Gill developmental program in the teleost mandibular arch
Figures
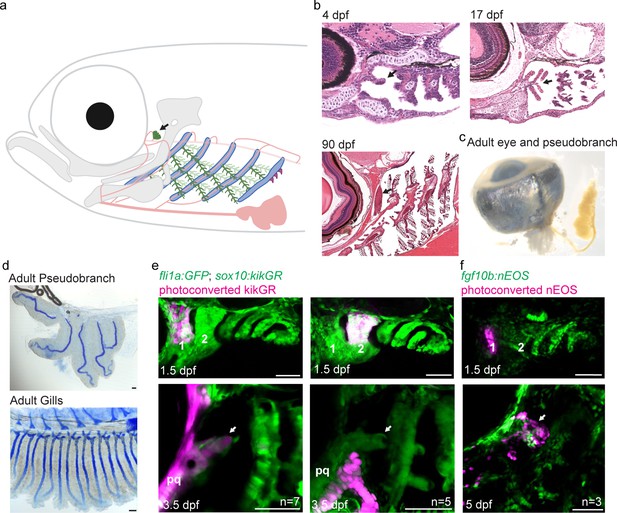
The zebrafish pseudobranch derives from mandibular arch mesenchyme and first pouch epithelia.
(a), Schematic showing the pseudobranch (arrows), gill filaments (branched green structures) connected to gill bars (blue), teeth (purple), vasculature (pink), and jaw and jaw-support skeleton (gray). (b) Hematoxylin and Eosin-stained sections show emergence of the pseudobranch bud at 4 dpf (adapted from https://bio-atlas.psu.edu/zf/view.php?atlas=5&s=41), five filaments at 17 dpf (adapted from https://bio-atlas.psu.edu/zf/view.php?atlas=65&s=1738), and the fused pseudobranch at 90 dpf (adapted from https://bio-atlas.psu.edu/zf/view.php?atlas=29&s=312). (c) Dissected adult pseudobranch shows the ophthalmic artery connecting it to the eye. (d) Alcian staining shows five cartilage rods in the pseudobranch and similar cartilage in gill primary filaments. (e) Photoconverted kikGR-expressing mesenchyme (red) from the dorsal first arch (numbered) at 1.5 dpf contributes to the palatoquadrate cartilage (pq) and pseudobranch mesenchyme (arrow) at 3.5 dpf. Photoconverted dorsal second arch cells do not contribute to the pseudobranch. In green, fli1a:GFP labels the vasculature and neural crest-derived mesenchyme, with mesenchyme also labeled by unconverted sox10:kikGR. (f) In fgf10:nEOS embryos, photoconversion of first pouch endoderm (numbered) at 1.5 dpf labels the pseudobranch epithelium (arrow) at 5 dpf. n numbers denote experimental replicates in which similar contributions were observed. Scale bars, 50 µm.
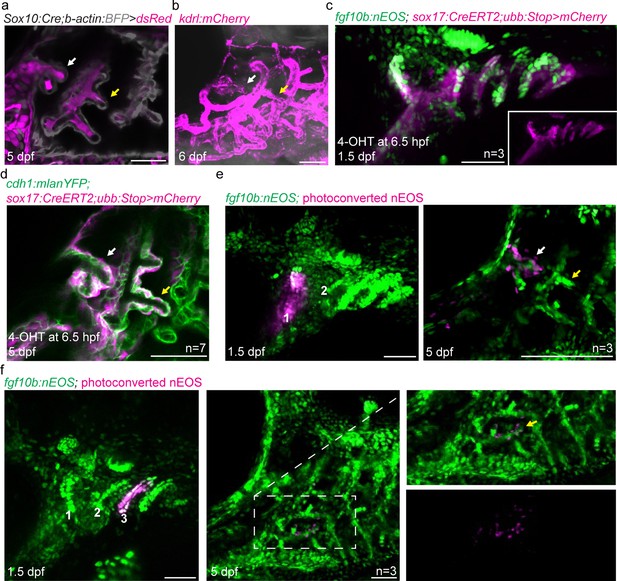
Development of zebrafish pseudobranch and lineage analysis of gill filament epithelia.
(a) In Sox10:Cre; acta2:loxP-BFP-Stop-loxP-dsRed fish at 5 dpf, the developing pseudobranch (white arrow) and gill buds (yellow arrow) consist of Cre-converted dsRed+ neural crest-derived mesenchyme (magenta) and unconverted BFP+ epithelia (gray). (b) At 6 dpf, kdrl:mCherry labeling of vasculature reveals a branch of the first aortic arch in the position of the pseudobranch (white arrow), and branches of the posterior aortic arches in the positions of the gills (yellow arrow). (c) Endoderm is labeled in red by adding 4OH-tamoxifen to sox17:CreERT2; ubb:loxP-Stop-loxP-mCherry fish at 6.5 hr post-fertilization to induce Cre recombination that removes the Stop cassette and allows mCherry expression (mCherry channel alone shown in inset). Co-localization shows fgf10b:nEOS expression (green) in endodermal pouches. (d), Endoderm labeling by addition of 4OH-tamoxifen to sox17:CreERT2; ubb:loxP-Stop-loxP-mCherry fish at 6.5 hr post-fertilization results in contribution to cdh1:mlanYFP+ pseudobranch epithelium at 5 dpf. (e,f) In fgf10:nEOS embryos, photoconversion of first pouch endoderm (and some more ventral mandibular cells) at 1.5 dpf labels pseudobranch (white arrow) but not gill epithelia (yellow arrow) at 5 dpf in (e), and conversion of third pouch cells labels the first gill filament epithelium (yellow arrow, boxed region magnified to right and shown in merged and red-only channels) in (f). Scale bars, 50 µM.
