Single-cell profiling of lncRNAs in human germ cells and molecular analysis reveals transcriptional regulation of LNC1845 on LHX8
Figures
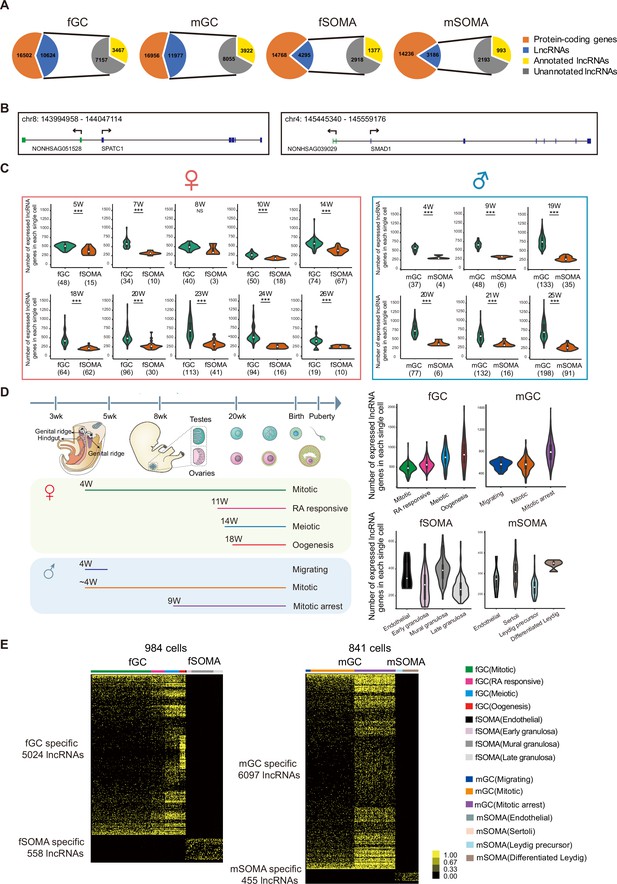
Figure numerous.
(A) Long non-coding RNAs (lncRNAs) are differentially expressed during human germ cell (hGC) development. (B) Two unannotated lncRNAs and their neighboring protein-coding genes with their genomic positions. (C) Number of lncRNAs in an individual cell of hGCs and gonadal somatic cells from different developmental weeks. Student’s t-test was used for the comparisons. ***p < 0.001. (D) The composition of different cell types from different developmental weeks and the numbers of lncRNAs in an individual cell from different cell types. The color lines below indicate cell collections from different developmental weeks. (E) Expression heatmap of hGC-specific lncRNAs and gonadal somatic cell-specific lncRNAs.
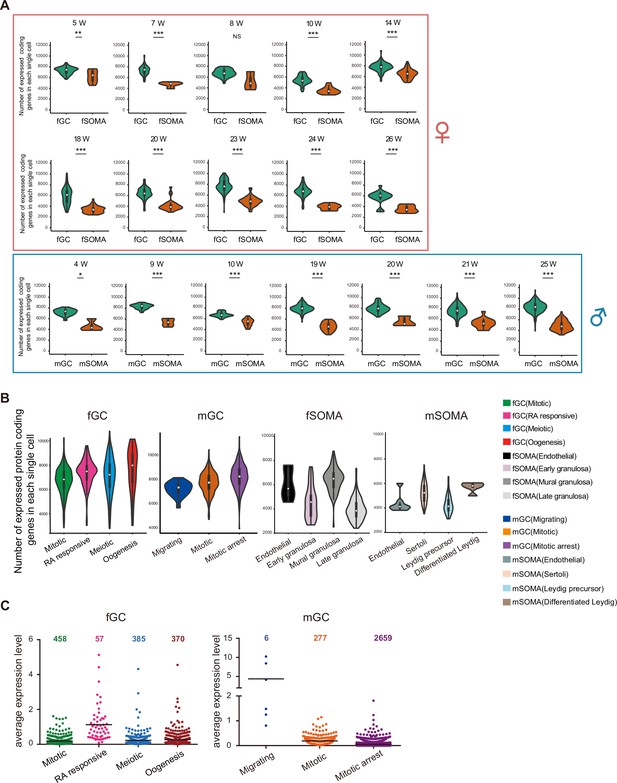
Numerous long non-coding RNAs (lncRNAs) are differentially expressed during human germ cell development.
(A) Numbers of protein-coding genes expressed in individual female (upper) and male (bottom) germ cells or somatic cells from different developmental week. Student’s t-test was used for the comparisons. *p < 0.05, **p < 0.01, ***p < 0.001. (B) Numbers of protein-coding genes expressed in an individual cell from different cell types. (C) lncRNA numbers and average expression level of different developmental stages. Data information: The same color markings are used in (B) and (C).
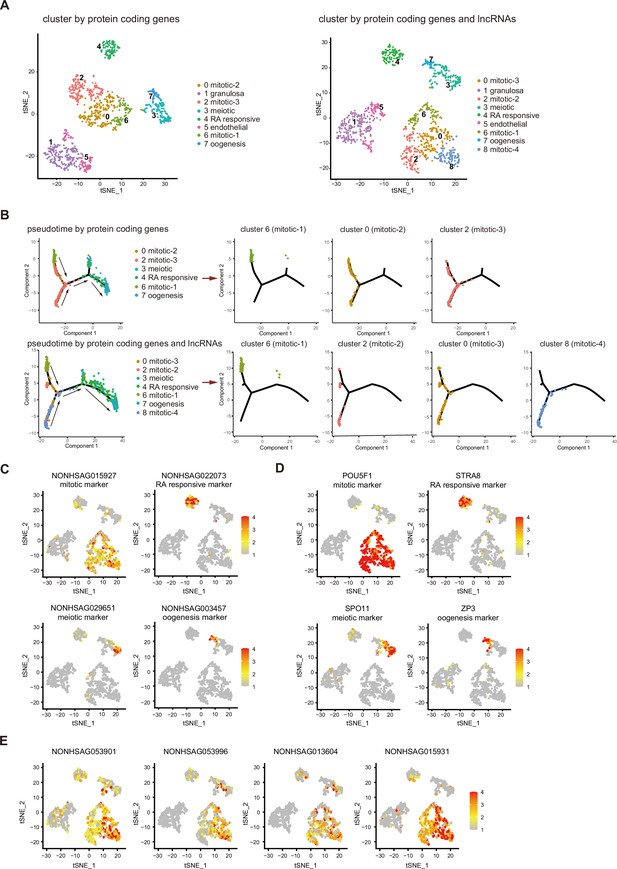
Integrating long non-coding RNA (lncRNA) expression enhances cell-type classifications of human female germ cells.
(A) tSNE plot of female germ cells and somatic cells colored by identified cell types. Left: cluster according to protein-coding genes. Right: cluster according to protein-coding genes and lncRNAs. (B) Single-cell trajectories of female germ cell states through the pseudotime according to protein-coding genes or protein-coding genes with lncRNAs. The different subtypes of mitotic stage germ cells are shown through the pseudotime separately. Arrows indicate the developmental order of these cells. (C) Expression pattern of identified cell-type marker lncRNAs exhibited on t-SNE plots, including mitotic, RA responsive, meiotic, and oogenesis markers. (D) Expression pattern of identified cell-type marker coding genes exhibited on t-SNE plots, including mitotic, RA responsive, meiotic, and oogenesis markers. (E) Expression pattern of newly identified mitotic-4 cell lncRNA markers exhibited on t-SNE plots. Data information: In (C–E), a gradient of gray, yellow, orange, and red indicates low to high expression.
