Age acquired skewed X chromosome inactivation is associated with adverse health outcomes in humans
Figures
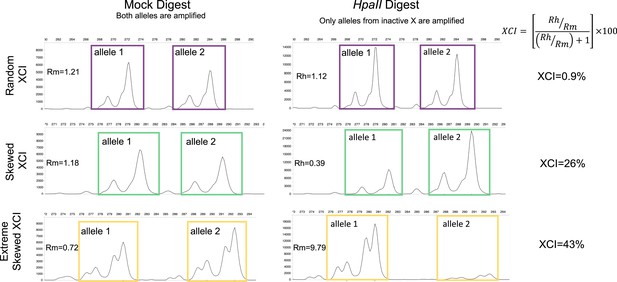
Measuring XCI with the HUMARA assay.
The Human Androgen Receptor Assay (HUMARA) uses methylation-sensitive restriction enzyme digest and PCR to measure skewed X-inactivation. The assay estimates XCI-skew by comparing the relative abundance of allele specific fragments from a mock digest to a methylation-sensitive HpaII digest in which only the alleles from the inactive X are amplified. Representative examples are displayed of fragment analysis of the PCR products for samples with random XCI (top), skewed XCI (middle), and extreme skewed XCI (bottom). The x-axis shows the size, and the y-axis represents the abundance, of the PCR products, respectively. The left panel shows the PCR products after a mock digest with water, resulting in amplification of both alleles regardless of chromosomal inactivation. The right panel shows the PCR products after a restriction enzyme digest with methylation-sensitive enzyme HpaII, resulting in amplification of only the alleles deriving from inactive chromosomes. For each sample, the ratio of the HpaII digested allele products (Rh = allele 1/allele 2) is divided by the ratio of the Mock digest allele products (Rm = allele 1/allele 2) to create a Normalized Ratio (Rn). The XCI percentage is then calculated using the formula [Rn/(Rn +1)] * 100. Images were generated using the Microsatellite Analysis Software on the Thermo Fisher Cloud.
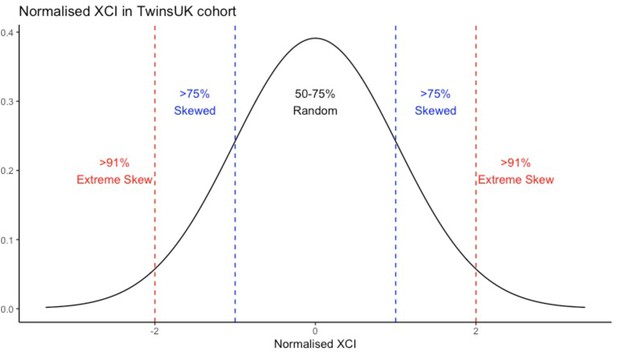
A histogram showing the calculation of the XCI-skew categorical variable from the normalised distribution of XCI scores across the TwinsUK cohort (n=1575).
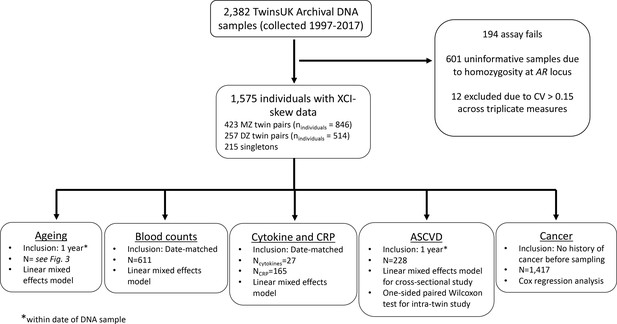
A flowchart of sample processing and inclusion criteria for each results group.
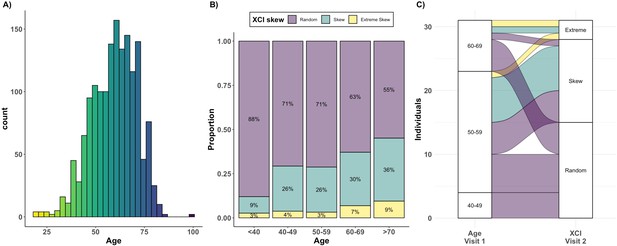
Age acquired XCI-skew across age groups and time.
