Structure of the IL-27 quaternary receptor signaling complex
Figures
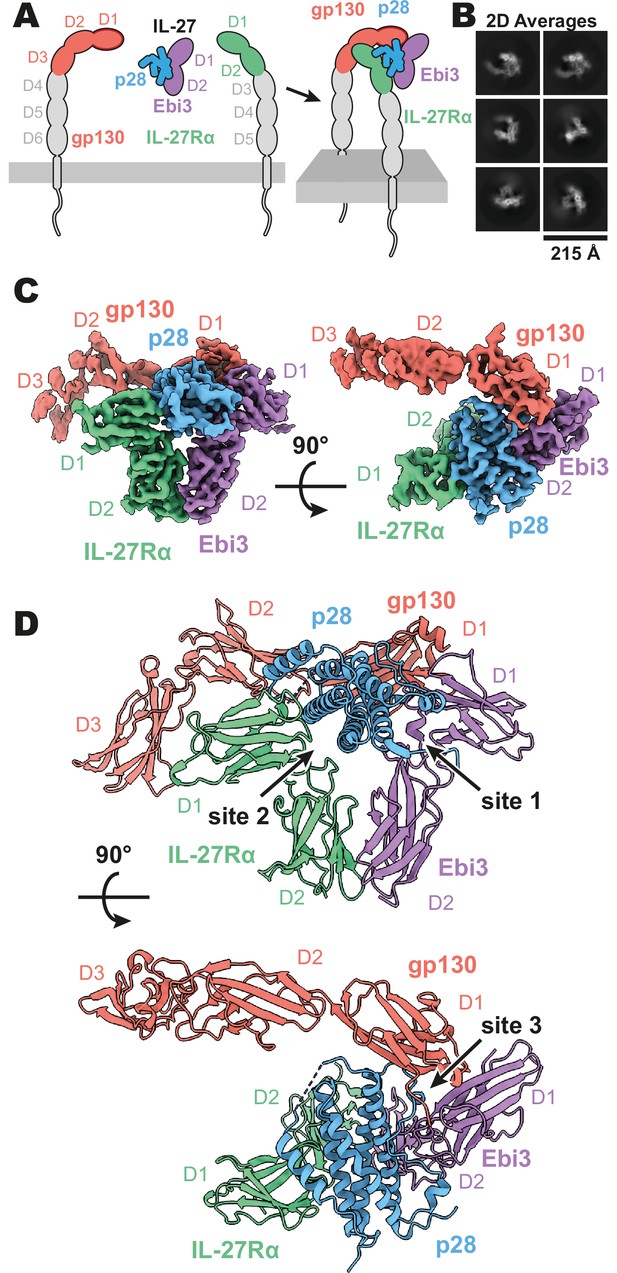
Composition and cryogenic-electron microscopy (cryoEM) structure of human interleukin 27 (IL-27) quaternary complex.
(A) Cartoon representation of the components of the IL-27 quaternary signaling complex. gp130 (red), p28 (blue), Epstein-Barr Virus-Induced 3 (Ebi3; purple), IL-27Rα (green), with domains excluded from the imaged constructs in gray. The D1 Ig domain of gp130 is represented by red, with an additional dark red outline to distinguish it from FNIII domains in gp130, Ebi3, and IL-27Rα. (B) Reference-free 2D averages from cryoEM of the IL-27 quaternary complex. (C) Refined and sharpened cryoEM density maps of IL-27 quaternary complex, colored as in (A). (D) Ribbon representation of the atomistic modeling of IL-27 quaternary complex, colored as in (A).