Integrating long non-coding RNA (lncRNA) expression enhances cell-type classifications of human female germ cells.
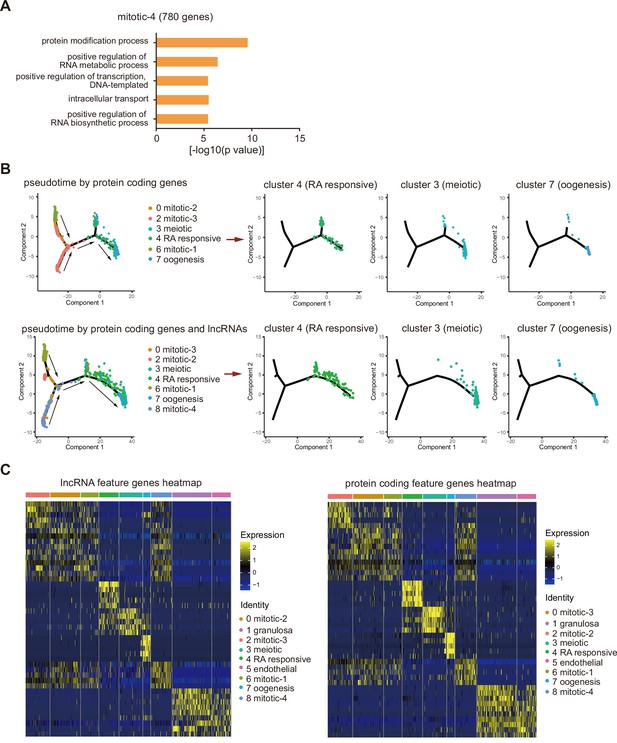
(A) Gene ontology biological processes enrichment of mitotic-4 cells feature genes. (B) Single-cell trajectories of female germ cell states through the pseudotime according to protein-coding genes or protein-coding genes with lncRNAs. Germ cells from different developmental stages are shown through the pseudotime separately. (C) Expression heatmap of feature lncRNAs or protein-coding genes of the nine clusters.
Genomic distributions and biotypes of the long non-coding RNAs (lncRNAs) expressed during human gonadal development.
(A) The genomic position distribution of lncRNAs expressed in human germ cells (hGCs) and gonadal somatic cells. (B) The distribution and percentage of the six locus biotypes of each developmental stage-specific lncRNAs. (C) Expression correlations of lncRNA–mRNA pairs of the six biotypes in different developmental stages.
Genomic distributions and biotypes of the long non-coding RNAs (lncRNAs) expressed during human gonadal development.
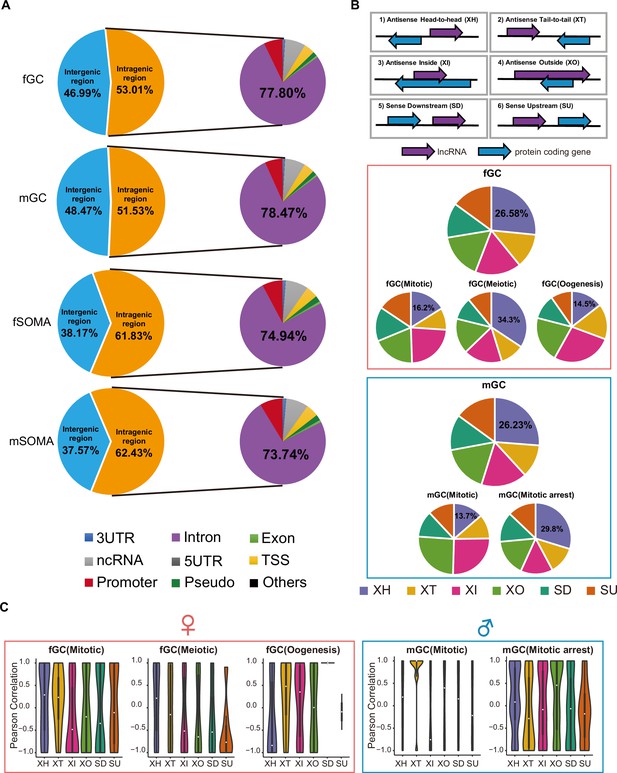
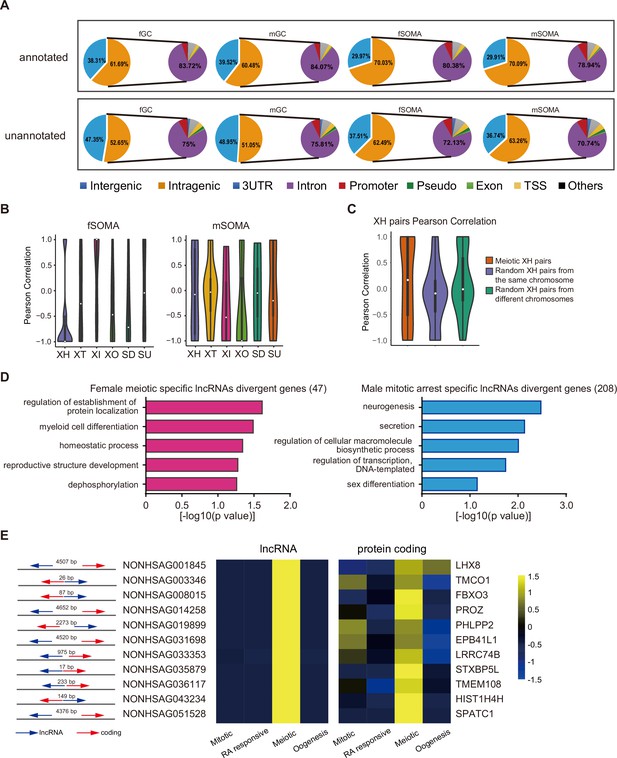
(A) The genomic position distribution of annotated and unannotated lncRNAs expressed in human germ cells (hGCs) and gonadal somatic cells. (B) Expression correlations of lncRNA–mRNA pairs of the six biotypes in fSOMA and mSOMA. (C) Expression correlations of XH lncRNA–mRNA pairs from meiotic specific expressed pairs or randomly selected pairs on the same or different chromosomes. (D) Gene ontology biological processes enrichment of female meiotic specific lncRNAs divergent genes and male mitotic arrest-specific lncRNAs divergent genes. (E) Relative position of meiotic specific expressed divergent lncRNA–mRNA pairs and their relative expression in different developmental stages.
Characterization of a divergent long non-coding RNA (lncRNA), LNC1845, expressed during the meiotic stage and is required for normal expression of LHX8.