(A) A histogram displaying the age distribution of the TwinsUK HUMARA cohort (age range: 19–99; median age = 61). (B) The proportions of individuals (y-axis) in each of three XCI-skew categories across increasing age groups (x-axis) are shown (N=1575). (C) A Sankey plot shows the longitudinal changes to XCI in 31 individuals across two measures 15–17 years apart. Colours indicate XCI at visit 1, axis 1 displays the age group of individuals at visit 1, and axis 2 displays XCI at visit 2.
-
Figure 2—source data 1
XCI-skew across age groups.
Categorical variable of XCI-skew and corresponding age group of each individual in the study (n=1575).
- https://cdn.elifesciences.org/articles/78263/elife-78263-fig2-data1-v1.txt
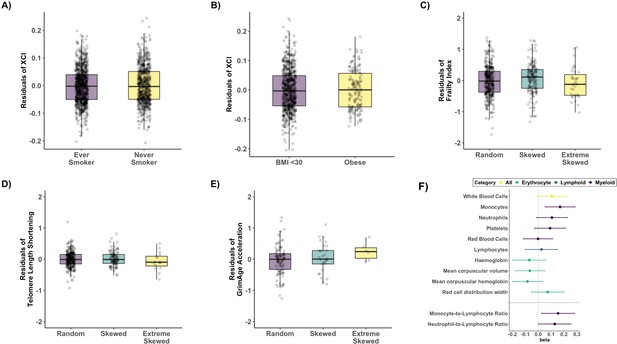
Age acquired XCI-skew and markers of biological ageing and blood cell counts.
Box plots representing the results of the linear regression mixed effects models to assess (A) Smoking status (p=0.33, Nnever_smoker = 879; Never_smoker = 673) and (B) obesity (p=0.88, Nnot_obese = 726; Nobese = 165) after correcting for age with XCI as the dependent variable, and (C) frailty index (p=0.59, Nranodm XCI = 398; nSkewed XCI = 177; Nextreme skew = 36), after correcting for age and BMI, (D) Leukocyte telomere length shortening (p=0.9, Nranodm XCI = 278; nSkewed XCI = 103; Nextreme skew = 16) after correcting for age and smoking status, and (E) DNAm GrimAge acceleration (p=0.22, Nranodm XCI = 101; nSkewed XCI = 30; Nextreme skew = 6), with XCI-skew as the dependent variable. All boxplots display the median and IQR, and have the residuals of the models on the y-axis. (F) A forest plot of associations with data-matched Complete Blood Count data (top panel) and Myeloid-to-lymphoid ratios (bottom) with effect size and lower and upper confidence intervals indicated. Associations were tested with XCI-skew as an independent variable after controlling for age, BMI, seasonality, and smoking status as fixed effects in a linear mixed effects model (Nranodm XCI = 445; nSkewed XCI = 183; Nextreme skew = 43). The significance threshold after Bonferroni correction was p<0.005 and p<0.023 to account for multiple testing across the 10 tests and 2 tests, respectively.
-
Figure 3—source data 1
XCI-skew and DNAm GrimAge Acceleration.
Normalised DNAm GrimAge Acceleration variable and categorical XCI-skew data (n=137).
- https://cdn.elifesciences.org/articles/78263/elife-78263-fig3-data1-v1.txt
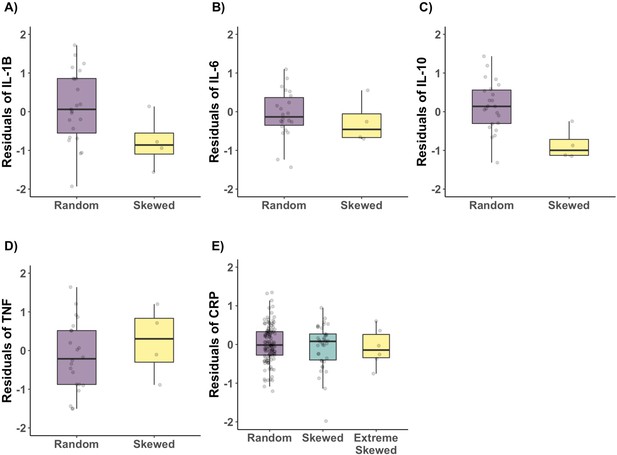
Age acquired XCI-skew and Cytokine and CRP measures.
