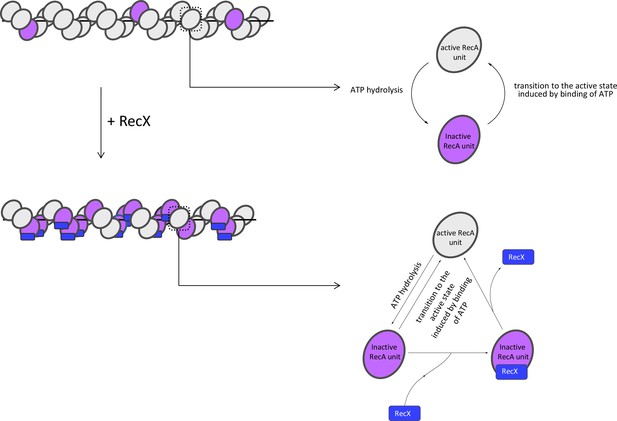
A new insight into RecA filament regulation by RecX from the analysis of conformation-specific interactions
Figures
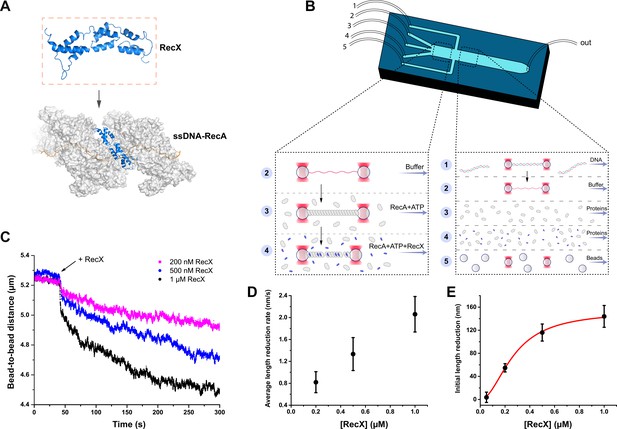
The study of the RecX effect on the RecA-ssDNA filaments.
(A) A schematic of RecX binding along the groove of the active RecA-ssDNA. Atomic structure model for RecA::RecX::ssDNA is adopted from Shvetsov et al., 2014. (B) A schematic of a five-channel microfluidic flow cell (Lumicks). Dash line highlights two working regions. The three-channel region was used to study the effect of RecX on the RecA-ssDNA filament. In the five-channel region, the beads trapping, DNA tether formation, and generation of ssDNA by force-induced melting were performed. (C) The change in the length of RecA-ssDNA filament upon transition from the channel containing 1 μM RecA and 1 mM ATP to the channel containing 1 μM RecA, 1 mM ATP, and various concentrations of RecX. During incubation, a constant tension of 3 pN was applied to the tether. (D) The impact of RecX concentration on the average rate of reduction in the RecA-ssDNA filament length over 250 s after initial steep decrease. (E) The dependence of the RecX induced initial sharp decrease in RecA-ssDNA filament on the RecX concentration. Solid curve - fit of experimental data with Hill equation with a Hill coefficient of 2.0±0.3. Each data point in (D) and (E) is a mean value of at least three measurements, bars represent SD.
-
Figure 1—source data 1
Source data for traces of RecA-ssDNA filaments, average length reduction rate, and initial length reduction values.
