Maternal | Germline | 3.2 kb | 4 | 9.1 |
| 12 | FAM83H | Maternal | Germline | 149 kb | 48.8 | 12 |
| 20 | OPCML | Maternal | Germline | 744.1 kb | 45.1 | 25 |
| 19 | YBX2P1 | Maternal | Germline | >2 Mb | 3.7 | 7.7 |
| 26 | NALCN-AS1 | Maternal | Germline | >2 Mb | 30.5 | 10 |
| 28 | PTGDR | Maternal | Germline | >2 Mb | 8.4 | 3.4 |
| 32 | AATK | Maternal | Germline | >2 Mb | 23.2 | 9.1 |
| 34 | MIR7-3HG | Maternal | Germline | >2 Mb | 8.1 | 3.6 |
| 2 | AC007391.3 | Maternal | Germline | >2 Mb | 37.2 | 60.7 |
| 5 | C4orf50 | Maternal | Germline | >2 Mb | 14.5 | 22.2 |
| 7 | CPLANE1 | Maternal | Germline | >2 Mb | 2.3 | 7.1 |
| 14 | SPTLC1 | Maternal | Germline | >2 Mb | 5.8 | 48.1 |
| 1 | BMP8A | Maternal | Somatic | 0 | 4.5 | 26.1 |
| 24 | LPAR6;RB1 | Maternal | Somatic | 0 | 2.3 | 10.3 |
| 36 | ZNF714 | Maternal | Somatic | 0 | 43.9 | 29.6 |
| 17 | BTBD7P1 | Maternal | Somatic | >2 Mb | 73.5 | 55.6 |
| 18 | ANKRD2 | Maternal | Somatic | >2 Mb | 34.1 | 3.8 |
| 23 | GJA3 | Maternal | Somatic | >2 Mb | 25.6 | 21.4 |
| 27 | RANBP20P | Maternal | Somatic | >2 Mb | 28 | 32 |
| 33 | NFE2L3P1 | Maternal | Somatic | >2 Mb | 29.3 | 44.4 |
| 39 | AL096828.3 | Maternal | Somatic | >2 Mb | 50 | 7.7 |
| 41 | SLC5A1 | Maternal | Somatic | >2 Mb | 56.1 | 25.9 |
| 8 | TMEM232 | Maternal | Somatic | >2 Mb | 25.6 | 37.9 |
| 9 | NME5 | Maternal | Somatic | >2 Mb | 22.1 | 10.7 |
| 11 | AC092634.8 | Maternal | Somatic | >2 Mb | 6.1 | 3.8 |
| 13 | CDRT15P6 | Maternal | Somatic | >2 Mb | 12.7 | 5.6 |
| 15 | TMOD1 | Maternal | Somatic | >2 Mb | 35.4 | 14.8 |
| 16 | LHX6 | Maternal | Somatic | >2 Mb | 44.6 | 25 |
| 30 | MIR6085 | Paternal | Germline | >2 Mb | 27.1 | 25.9 |
| 3 | PAX8;PAX8-AS1 | Paternal | Somatic | 0 | 24.4 | 32.1 |
| 4 | ZDBF2 | Paternal | Somatic | 0 | 53.6 | 58.6 |
| 29 | SNHG14 | Paternal | Somatic | 3 kb | 4.7 | 37.9 |
| 37 | AC092296.3 | Paternal | Somatic | 90 kb | 1.2 | 7.7 |
| 40 | RIMBP3 | Paternal | Somatic | 296 kb | 17.4 | 11.1 |
| 10 | RNU6-293P | Paternal | Somatic | 1.03 Mb | 65.1 | 37 |
| 21 | PARP11 | Paternal | Somatic | >2 Mb | 10.8 | 22.2 |
| 25 | UBAC2 | Paternal | Somatic | >2 Mb | 1.2 | 6.9 |
| 31 | CFAP161 | Paternal | Somatic | >2 Mb | 22.4 | 6.9 |
| 6 | MIR4456 | Paternal | Somatic | >2 Mb | 11.3 | 22.2 |
Additional files
-
Supplementary file 1
List of the reported imprinted differentially methylated regions (DMRs).
- https://cdn.elifesciences.org/articles/77898/elife-77898-supp1-v1.xlsx
-
Supplementary file 2
The list of 200 detected imprinted differentially methylation regions from nanopore data.
This list demonstrates the results of differential methylation analysis (DMA) using nanomethphase dma module that uses DSS R package for DMA.
- https://cdn.elifesciences.org/articles/77898/elife-77898-supp2-v1.xlsx
-
Supplementary file 3
Mapping detected differentially methylated regions (DMRs) using nanopore sequencing across 12 lymphoblastoid cell lines (LCLs) to reported DMRs.
- https://cdn.elifesciences.org/articles/77898/elife-77898-supp3-v1.xlsx
-
Supplementary file 4
Detected differentially methylated regions (DMRs) using nanopore sequencing across 12 lymphoblastoid cell lines (LCLs) that mapped to reported DMRs or confirmed in whole-genome bisulfite sequencing (WGBS) data.
- https://cdn.elifesciences.org/articles/77898/elife-77898-supp4-v1.xlsx
-
Supplementary file 5
The results for examining inter-individual variations of the detected differentially methylated regions (DMRs) in whole-genome bisulfite sequencing (WGBS) datasets from blood.
- https://cdn.elifesciences.org/articles/77898/elife-77898-supp5-v1.xlsx
-
Supplementary file 6
The allelic read counts of H3K4me3 chromatin immunoprecipitation sequencing (ChIP-seq) for each heterozygous single-nucleotide variant (SNV) at the detected differentially methylated regions (DMRs) in six lymphoblastoid cell line (LCL) samples.
- https://cdn.elifesciences.org/articles/77898/elife-77898-supp6-v1.xlsx
-
Supplementary file 7
List of the known imprinted genes.
- https://cdn.elifesciences.org/articles/77898/elife-77898-supp7-v1.xlsx
-
Supplementary file 8
The allelic read counts of H3K36me3 and H3K27me3 chromatin immunoprecipitation sequencing (ChIP-seq) for the seven parent-of-origin (PofO)-biased methylation blocks in six lymphoblastoid cell line (LCL) samples.
- https://cdn.elifesciences.org/articles/77898/elife-77898-supp8-v1.xlsx
-
Supplementary file 9
The allelic read counts of H3K36me3 and H3K27me3 chromatin immunoprecipitation sequencing (ChIP-seq) for the seven test blocks in six lymphoblastoid cell line (LCL) samples.
- https://cdn.elifesciences.org/articles/77898/elife-77898-supp9-v1.xlsx
-
Supplementary file 10
List of the whole-genome bisulfite sequencing (WGBS) tissues and WGBS blood samples used in our study.
- https://cdn.elifesciences.org/articles/77898/elife-77898-supp10-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/77898/elife-77898-transrepform1-v1.pdf
-
Appendix 1—figure 1—source data 1
Number of ZFP57-binding motif (TGCCGC) at differentially methylated regions (DMRs).
- https://cdn.elifesciences.org/articles/77898/elife-77898-app1-fig1-data1-v1.zip