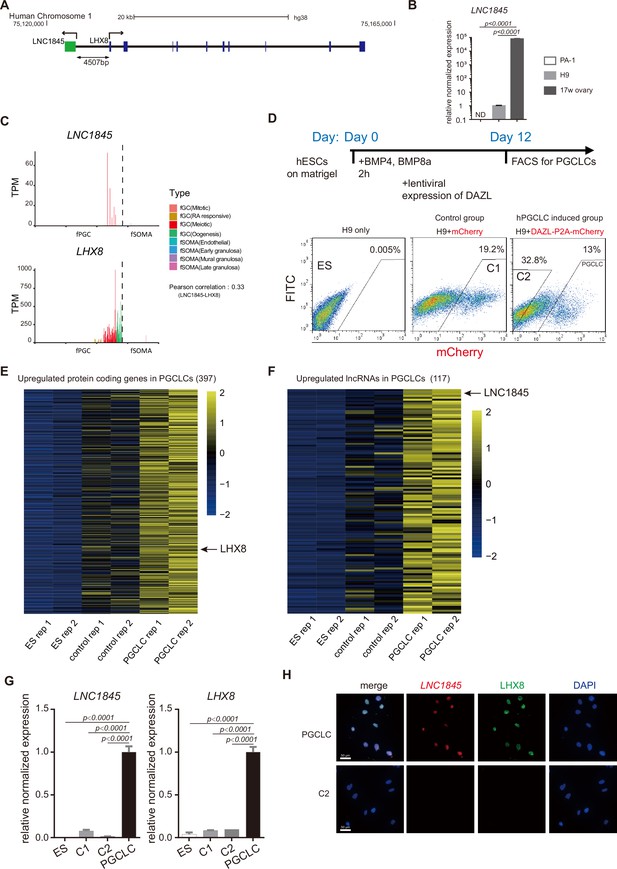
(A) Genomic locus and relative position of LNC1845 (in green) and LHX8 (in blue). (B) Expression analysis by RT-qPCR of PA-1 cells, undifferentiated H9 cells, and RNA sample extracted from human 17-week ovary (n = 3 technical replicates). (C) LNC1845 and LHX8 expression level in single hPGC or gonadal somatic cells from different developmental stages. (D) Schematic timelines for primordial germ cell-like cell (PGCLC) induction. FACS plots showing distinct populations expressing mCherry fluorescent proteins, indicating control groups or PGCLCs. (E) Expression heatmap of female germ cell (fGC)-specific protein-coding genes upregulated in PGCLCs. (F) Expression heatmap of fGC-specific lncRNAs upregulated in PGCLCs. (G) Expression analysis by RT-qPCR of undifferentiated H9 (ES) cells (n = 3), mCherry-positive control (control-1) cells (n = 3), DAZL-negative (control-2) cells (n = 3), and DAZL-positive PGCLCs. (H) Immunofluorescence of LHX8 (green) co-stained with RNA fluorescent in situ hybridization (RNA FISH) of LNC1845 (red) in PGCLCs or control cells. Scale Bar, 50 μm.
In (B) and (G), the y-axis represents relative mean expression normalized to GADPH and control cells. Error bars indicate the mean ± standard deviation (SD), three independent experiments were carried out for (G). One-way analysis of variance (ANOVA) was used for the comparisons.
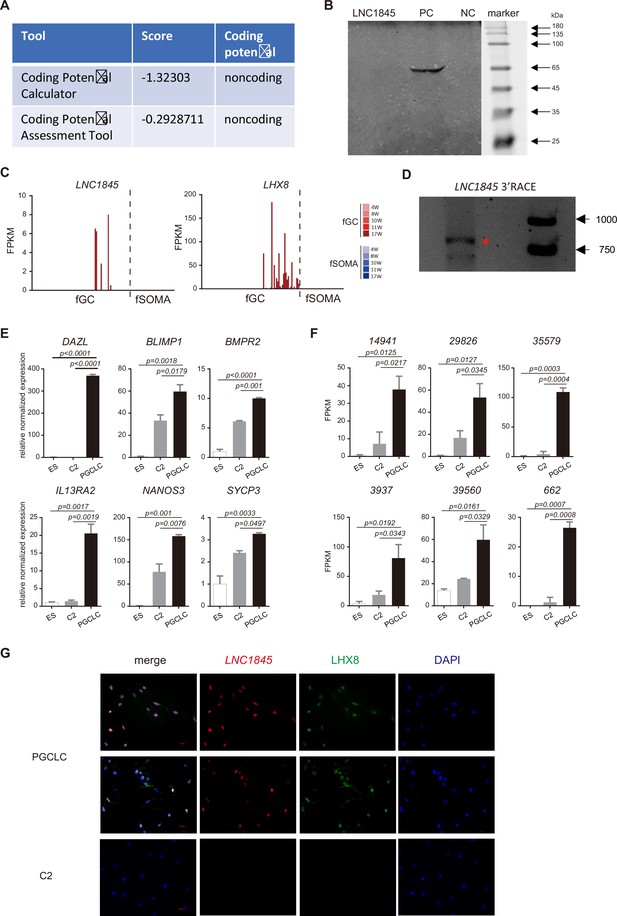
Characterization of a divergent long non-coding RNA (lncRNA), LNC1845, expressed during the meiotic stage and is required for normal expression of LHX8.
(A) LNC1845 was predicted to be non-coding RNA. The RNA sequences of LNC1845 were put into the Coding Potential Calculator (CPC) program and the Coding Potential Assessment Tool. (B) In vitro transcription and translation of LNC1845. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) showed that there was no band detected in the LNC1845 lane. NC, plasmid with empty T7 promoter was used as the negative control. PC, plasmids with T7 promoter driving luciferase, was used as the positive control. (C) LNC1845 and LHX8 expression level in single human germ cells (hGCs) or gonadal somatic cells from different developmental weeks. (D) 3′RACE of LNC1845 in primordial germ cell-like cells (PGCLCs). (E) RT-qPCR analysis of germ cell marker genes in undifferentiated H9 (ES) cells, DAZL-negative (control-2) cells, and DAZL-positive PGCLCs. The y-axis represents relative mean expression normalized to GADPH and control cells. Error bars indicate the mean ± standard deviation (SD), three independent experiments were carried out. One-way analysis of variance (ANOVA) was used for the comparisons.(F) RNA-seq results of germ cell-specific lncRNAs in PGCLCs and control cells. One-way analysis of variance (ANOVA) was used for the comparisons.(G) Immunofluorescence of LHX8 (green) co-stained with RNA fluorescent in situ hybridization (RNA FISH) of LNC1845 (red) in PGCLCs or control cells. Scale Bar, 100 μm.
-
Figure 4—figure supplement 1—source data 1
Characterization of a divergent long non-coding RNA (lncRNA), LNC1845, expressed during the meiotic stage and is required for normal expression of LHX8.
- https://cdn.elifesciences.org/articles/78421/elife-78421-fig4-figsupp1-data1-v2.zip
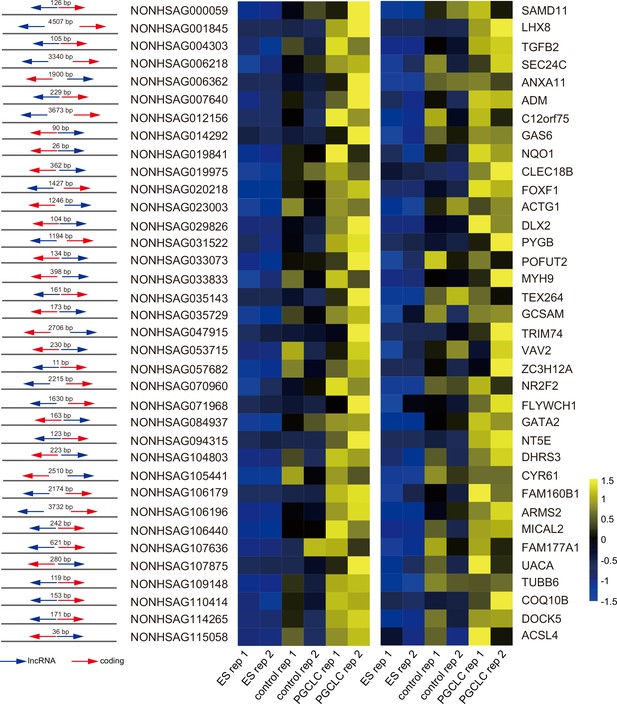
Upregulated XH long non-coding RNA (lncRNA)–mRNA pairs in hPGCLCs.
Relative position of upregulated divergent lncRNA–mRNA pairs and their relative expression in primordial germ cell-like cells (PGCLCs) and control cells.