- https://cdn.elifesciences.org/articles/78409/elife-78409-fig1-data1-v2.xlsx
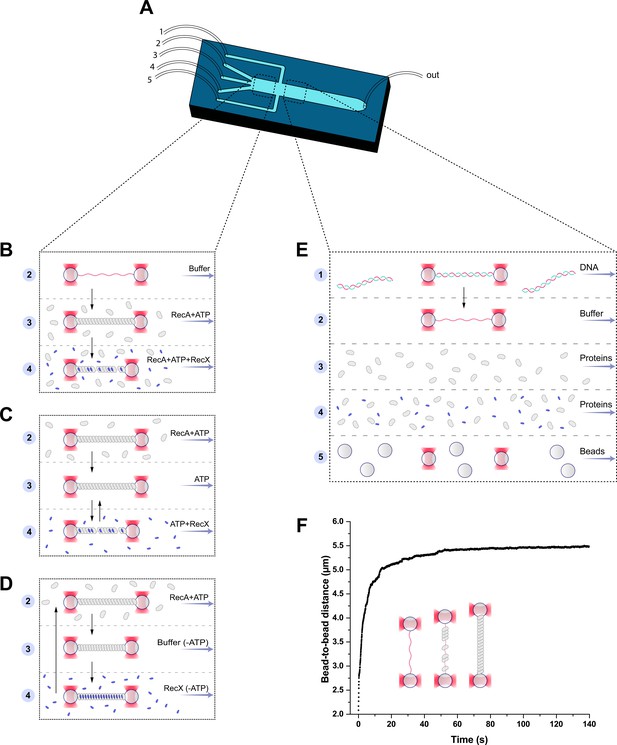
Single-molecule assay.
Single-molecule manipulations were performed within a five-channel microfluidic flow chip (A). Two working regions are highlighted with a dash line. The three-channel region (left) was used to study the effect of RecX on the RecA-ssDNA filament (B–D). In the five-channel region, the beads trapping, DNA tether formation, and generation of ssDNA by force-induced melting were performed (E). The RecA-ssDNA filaments were assembled by applying a stretching force of 12 pN to the ssDNA molecule in the channel containing 1 μM RecA and 1 mM ATP. Binding of RecA to ssDNA was followed by an increase in the end-to-end distance (F).
-
Figure 1—figure supplement 1—source data 1
Source data for Figure 1—figure supplement 1E.
- https://cdn.elifesciences.org/articles/78409/elife-78409-fig1-figsupp1-data1-v2.xlsx
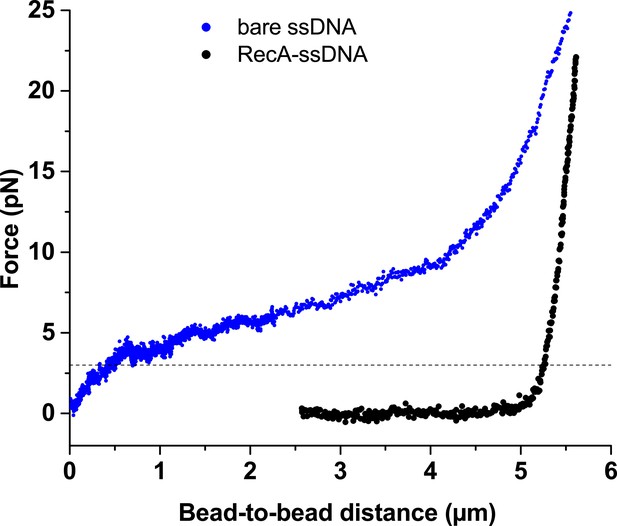
The comparison of force-extension behavior of bare ssDNA (blue) and the ATP-bound RecA-ssDNA filament (black).
Adopted from Alekseev et al., 2020b. The dash line indicates 3 pN tension.
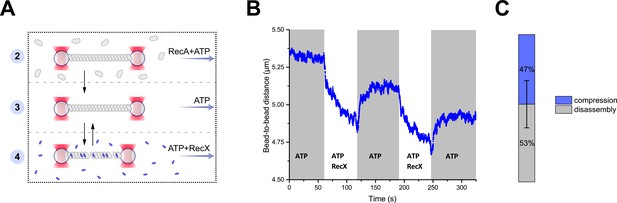
RecX-induced reversible changes in the RecA-ssDNA filament structure.
(A) A schematic of the experiment revealing that RecX is able to induce reversible structural changes in the RecA-ssDNA filaments. (B) RecX induces reversible changes in the RecA-ssDNA filament structure in the presence of ATP. (C) A comparison of the reversible (compression) and the irreversible (disassembly) reduction in RecA-ssDNA filament length. Stacked histogram represents multiple measurements for six different molecules. Bars represent SD.