Using a linear regression mixed effects model to control for batch effects, age, seasonality, family structure and zygosity, XCI-skew was assessed for its association with (A) IL-1β (p=0.02); (B) IL-6 (p=0.41); (C) IL-10 (p=0.0008); (D) TNF (p=0.61); (E) CRP (p=0.41).
All boxplots display the median and IQR, and have the residuals of the models on the y-axis. The significance threshold after Bonferroni correction for 5 tests was p=0.01. The sample sizes are (A-D) Nranodm XCI=23; nSkewed XCI = 4; (E) Nranodm XCI=121; nSkewed XCI = 38; Nextreme skew = 6.
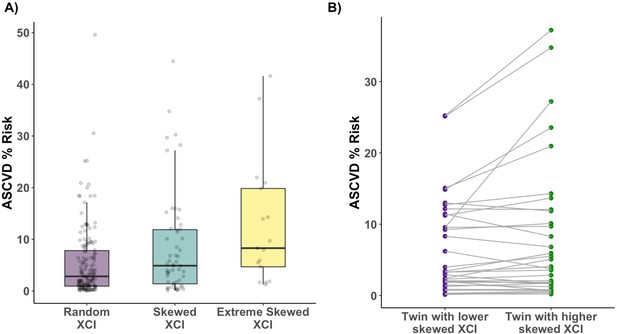
Age acquired XCI-skew and cardiovascular disease risk score.
(A) Using a linear regression mixed effects model to control for BMI, monocyte count, relatedness and zygosity, XCI-skew is associated with increased atherosclerotic cardiovascular disease risk score (Nranodm XCI = 155; nSkewed XCI = 56; Nextreme skew = 17; p=0.01). The boxplot displays the median and IQR. (B) Using age-matched twin pairs discordant for their XCI-skew status (Npairs = 34), XCI-skew is associated with increased atherosclerotic cardiovascular disease risk score in the intra-twin analysis (one-sided paired samples Wilcoxon test; p=0.025).
-
Figure 4—source data 1
XCI-skew and ASCVD Risk Score.
ASCVD Risk Score, log ASCVD Risk Score, and categorical XCI-skew data (n=228).
- https://cdn.elifesciences.org/articles/78263/elife-78263-fig4-data1-v1.txt
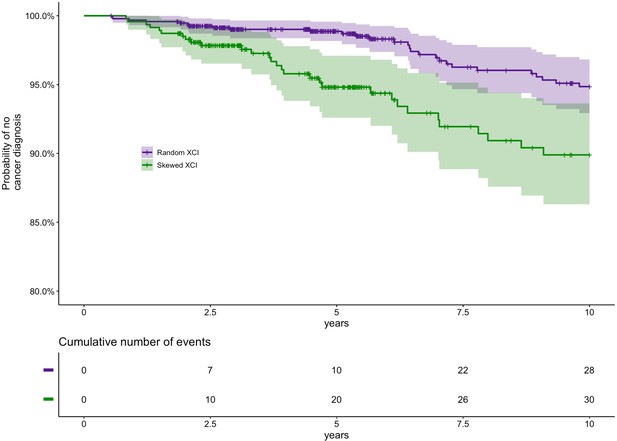
Prospective study of XCI-skew and future cancer diagnosis.
Using a Cox regression analysis and controlling for age, relatedness and zygosity, XCI-skew is predictive of future cancer incidence in a 10 year follow-up (p=0.012; HR = 1.95). Each cancer event represents the first cancer diagnosis (excluding non-melanoma skin cancer) in each individual. A Kaplan-Meier plot (top) and cumulative events (bottom) of cancer diagnosis in individuals with XCI-skew (N=469) and random XCI (N=948) are shown. 2.9% (28/948) of individuals with random XCI, and 6.4% (30/469) of individuals with XCI-skew, developed cancer in the 10 years follow-up.
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/78263/elife-78263-transrepform1-v1.docx
-
Supplementary file 1
Number of cancer diagnoses recorded in 10-year follow-up by organ/site.
- https://cdn.elifesciences.org/articles/78263/elife-78263-supp1-v1.docx
-
Reporting standard 1
STROBE statement.
- https://cdn.elifesciences.org/articles/78263/elife-78263-repstand1-v1.docx