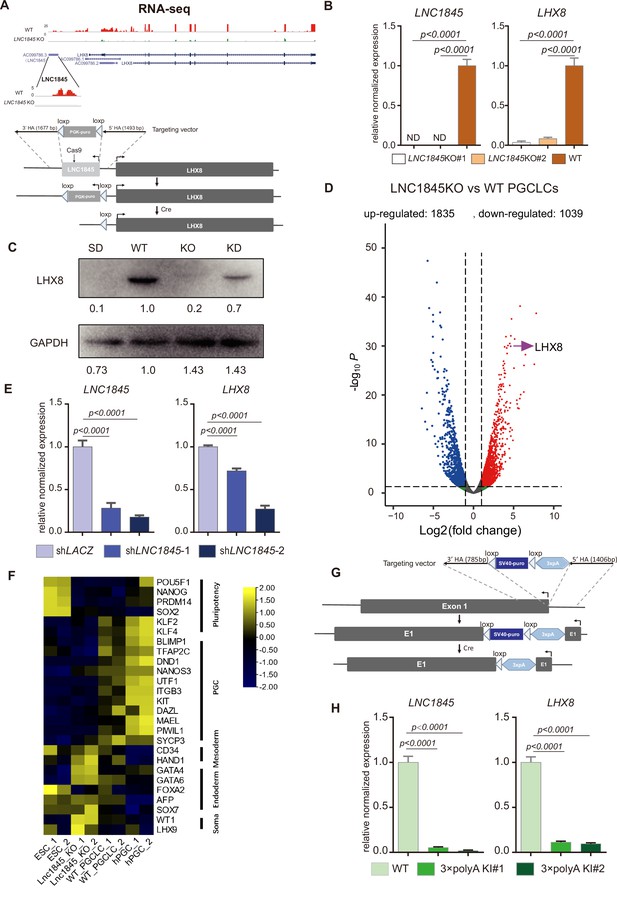
LNC1845 RNA transcripts are required for normal expression of LHX8.
(A) Schematic diagram of knockout strategies at the LNC1845 loci. (B) Expression analysis by RT-qPCR of WT primordial germ cell-like cells (PGCLCs) (n = 3) and LNC1845 KO PGCLCs (n = 3). (C) Western blot analysis of LHX8 in PGCLCs from WT, LNC1845 KO, or RNA interfering LNC1845 (KD) human embryonic stem cells (hESCs). SD, spontaneous differentiation cells as the negative control. (D) Volcano plots of the differentially expressed protein-coding genes between LNC1845KO and WT PGCLCs. The screening threshold of differentially expressed genes (fold change) is ≧2, p < 0.05. (E) Expression analysis by RT-qPCR analysis of LNC1845 and LHX8 in LNC1845 RNAi cells (n = 3). (F) Heatmap of gene expression of key primordial germ cell (PGC)-associated genes and of pluripotency, mesoderm, endoderm, and somatic markers. (G) Schematic diagram of LNC1845-upstream 3×polyA knock-in and the targeted allele after 3×polyA insertion. (H) Expression analysis by RT-qPCR of LNC1845 and LHX8 in WT PGCLCs (n = 3) and LNC1845 3×polyA KI cells (n = 3).
In (B), (E), and (H), the y-axis represents relative mean expression normalized to GADPH and control cells. Error bars indicate the mean ± standard deviation (SD), three independent experiments were carried out. One-way analysis of variance (ANOVA) was used for the comparisons.
-
Figure 5—source data 1
Western blot analysis of LHX8 in PGCLCs from WT, LNC1845 KO, or RNA interfering LNC1845 (KD) hESCs. SD, spontaneous differentiation cells as the negative control.
- https://cdn.elifesciences.org/articles/78421/elife-78421-fig5-data1-v2.zip
-
Figure 5—source data 2
Western blot analysis of GAPDH in PGCLCs from WT, LNC1845 KO, or RNA interfering LNC1845 (KD) hESCs. SD, spontaneous differentiation cells as the negative control.
- https://cdn.elifesciences.org/articles/78421/elife-78421-fig5-data2-v2.zip
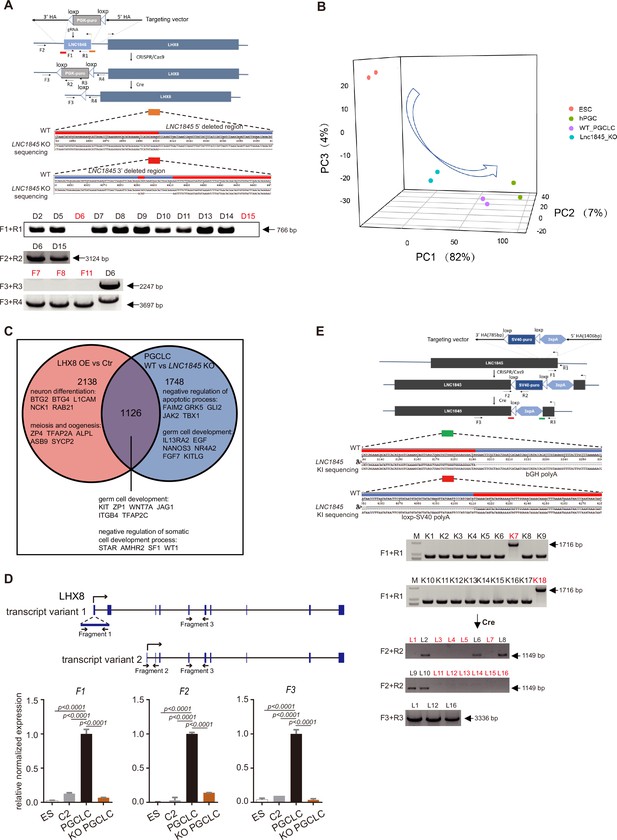
LNC1845 RNA transcripts are required for normal expression of LHX8.
(A) Genotyping and sequencing of LNC1845 KO single colonies to confirm the deletion of LNC1845. (B) Principle component analysis (PCA) of RNA-seq datasets from ESCs, LNC1845 KO primordial germ cell-like cells (PGCLCs), WT PGCLCs, and in vivo hPGCs. (C) Venn diagram depicting the overlap of differential expression genes examined in LNC1845 KO and LHX8 overexpressed. (D) RT-qPCR analysis of LHX8 transcript variants in control cells (n = 3), WT PGCLCs (n = 3), or LNC1845 KO PGCLCs (n = 3). The y-axis represents relative mean expression normalized to GADPH and control cells. Error bars indicate the mean ± standard deviation (SD), three independent experiments were carried out. One-way analysis of variance (ANOVA) was used for the comparisons. (E) Genotyping and sequencing of LNC1845-upstream 3×polyA insertion single colonies.
-
Figure 5—figure supplement 1—source data 1
Genotyping of LNC1845 KO single colonies to confirm the deletion of LNC1845.
- https://cdn.elifesciences.org/articles/78421/elife-78421-fig5-figsupp1-data1-v2.zip
-
Figure 5—figure supplement 1—source data 2
Genotyping of LNC1845-upstream 3×polyA insertion single colonies.