-
Figure 2—source data 1
Source data for RecX-induced reversible changes in the length of RecA-ssDNA filament.
- https://cdn.elifesciences.org/articles/78409/elife-78409-fig2-data1-v2.xlsx
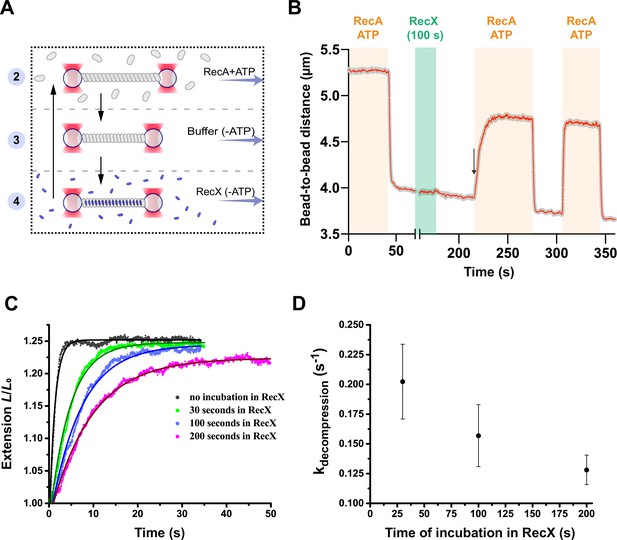
RecX affects the conformational transition of RecA-ssDNA filament from the inactive state to the active state.
(A) A schematic of the experiment revealing that RecX binds inactive RecA-ssDNA filaments. (B) The change of the RecA-ssDNA filament length upon conformational transitions between apo and ATP-bound states. Incubation of apo RecA-ssDNA filament with 500 nM RecX (green area) leads to a slowdown of the subsequent decompression of the RecA-ssDNA filament (black arrow points the beginning of the slowed down decompression). A constant tension of 3 pN was applied to the tether during incubation and transitions. (C) Relative extension of the RecA-ssDNA filament in the course of decompression after incubation of inactive RecA-ssDNA filament with 500 nM RecX for 30, 100, and 200 s. (D) Corresponding rate constants of the decompression obtained by exponential fitting (solid line in (C)) of the elongation profiles. Each point is a mean of at least six measurements. Bars represent SD.
-
Figure 3—source data 1
Source data for slowdown decompression of apo RecA-ssDNA filament caused by incubation with RecX.
- https://cdn.elifesciences.org/articles/78409/elife-78409-fig3-data1-v2.xlsx
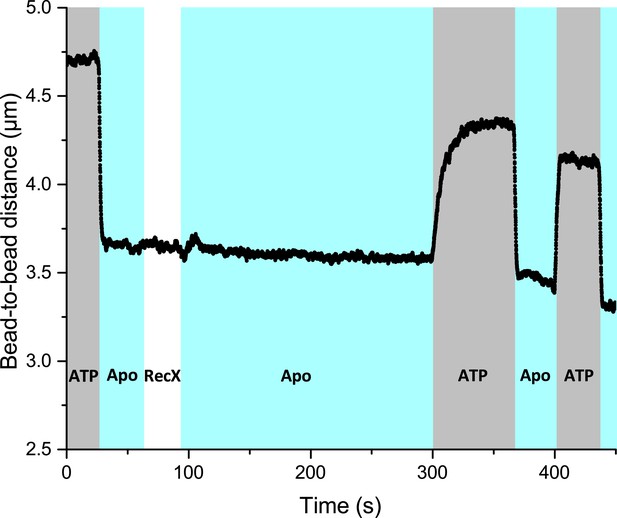
The effect of the slowed down decompression retains when RecA-ssDNA filament is incubated in the RecX-free buffer after short incubation with RecX.