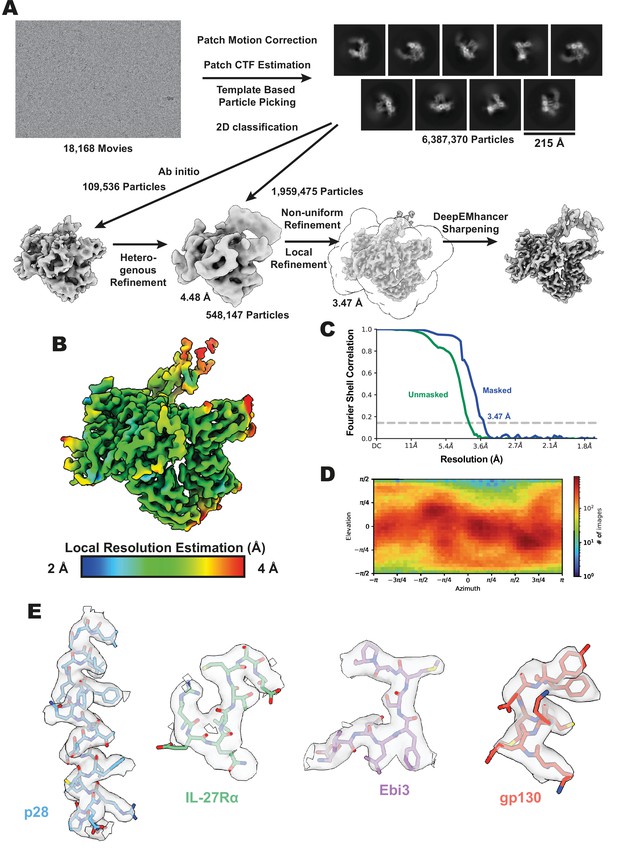
Interleukin 27 (IL-27) quaternary complex cryogenic-electron microscopy (cryoEM) data processing.
(A) Workflow for cryoEM data processing. Representative micrograph, reference free 2D averages, and cryoEM maps at the various stages of processing. Depicted maps were z-flipped prior to model building. (B) Local resolution estimation of the finalized cryoEM map of IL-27 quaternary complex (Punjani et al., 2017). (C) FSC curve of the IL-27 quaternary complex reconstruction using gold-standard refinement calculated from unmasked and masked half maps. (D) Orientational distribution of the IL-27 quaternary complex reconstruction. (E) Representative cryoEM density regions of p28, IL-27Rα, Epstein-Barr Virus-Induced 3 (Ebi3), and gp130.
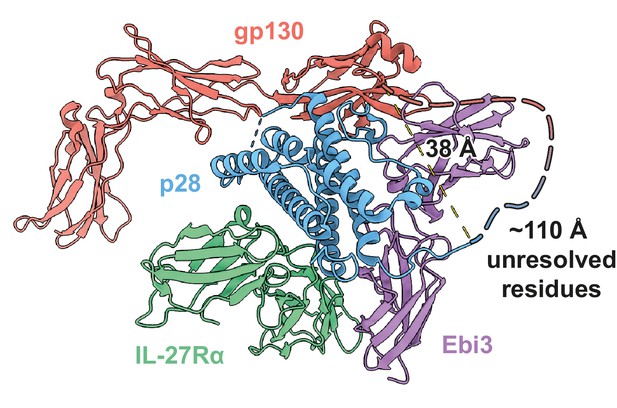
Distances and constraints of the Ebi3-gp130 GS-linker.
38 Å separate resolved residues at the C-terminus of p28 and the N-terminus of gp130; a distance easily accommodated by the 20 a.a. GS linker and unresolved residues (~110 Å).
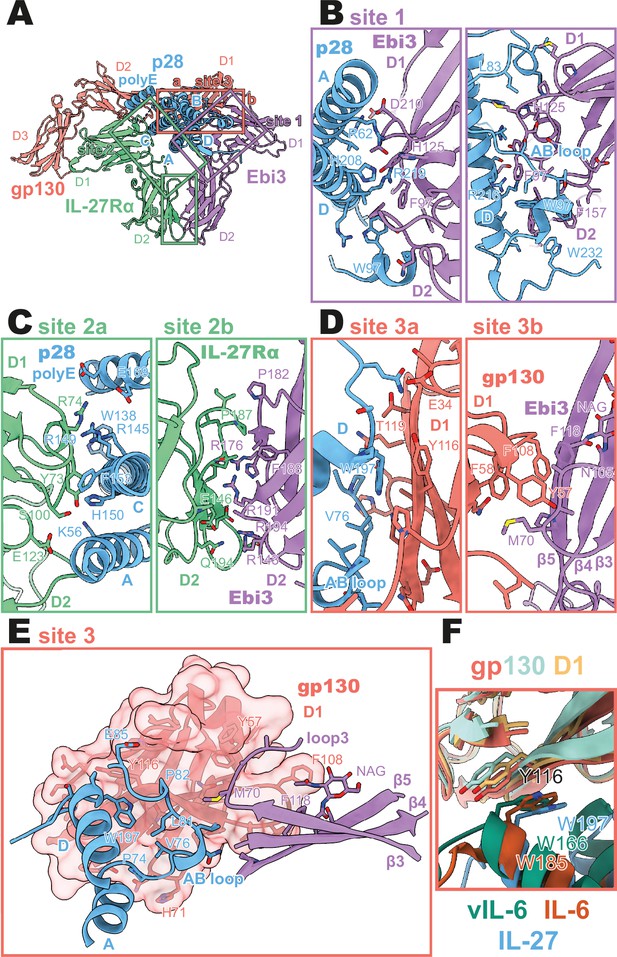
Binding interfaces of the human interleukin 27 (IL-27) quaternary complex.