- https://cdn.elifesciences.org/articles/78421/elife-78421-fig5-figsupp1-data2-v2.zip
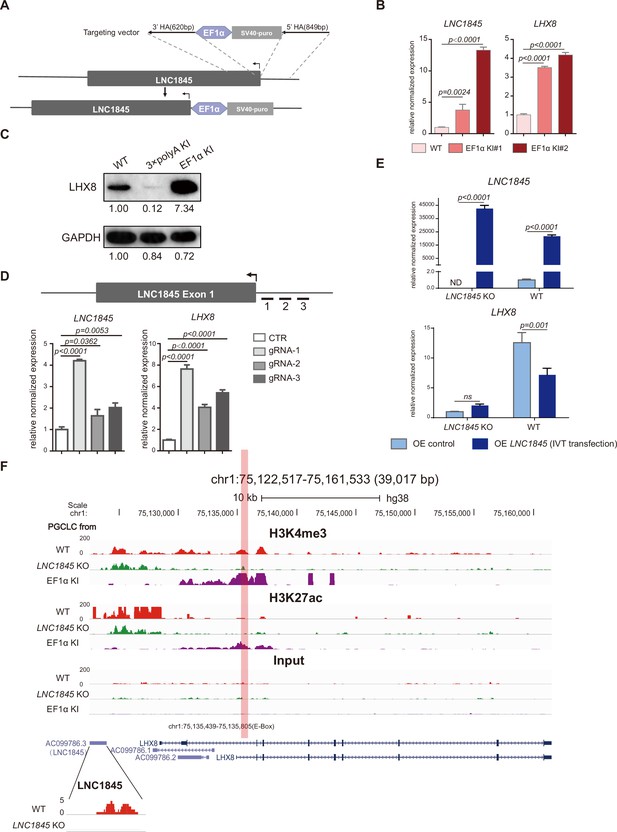
LNC1845 regulates LHX8 expression in cis by changing chromatin modifications.
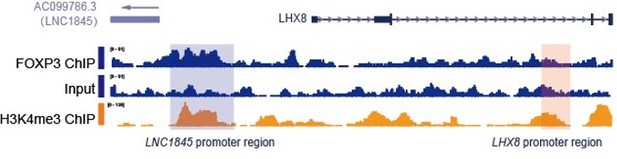
(A) Schematic diagram of LNC1845-upstream EF1α promoter knock-in and the targeted allele after EF1α promoter insertion. (B) Expression analysis by RT-qPCR of LNC1845 and LHX8 in WT primordial germ cell-like cells (PGCLCs) (n = 3) and LNC1845-upstream EF1α promoter knock-in cells (n = 3). (C) Western blot analysis of LHX8 in PGCLCs from WT, LNC1845-upstream 3×polyA knock-in, or LNC1845-upstream EF1α promoter knock-in cells. (D) CRISPR-ON-mediated LNC1845 activation and LHX8 upregulation. RT-qPCR analysis of LNC1845 and LHX8, with or without gRNAs (n = 3). Short lines with numbers indicate the relative locations of sgRNAs. (E) RT-qPCR analysis of LNC1845 and LHX8 in cells overexpressing LNC1845 transcripts (n = 3). (F) H3K4me3 and H3K27Ac levels at the LNC1845/LHX8 locus. These tracks show normalized read densities of H3K4me3 and H3K27Ac in PGCLCs differentiated from WT, LNC1845 KO, or LNC1845-upstream EF1α promoter knock-in human embryonic stem cells (hESCs). The E-BOX region of LHX8 locus is boxed in orange. Some peaks will exceed the largest scale for better results presenting.
In (B), (D), and (E), the y-axis represents relative mean expression normalized to GADPH and control cells. Error bars indicate the mean ± standard deviation (SD), three independent experiments were carried out. One-way analysis of variance (ANOVA) was used in (B) and (D), and two-way ANOVA was used in (E) for the comparisons.
-
Figure 6—source data 1
Western blot analysis of LHX8 in PGCLCs from WT, LNC1845-upstream 3×polyA knock-in, or LNC1845-upstream EF1α promoter knock-in cells.
- https://cdn.elifesciences.org/articles/78421/elife-78421-fig6-data1-v2.zip
-
Figure 6—source data 2
Western blot analysis of GAPDH in PGCLCs from WT, LNC1845-upstream 3×polyA knock-in, or LNC1845-upstream EF1α promoter knock-in cells.
- https://cdn.elifesciences.org/articles/78421/elife-78421-fig6-data2-v2.zip
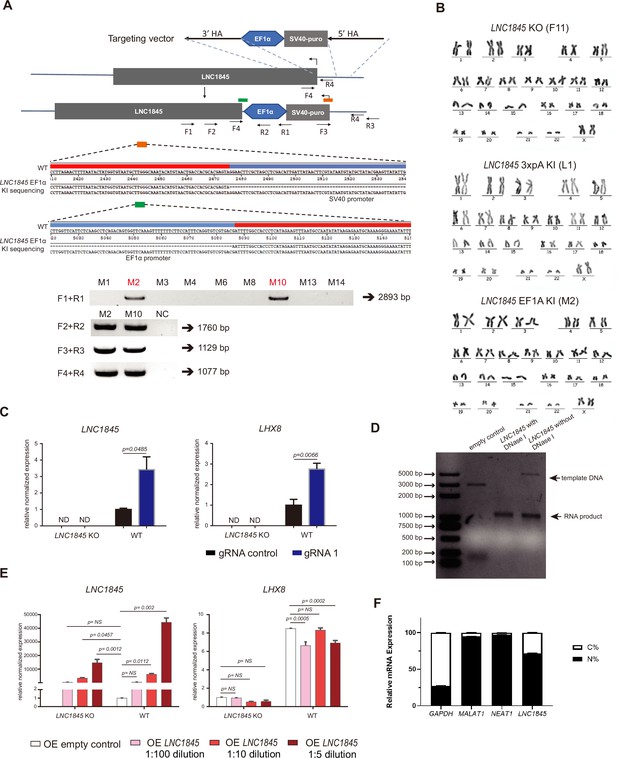
LNC1845 regulates LHX8 expression in cis by changing chromatin modifications.
(A) Genotyping and sequencing of LNC1845-upstream EF1α promoter insertion single colonies. (B) Karyotype analysis of LNC1845 KO human embryonic stem cell (hESC) clone F11, LNC1845 3xpolyA knock in hESC clone L1, and LNC1845 EF1α knock in hESC clone M2. All three clones showed normal karyotype with 46, XX after CRISPR/Cas9 editing. (C) CRISPR-ON-mediated LNC1845 activation and LHX8 upregulation in wild-type or LNC1845 KO ES cells (n = 3). RT-qPCR analysis of LNC1845 and LHX8, with or without gRNAs. (D) Agarose gel analysis of LNC1845 transcripts by in vitro transcription. (E) RT-qPCR analysis of LNC1845 and LHX8 in WT or LNC1845 KO cells overexpressing LNC1845 by different concentration lentiviral infection (n = 3). The LNC1845 sequences were inserted into the lentiviral vector p2k7 reported in our previous study, and after the lentiviral packaging, the lentivirus were diluted from 5- to 100-fold for different concentration lentiviral infection. (F) LNC1845 expression in subcellular fractions of oeLNC1845 primordial germ cell-like cells (PGCLCs) was verified by qRT-PCR. In (C) and (E), the y-axis represents relative mean expression normalized to GADPH and control cells. Error bars indicate the mean ± standard deviation (SD) from three independent biological replicates. Two-way analysis of variance (ANOVA) was used for the comparisons.
-
Figure 6—figure supplement 1—source data 1
Genotyping of LNC1845-upstream EF1α promoter insertion single colonies.
- https://cdn.elifesciences.org/articles/78421/elife-78421-fig6-figsupp1-data1-v2.zip
-
Figure 6—figure supplement 1—source data 2
Agarose gel analysis of LNC1845 transcripts by in vitro transcription.