ATP-containing channel was also supplemented with free RecA.
-
Figure 3—figure supplement 1—source data 1
Source data for Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/78409/elife-78409-fig3-figsupp1-data1-v2.xlsx
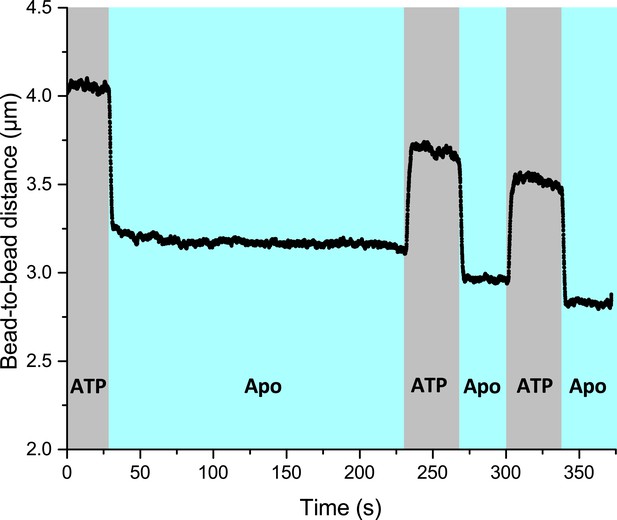
The effect of the slowed down decompression is independent of incubation time of the RecA-ssDNA filament in the apo channel in the absence of RecX.
ATP-containing channel was also supplemented with free RecA.
-
Figure 3—figure supplement 2—source data 1
Source data for Figure 3—figure supplement 2.
- https://cdn.elifesciences.org/articles/78409/elife-78409-fig3-figsupp2-data1-v2.xlsx
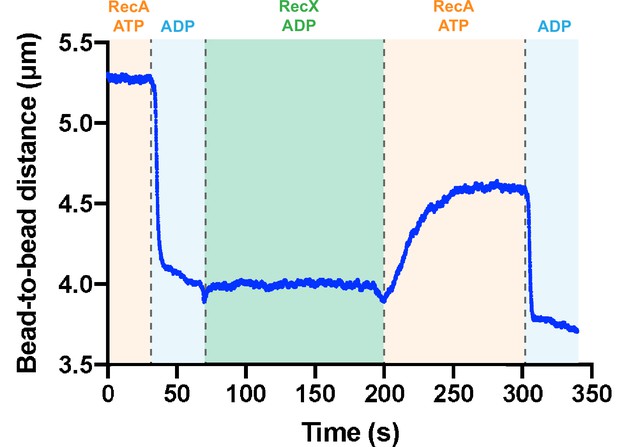
Incubation of ADP-bound form of the RecA-ssDNA filament with RecX results in the slowdown of the following decompression.
-
Figure 3—figure supplement 3—source data 1
Source data for Figure 3—figure supplement 3.
- https://cdn.elifesciences.org/articles/78409/elife-78409-fig3-figsupp3-data1-v2.xlsx
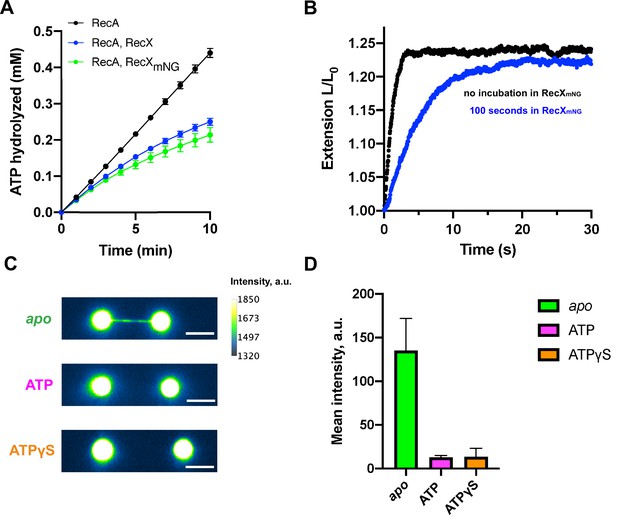
Fluorescent visualization reveals that RecX dissociates from the ATP-bound state of the RecA-ssDNA.