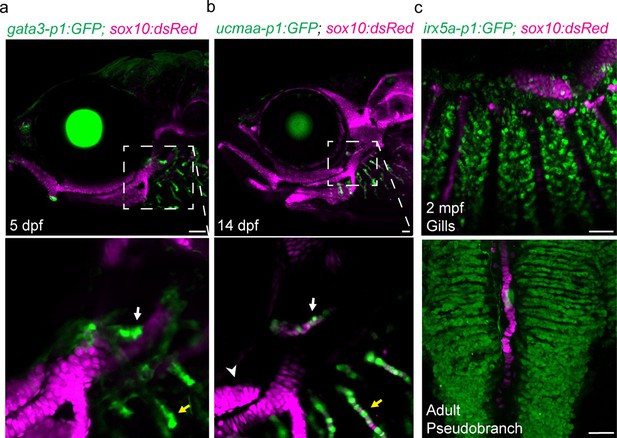
Shared regulatory program for pseudobranch and gill development.
(a-c) In the pseudobranch (white arrows) and gill filaments (yellow arrows), gata3-p1:GFP labels growing buds, ucmaa-p1:GFP labels cellular cartilage (distinct from hyaline cartilage, arrowhead), and irx5a-p1:GFP labels pillar cells. sox10:dsRed labels cartilage for reference. Images in (b) and (c) are confocal projections, with magnified regions shown below in single sections for gata3-p1:GFP and ucmaa-p1:GFP. Scale bars, 50 µM.
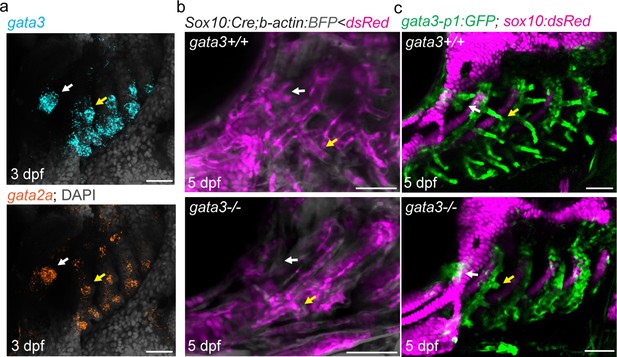
Pseudobranch and gill development requires gata3 function.
(a) Similar expression of gata3 and gata2a in developing pseudobranch (white arrows) and gill regions (yellow arrows). (b) Sox10:Cre; acta2:loxP-BFP-Stop-loxP-dsRed labels Cre-converted dsRed+ neural crest-derived mesenchyme (magenta) and unconverted BFP+ epithelia (gray). (c) gata3-p1:GFP labels pseudobranch and gill filament buds, and sox10:dsRed labels cartilage. For both (b) and (c), 3/3 gata3 mutants displayed reduced formation of the pseudobranch (white arrows) and gill filaments (yellow arrows), compared to 3 controls each. Scale bars, 50 µM.
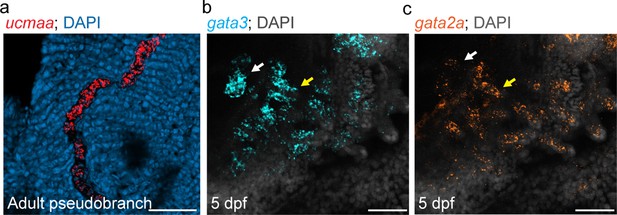
Pseudobranch shares gene expression with gill filaments.
(a) Expression of ucmaa in pseudobranch cartilage from one-year-old fish. (b,c) Developing pseudobranch (white arrows) and gill buds (yellow arrows) express gata3 and gata2a at 5 dpf. DAPI labels nuclei in blue (a) or white (b,c). Scale bars, 50 µM.