- https://cdn.elifesciences.org/articles/78421/elife-78421-fig6-figsupp1-data2-v2.zip
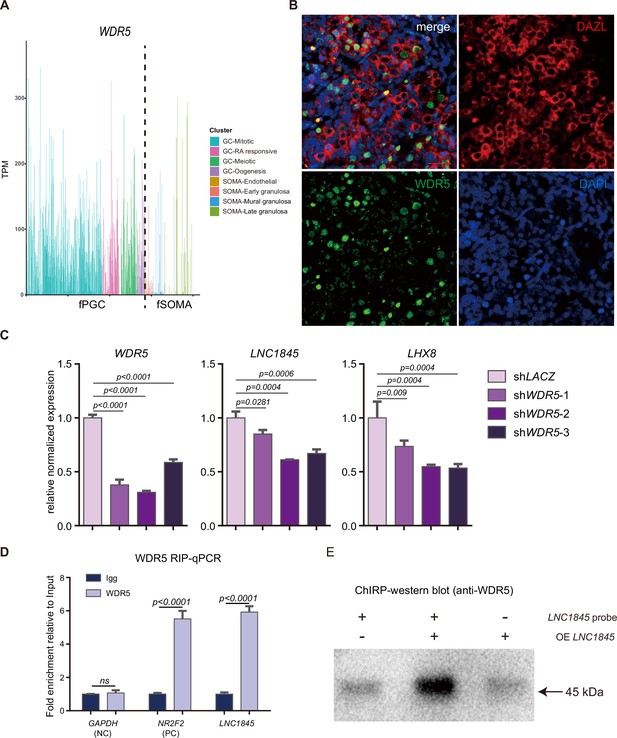
LNC1845 regulates LHX8 expression in cis by physically interacting with WDR5.
(A) WDR5 expression level in single hPGC or gonadal somatic cells at different developmental stages. (B) Immunofluorescence of WDR5 (green) co-stained with DAZL (red) in 14-week ovary section samples. Bar, 100 μm. (C) Expression analysis by RT-qPCR of LNC1845 and LHX8 in WDR5 knockdown cells (n = 3). (D) RIP-qPCR analysis of WDR5 binding to LNC1845 transcripts in primordial germ cell-like cells (PGCLCs). GAPDH and NR2F2 transcripts were used as negative and positive control. (E) LNC1845 binding for WDR5 was verified by CHIRP-western blot.
In (C) and (D), the y-axis represents relative mean expression normalized to control cells. Error bars indicate the mean ± standard deviation (SD) from three independent biological replicates. One-way analysis of variance (ANOVA) was used in (C) and Student’s t-test was used in (D) for the comparisons.
-
Figure 6—figure supplement 2—source data 1
LNC1845 binding for WDR5 was verified by CHIRP-western blot.
- https://cdn.elifesciences.org/articles/78421/elife-78421-fig6-figsupp2-data1-v2.zip
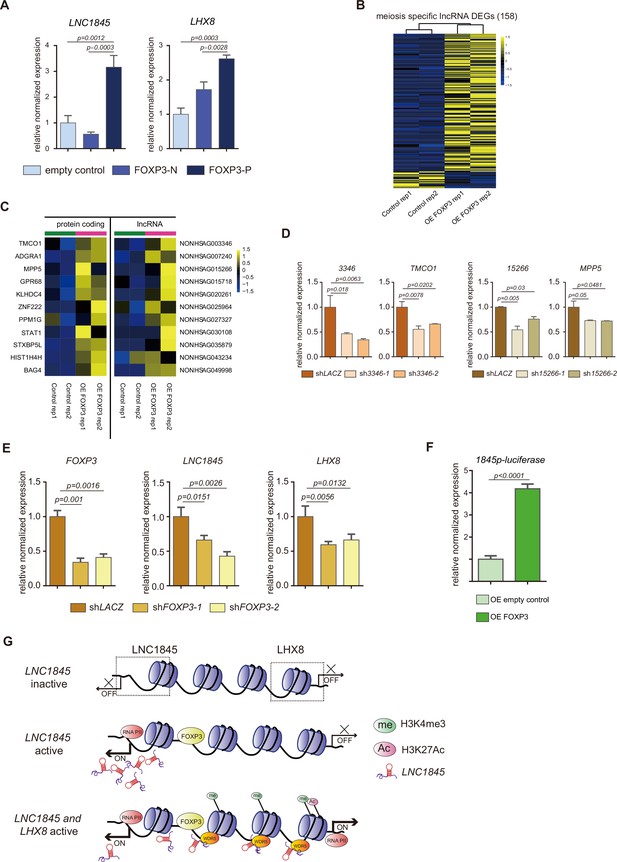
FOXP3 upregulates expression of LNC1845 and other long non-coding RNAs (lncRNAs).
(A) RT-qPCR analysis of LNC1845 and LHX8 in primordial germ cell-like cells (PGCLCs) overexpresses FOXP3. (B) Heatmap of differentially expressed ms-lncRNAs in control or OE FOXP3 groups, rep1 and rep2 represent two independent populations. (C) Heatmap of upregulated divergent lncRNA–mRNA pairs in OE FOXP3 groups, rep1and rep2 represent two independent populations. (D) Expression analysis by RT-qPCR of lncRNA and their divergent protein-coding genes in lncRNA knockdown cells (n = 3). (E) Expression analysis by RT-qPCR of LNC1845 and LHX8 in FOXP3 knockdown PGCLCs (n = 3). (F) Luciferase assay of FOXP3 on LNC1845 promotor activity. 293 FT cells were transiently transfected with empty control or FOXP3 expression vector in combination with luciferase vectors, n = 3. (G) The FOXP3-LNC1845-LHX8 regulatory model. The upstream transcription factors, including FOXP3, could upregulate LNC1845, and then LNC1845 could induce H3K4me3 and H3K27Ac in the regions near the LHX8 transcription start site, which in turn helps activate LHX8 expression.
In (A), (D), (E), and (F), the y-axis represents relative mean expression normalized to GADPH and control cells. Error bars indicate the mean ± standard deviation (SD), three independent experiments were carried out. One-way analysis of variance (ANOVA) was used in (A), (D), and (E), and Student’s t-test was used in (F) for the comparisons.
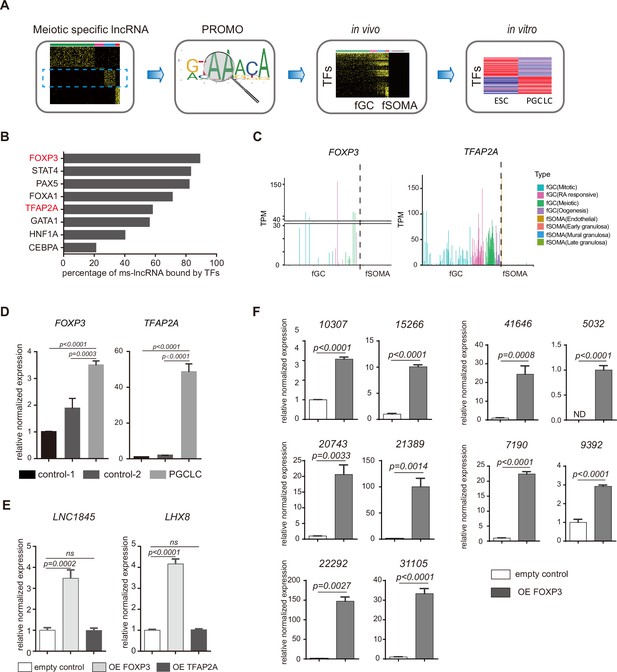
FOXP3 upregulates expression of LNC1845 and other long non-coding RNAs (lncRNAs).