(A) Inhibition of RecA ATPase activity by wild-type RecX (blue) and fluorescent mNeonGreen-RecX (RecXmNG) (green). ATP hydrolysis by RecA in the absence of RecX is shown in black. Each data point represents the average of three independent experiments (error bars – SD). (B) Relative extension of the RecA-ssDNA filament in the course of apo-ATP transition without incubation in RecXmNG (black curve) and after incubation of apo RecA-ssDNA filament with 500 nM RecXmNG for 100 s (blue curve). (C) Fluorescent images of: RecA-ssDNA filament in apo (top) and ATP-bound state (middle) after incubation with 1 μM RecXmNG for 30 s; RecA-ssDNA filament assembled in the presence of ATPgS (bottom) after incubation with 1 μM RecXmNG for 30 s. Scale bar is 5 μm. (D) Comparison of the average intensity of the tether after incubation with RecXmNG for apo (N=6 molecules), ATP-bound RecA-ssDNA filament (N=3 molecules), and the filament assembled in the presence of ATPgS (N=6 molecules) (consistently with (B)). Data are representative of three independent experiments, and values are expressed in mean ± SD.
-
Figure 4—source data 1
Source data for RecXmNG-induced slowdown decompression of RecA-ssDNA filament and average intensity values for apo, ATP-bound, and ATPγS-bound RecA-ssDNA filaments after incubation with RecXmNG.
- https://cdn.elifesciences.org/articles/78409/elife-78409-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Raw tiff images of apo, ATP-bound, and ATPγS-bound RecA-ssDNA filaments after incubation with RecXmNG.
- https://cdn.elifesciences.org/articles/78409/elife-78409-fig4-data2-v2.zip
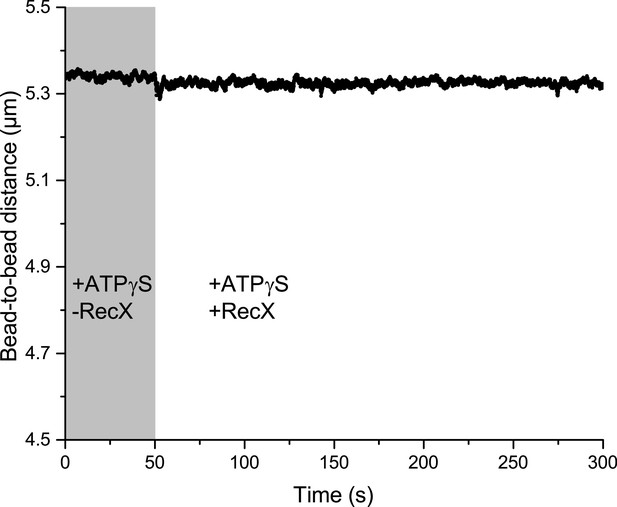
The effect of 1 µM RecX on the dynamics of RecA-ssDNA filament formed in the presence of 0.5 mM ATPγS.
-
Figure 4—figure supplement 1—source data 1
Source data for Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/78409/elife-78409-fig4-figsupp1-data1-v2.xlsx
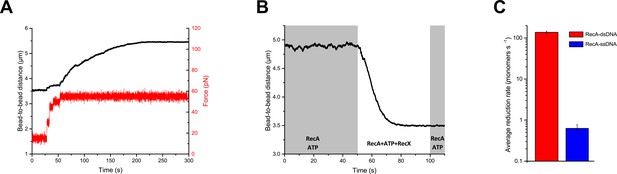
RecX effectively promotes disassembly of RecA-dsDNA filaments.