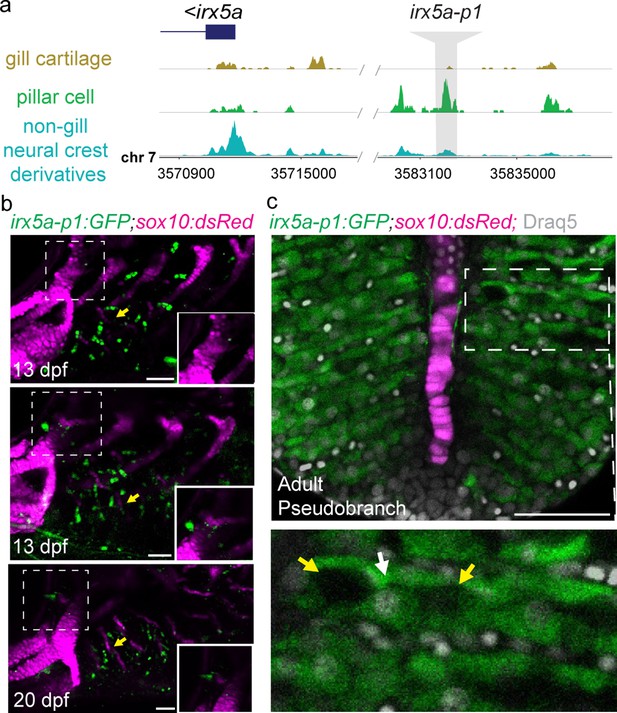
The pseudobranch shares irx5a-p1 pillar cell enhancer activity with gill filaments.
(a) An intergenic irx5a-p1 region displays accessible chromatin specifically in pillar cells at 60 dpf. (b) irx5a-p1 drives GFP expression in pillar cells of the developing gills (yellow arrows) and pseudobranch (insets). (c,d) Dissected pseudobranch from a 60 dpf irx5a-p1:GFP adult shows GFP-positive pillar cells adjacent to a core of filament cartilage. The boxed region magnified below depicts an individual pillar cell (white arrow) flanked by characteristic lacunae (yellow arrows). Cartilage is labeled by sox10:dsRed in (b) and (c). Scale bars, 50 µM.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Danio rerio) | ucmaa | Ensembl: ENSDARG00000027799 | ||
| Gene (Danio rerio) | gata3 | Ensembl: ENSDARG00000016526 | ||
| Gene (Danio rerio) | gata2a | Ensembl: ENSDARG00000059327 | ||
| Gene (Danio rerio) | irx5a | Ensembl: ENSDARG00000034043 | ||
| Genetic reagent (Danio rerio) | Tübingen | ZIRC | RRID:ZIRC_ZL57 | Wildtype strain of zebrafish |
| Genetic reagent (Danio rerio) | Tg(fli1a:eGFP)y1 | Lawson and Weinstein, 2002 | ||
| Genetic reagent (Danio rerio) | Tg(sox10:kikGR)el2 | Balczerski et al., 2012 | ||
| Genetic reagent (Danio rerio) | Tg(ucmaa_p1:GFP, cryaa:Cerulean)el851 | Fabian et al., 2022 | ||
| Genetic reagent (Danio rerio) | Tg(gata3_p1:GFP, cryaa:Cerulean)el858 | Fabian et al., 2022 | ||
| Genetic reagent (Danio rerio) | Tg(fgf10b:nEOS)el865 | Fabian et al., 2022 | ||
| Genetic reagent (Danio rerio) | Tg(–3.5ubb:loxP-STOP-loxP-mCherry)el818 | Fabian et al., 2020 | ||
| Genetic reagent (Danio rerio) | Tg(Mmu.Sox10-Mmu.Fos:Cre)zf384 | Kague et al., 2012 | ||
| Genetic reagent (Danio rerio) | Tg(actab2:loxP-BFP-STOP-loxP-dsRed)sd27 | Kobayashi et al., 2014 | ||
| Genetic reagent (Danio rerio) | Tg(−6.5kdrl:mCherry)ci5 | Proulx et al., 2010 | ||
| Genetic reagent (Danio rerio) | Tg(–5.0sox17:Cre-ERT2,myl7:DsRed)sid1Tg | Hockman et al., 2017 | ||
| Genetic reagent (Danio rerio) | Tg(cdh1:mlanYFP)xt17Tg | Cronan and Tobin, 2019 | ||
| Genetic reagent (Danio rerio) | gata3b1075 | Sheehan-Rooney et al., 2013 | ||
| Genetic reagent (Danio rerio) | Tg(irx5a-p1:GFP, cryaa:Cerulean)el859 | This paper | See Materials and Methods, Section Zebrafish Lines | |
| Recombinant DNA reagent | PCS2FA-transposase | Tol2Kit | PUBMED: 17937395 396.pCS2-transposase | |
| Recombinant DNA reagent | pDestTol2AB2-irx5a-p1-E1B:GFP_pA | This paper | See Materials and Methods, Section Zebrafish Lines | |
| Sequence-based reagent | ucmaa RNAScope probe (Danio rerio); Channel 1 | ACD Bio | ||
| Sequence-based reagent | gata2a RNAScope probe (Danio rerio); Channel 1 | ACD Bio | ||
| Sequence-based reagent | gata3 RNAScope probe (Danio rerio); Channel 2 | ACD Bio | ||
| Commercial assay or kit | In-Fusion HD Cloning Plus | Takara | Takara:638,910 | |
| Commercial assay or kit | RNAScope Multiplex Fluorescent v2 Assay | ACD Bio | ACD Bio:323,100 | |
| Other | Draq5 nuclear dye | Abcam | Abcam:Ab108410 | See Materials and Methods, Section Imaging |