(A) Schematic diagram of input features for the computational prediction used to predict transcription factors involved in female meiotic stage-specific lncRNAs upregulation. (B) Bar chart displaying percentage of ms-lncRNAs that contained the binding motif of different germ cell-specific transcription factors in promoter region. In vitro upregulated transcription factors are in red. (C) FOXP3 and TFAP2A expression level in single hPGC or gonadal somatic cell from different developmental stages. (D) Expression analysis of FOXP3 and TFAP2A by RT-qPCR of spontaneous differentiation cells (control-1), mCherry-positive control cells (control-2), and DAZL-positive primordial germ cell-like cells (PGCLCs). (E) RT-qPCR analysis of LNC1845 and LHX8 in 293 FT cells overexpress FOXP3 or TFAP2A. (F) RT-qPCR analysis of 10 ms-lncRNAs in 293 FT cells after empty control vector or FOXP3 overexpression vector transfection.
In (D–F), the y-axis represents relative mean expression normalized to GADPH and control group cells. Error bars indicate mean ± standard deviation (SD), n = 3, three independent experiments were carried out. One-way analysis of variance (ANOVA) was used in (D) and (E) and Student’s t-test was used in (F) for the comparisons.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | LIM homebox 8(LHX8) | Human genome informatics | Ensembl:ENSG 00000162624 | |
| Gene (Homo sapiens) | LNC1845(AC099786.3) | Human genome informatics | NONHSAG001845. 2/ENSG00000261213.1 | |
| Cell line (Homo sapiens) | H9 LNC1845 KO stem cells | This paper | LNC1845 KO cell line generated by CRISPR/Cas9 | |
| Cell line (Homo sapiens) | H9 LNC1845 polyA KI stem cells | This paper | LNC1845 polyA KI cell line generated by CRISPR/Cas9 | |
| Cell line (Homo sapiens) | H9 LNC1845 EF1α KI stem cells | This paper | LNC1845 EF1α KI cell line generated by CRISPR/Cas9 | |
| Cell line (Homo sapiens) | PA-1 cells | This paper | Human ovary teratocarcinoma Cell Line | |
| Cell line (Homo sapiens) | 293 FT cells | Thermo Fisher Scientific | Cat# R70007 | |
| Cell line (Mus musculus) | Mouse embryonic fibroblasts | This paper | Mouse embryonic fibroblasts isolated from embryonic day 13.5 | |
| Recombinant DNA reagent | P2k7-EF1α-DAZL2- P2A-mCherry | This study | Methods | |
| Recombinant DNA reagent | P2k7-EF1α-mCherry | This study | Methods | |
| Recombinant DNA reagent | P2k7-EF1α-1845 | This study | Methods | |
| Recombinant DNA reagent | P2k7-EF1α-FOXP3 | This study | Methods | |
| Recombinant DNA reagent | P2k7-EF1α-TFAP2A | This study | Methods | |
| Recombinant DNA reagent | P2k7-EF1α-FOXP3- P2A-mCherry | This study | Methods | |
| Recombinant DNA reagent | P2k7-EF1α-FOXP3- 3xFLAG-P2A-mCherry | This study | Methods | |
| Recombinant DNA reagent | pEASY-T7 promoter | This study | Methods | |
| Recombinant DNA reagent | pEASY-T7-1845 | This study | Methods | |
| Recombinant DNA reagent | pX335-U6-Chimeric_BB-CBh-hSpCas9n(D10A) | Addgene | # 42335 | |
| Recombinant DNA reagent | PGK-puro-1845KO donor | This study | Methods | |
| Recombinant DNA reagent | SV40-puro-3xpolyA 1845 KI donor | This study | Methods | |
| Recombinant DNA reagent | SV40-puro-EF1α 1845 KI donor | This study | Methods | |
| Recombinant DNA reagent | H1-shLACZ-Ubc-GFP | This study | Methods | |
| Recombinant DNA reagent | H1-sh1845①-Ubc-GFP | This study | Methods | |
| Recombinant DNA reagent | H1-sh1845②-Ubc-GFP | This study | Methods | |
| Recombinant DNA reagent | H1-sh15266①-Ubc-GFP | This study | Methods | |
| Recombinant DNA reagent | H1-sh15266②-Ubc-GFP | This study | Methods | |
| Recombinant DNA reagent | H1-sh3346①-Ubc-GFP | This study | Methods | |
| Recombinant DNA reagent | H1-sh3346②-Ubc-GFP | This study | Methods | |
| Recombinant DNA reagent | H1-shFOXP3①-Ubc-GFP | This study | Methods | |
| Recombinant DNA reagent | H1-shFOXP3②-Ubc-GFP | This study | Methods | |
| Recombinant DNA reagent | H1-shWDR5①-Ubc-GFP | This study | Methods | |
| Recombinant DNA reagent | H1-shWDR5②-Ubc-GFP | This study | Methods | |
| Recombinant DNA reagent | H1-shWDR5③-Ubc-GFP | This study | Methods | |
| Recombinant DNA reagent | Lenti-dCAS-VP64-Blast | Addgene | # 61425 | |
| Recombinant DNA reagent | Lenti-sgRNA(MS2)-zeo backbone | Addgene | # 61427 | |
| Recombinant DNA reagent | Lenti MS2-P65-HSF1_Hygro | Addgene | # 61426 | |
| Recombinant DNA reagent | Lenti-sgRNA(MS2)–1845 gRNA 1 | This study | Methods | |
| Recombinant DNA reagent | Lenti-sgRNA(MS2)–1845 gRNA 2 | This study | Methods | |
| Recombinant DNA reagent | Lenti-sgRNA(MS2)–1845 gRNA 3 | This study | Methods | |
| Biological sample (Homo sapiens) | Human 17 w ovary RNA | BIOPIKE | ||
| Antibody | anti-DAZL (Mouse monoclonal) | Bio-Rad | Cat# MCA2336, RRID:AB_2292585 | (IF 1:50) |
| Antibody | anti-LHX8 (Rabbit polyclonal) | abcam | Cat# ab221882 | (IF 1:50; WB 1:1000) |
| Antibody | anti-WDR5 (Rabbit monoclonal) | Cell Signaling Technology | Cat# 13105S, RRID:AB_2620133 | (IF 1:200; WB 1:2000) |
| Antibody | anti-GAPDH (Mouse monoclonal) | Cwbio | Cat# CW0100M, RRID:AB_2801390 | (WB 1:5000) |
| Antibody | anti-FLAG (Rabbit monoclonal) | Sigma | Cat# F3165, RRID:AB_259529 | (IP 3 μg) |
| Antibody | anti-H3K27Ac (Rabbit monoclonal) | Cell Signaling Technology | Cat# 8173, RRID:AB_10949503 | (ChIP 1 μg/rxn) |
| Antibody | anti-H3K4me3 (Rabbit monoclonal) | Millipore | Cat# 04-745, RRID:AB_1163444 | (ChIP 1 μg/rxn) |
| Antibody | Goat anti-Rabbit IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (Goat polyclonal) | Thermo Fisher Scientific | Cat# A-11008, RRID:AB_143165 | (WB 1:5000) |
| Antibody | Goat anti-Mouse IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 594 (Goat polyclonal) | Thermo Fisher Scientific | Cat# A32742, RRID:AB_2762825 | (WB 1:5000) |
| Antibody | Horseradish-labeled goat anti-rabbit IgG (H + L) (Goat polyclonal) | ZSGB-Bio | Cat# ZB-2301, RRID:AB_2747412 | (WB 1:5000) |
| Antibody | Peroxidase-Conjugated Goat anti-Mouse IgG (H + L) (Goat polyclonal) | ZSGB-Bio | Cat# ZB-2305, RRID:AB_2747415 | (WB 1:5000) |