(A) The assembly of the RecA-dsDNA filament. (B) The disassembly of the RecA-dsDNA filament in the presence of 200 nM RecX. (C) The comparison of the average length reduction of RecA-dsDNA (N=4) and RecA-ssDNA (N=4) filament induced by 200 nM RecX. The data for RecA-ssDNA is consistent with Figure 1D. Data are representative of at least three independent experiments, and values are expressed in mean ± SD.
-
Figure 5—source data 1
Source data for RecA-dsDNA filament assembly profile, RecX-induced disassembly of RecA-dsDNA filament, and average length reduction rate for RecA-dsDNA and RecA-ssDNA filaments in the presence of RecX.
- https://cdn.elifesciences.org/articles/78409/elife-78409-fig5-data1-v2.xlsx
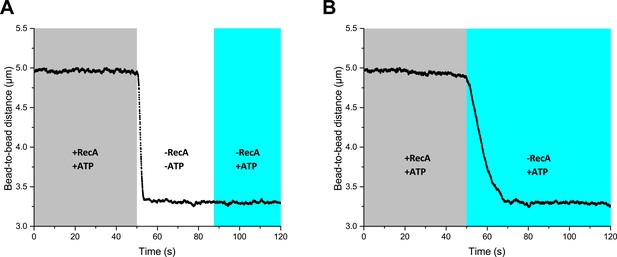
RecA-dsDNA filament is stable only in the presence of both free RecA and ATP.
(A) ATP elimination leads to rapid RecA-dsDNA filament disassembly. (B) The elimination of free RecA promotes RecA-dsDNA filament disassembly.
-
Figure 5—figure supplement 1—source data 1
Source data for Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/78409/elife-78409-fig5-figsupp1-data1-v2.xlsx
Videos
Continuous fluorescent visualization of the RecXmNG-RecA-ssDNA filament during transfer from the ATP-free to the ATP-containing channel.
Scale bar is 5 µm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Recombinant DNA reagent | prl574-rpoC (plasmid) | This paper | DNA manipulation, 11 kbp-long substrate; Khodorkovskii Lab, NanoBio, SPbPU, St.Petersburg, Russia | |
| Recombinant DNA reagent | pBAD-mng-recx (plasmid) | This paper | Histag-mNeonGreen-RecX purification (Ampicillin resistance); Khodorkovskii Lab, NanoBio, SPbPU, St.Petersburg, Russia | |
| Recombinant DNA reagent | Lambda DNA | New England BioLabs | New England BioLabs:N3011L | |
| Sequence-based reagent | Xbai_L_bio | Alkor Bio | 5’-CTAGCGAGTGXXXXX-3’ (X denotes biotin tag) | |
| Sequence-based reagent | SacI_L_bio | Alkor Bio | 5’-XXXXXCAGTCCAGCT-3’ (X denotes biotin tag) | |
| Sequence-based reagent | SacI_S | Alkor Bio | 5’-GGACTG-3’ | |
| Peptide, recombinant protein | Klenow fragment | Thermo Scientific | Thermo Scientific:EP0052 | |
| Chemical compound, drug | pyruvate kinase (from rabbit muscle) | Sigma | Sigma:P1506-5KU | |
| Chemical compound, drug | lactate dehydrogenase (from rabbit muscle) | Sigma | Sigma: L2500-25KU | |
| Chemical compound, drug | Biotin-16dCTP | Jena Bioscience | Jena Bioscience:NU-809-BIO16-S | |
| Chemical compound, drug | ATP | Sigma | Sigma:A7699-1G | |
| Chemical compound, drug | ATPγS | Sigma | Sigma:A1388-25MG | |
| Chemical compound, drug | poly(dT) | Sigma | Sigma:P6905-5UN | |
| Chemical compound, drug | PEP | Sigma | Sigma:P7252-1G |
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/78409/elife-78409-transrepform1-v2.docx
-
Source code 1
Average RecXmNG Intensity Calculation.
- https://cdn.elifesciences.org/articles/78409/elife-78409-code1-v2.zip