| Chemical compound, drug | Puromycin | Sigma | Cat# P9620 | |
| Chemical compound, drug | Geneticin | Thermo Fisher Scientific | Cat# 10131035 | |
| Chemical compound, drug | Blasticidin | Thermo Fisher Scientific | Cat# 461120 | |
| Chemical compound, drug | ROCK inhibitor | stemRD | Cat# Y-005 | |
| Peptide, recombinant protein | Recombinant human BMP-4 | R&D | Cat# 314 BP | |
| Peptide, recombinant protein | Recombinant human BMP-8a | R&D | Cat# 1073-BPC | |
| Peptide, recombinant protein | Recombinant human FGF basic | R&D | Cat# 233-FB-001MG/CF | |
| Commercial assay or kit | Gateway LR Clonase II Kit | Thermo Fisher Scientific | Cat# 11791100 | |
| Commercial assay or kit | PrimeScript RT reagent Kit with gDNA Eraser | TaKaRa | Cat# RR047A | |
| Commercial assay or kit | Fluorescent In Situ Hybridization Kit | RIBOBIO | Cat# C10910 | |
| Commercial assay or kit | RNAmax-T7 kit | RIBOBIO | Cat# R11073 | |
| Commercial assay or kit | TnT Quick Coupled Transcription/ Translation Systems | Promega | Cat# L1170 | |
| Commercial assay or kit | FirstChoice RLM-RACE kit | Ambion | Cat# AM1700 | |
| Commercial assay or kit | AMPure XP beads | Beckman Coulter | A63881 | |
| Commercial assay or kit | NEBNext DNA Library Prep Kit | NEB | #E7645S | |
| Commercial assay or kit | NEBNext Multiplex Oligos | NEB | #E7335S | |
| Commercial assay or kit | VAHTS mRNA-seq v2 Library Prep Kit | Vazyme | NR611-02 | |
| Software, algorithm | GraphPad Prism 6.0 | GraphPad Prism | https://www.graphpad.com/ | |
| Software, algorithm | FlowJo | FlowJo | https://www.flowjo.com/ | |
| Software, algorithm | DAVID Bioinformatics Resources | Huang et al., 2009 | https://david.ncifcrf.gov | |
| Software, algorithm | Coding Potential Calculator | Kong et al., 2007 | http://cpc.cbi.pku.edu.cn/ | |
| Software, algorithm | Coding Potential Assessment Tool | Wang et al., 2013 | http://rna-cpat.sourceforge.net/ | |
| Software, algorithm | PROMO | Messeguer et al., 2002 | http://alggen.lsi.upc.es/ | |
| Software, algorithm | TopHat (v2.1.1) | Trapnell et al., 2009 | http://ccb.jhu.edu/software/tophat/index.shtml | |
| Software, algorithm | HTSeq package | Anders et al., 2015 | https://htseq.readthedocs.io/en/release_0.9.1/ | |
| Software, algorithm | GO (DAVID) | Huang et al., 2009 | https://david.ncifcrf.gov/home.jsp | |
| Software, algorithm | HISAT2 (v2.1.0) | Kim et al., 2015 | http://ccb.jhu.edu/software/hisat2/manual.shtml | |
| Software, algorithm | NONCODE (v5) | Fang et al., 2017 | http://www.noncode.org/index.php | |
| Software, algorithm | Gencode (v29) | Uszczynska-Ratajczak et al., 2018 | https://www.gencodegenes.org/ | |
| Software, algorithm | Samtools (1.3.1) | Li et al., 2009 | http://samtools.sourceforge.net/ | |
| Software, algorithm | MACS2 | Zhang et al., 2008 | https://pypi.org/project/MACS2/2.1.1.20160309/ | |
| Software, algorithm | IGV | Thorvaldsdóttir et al., 2013 | http://www.igv.org/ | |
| Software, algorithm | Treeview | Saldanha, 2004 | http://jtreeview.sourceforge.net/ | |
| Software, algorithm | R (3.5.1) | N/A | https://www.r-project.org/ | |
| Software, algorithm | pheatmap | N/A | R software | |
| Software, algorithm | DESeq2 | Love et al., 2014 | R software | |
| Software, algorithm | ggplot2 | Wickham, 2016 | R software | |
| Software, algorithm | Seurat (3.2.3) | Satija et al., 2015 | R software | |
| Software, algorithm | Monocle (2.10.1) | Qiu et al., 2017 | R software | |
| Other | Matrigel | Corning | Cat# 354277 | Matrigel Basement Membrane Matrix |
| Other | DAPI | Invitrogen | Cat# D1306 | Blue-fluorescent DNA stain |
| Other | TrypLE Express | Invitrogen | Cat# 12605010 | Trypsin substitution |
| Other | Dynabeads Protein A | Invitrogen | Cat# 10002D | Dynabeads Protein A for Immunoprecipitation |
| Other | Dynabeads Protein G | Invitrogen | Cat# 10003D | Dynabeads Protein G for Immunoprecipitation |
| Other | ProLong Diamond Antifade Mountant | Invitrogen | Cat# P10144 | Antifade Solution |
| Other | TRIzol Reagent | Invitrogen | Cat# 15596026 | RNA extraction reagents |
| Other | TransStart Top Green qPCR Mix | TransGen | Cat# AQ131-02 | qPCR reagents |
| Other | Lipofectamine 2000 | Invitrogen | Cat# 11668019 | Transfection reagents |
| Other | Lipofectamine 3000 | Invitrogen | Cat# L3000001 | Transfection reagents |
Additional files
-
Supplementary file 1
Weekly expressed lncRNAs of female and male germ cells and gonadal somatic cells.
- https://cdn.elifesciences.org/articles/78421/elife-78421-supp1-v2.xlsx
-
Supplementary file 2
Human germ cells and gonadal somatic cells specific lncRNAs.
- https://cdn.elifesciences.org/articles/78421/elife-78421-supp2-v2.xlsx
-
Supplementary file 3
Human germ cell each stage-specific lncRNAs.
- https://cdn.elifesciences.org/articles/78421/elife-78421-supp3-v2.xlsx
-
Supplementary file 4
Feature protein-coding genes or lncRNAs of each cluster.
- https://cdn.elifesciences.org/articles/78421/elife-78421-supp4-v2.xlsx
-
Supplementary file 5
Female germ cell (fGC) meiotic and male germ cell (mGC) mitotic arrest lncRNA–mRNA pairs and expression correlation.
- https://cdn.elifesciences.org/articles/78421/elife-78421-supp5-v2.xlsx
-
Supplementary file 6
Upregulated lncRNA genes in primordial germ cell-like cells (PGCLCs).
- https://cdn.elifesciences.org/articles/78421/elife-78421-supp6-v2.xlsx
-
Supplementary file 7
Upregulated protein-coding genes in primordial germ cell-like cells (PGCLCs).
- https://cdn.elifesciences.org/articles/78421/elife-78421-supp7-v2.xlsx
-
Supplementary file 8
Differentially expressed protein-coding genes in LNC1845 KO primordial germ cell-like cells (PGCLCs).
- https://cdn.elifesciences.org/articles/78421/elife-78421-supp8-v2.xlsx
-
Supplementary file 9
Differentially expressed protein-coding genes in LHX8 OE primordial germ cell-like cells (PGCLCs).
- https://cdn.elifesciences.org/articles/78421/elife-78421-supp9-v2.xlsx
-
Supplementary file 10
Oligonucleotides.
- https://cdn.elifesciences.org/articles/78421/elife-78421-supp10-v2.xlsx