(A) Ribbon representation of the IL-27 quaternary signaling complex, containing gp130 (red), p28 (blue), Epstein-Barr Virus-Induced 3 (Ebi3; purple), and IL-27Rα (green). Regions containing sites 1, 2, and 3 are boxed in purple, green, and red, respectively. (B) Two views of the site 1 interface, colored as in (A). (C) The site 2 interface, composed of a site 2a, p28 to IL-27Rα, interaction, and a site 2b, Ebi3 to IL-27Rα, interaction. All proteins are colored as in (A). (D,E) The site 3 interface, composed of a site 3a, p28 to gp130, interaction, and a site 3b, Ebi3 to IL-27Rα, interaction. All proteins are colored as in (A). (F) Structural overlay of site 3 interacting domains from IL-27 complex, IL-6 complex (PDB 1P9M), and viral IL-6 (vIL-6) complex (PDB 1L1R). IL-27 quaternary complex colored as in (A), IL-6 ternary complex in orange (IL-6) and yellow (gp130), and vIL-6 binary complex in green (vIL-6) and teal (gp130).

Cryogenic-electron microscopy (CryoEM) density of binding interfaces of the human IL-27 quaternary complex.
(A) Ribbon representation of the IL-27 quaternary signaling complex, containing gp130 (red), p28 (blue), Epstein-Barr Virus-Induced 3 (Ebi3; purple), and IL-27Rα (green). Regions containing sites 1, 2, and 3 are boxed in purple, green, and red, respectively. (B) CryoEM density of the site 1 interaction. Colored as in (A). (C) CryoEM density of the site 2 interaction. Colored as in (A). (D) CryoEM density of the site 3 interaction. Colored as in (A).
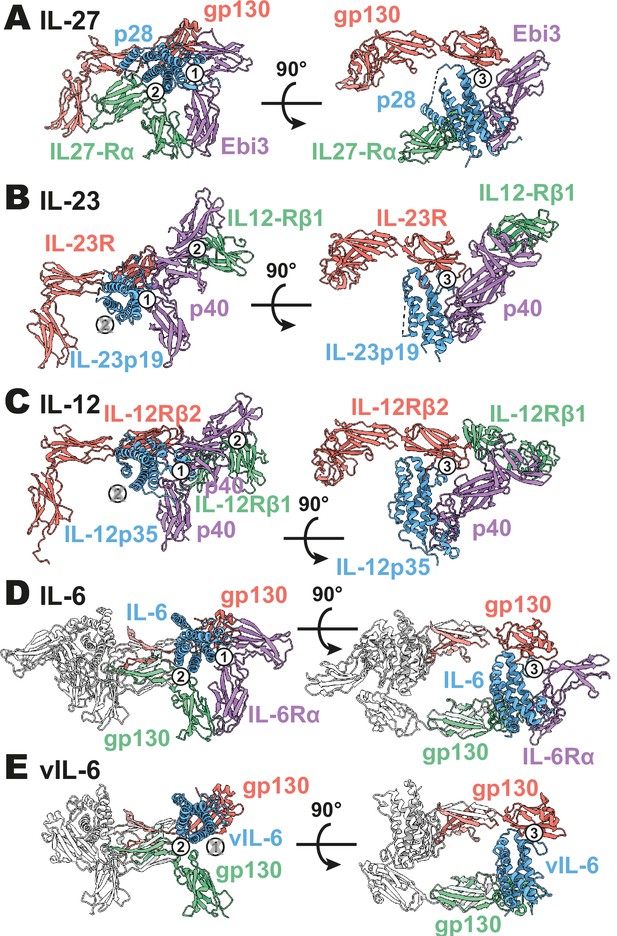
Comparison of IL-27 to IL-12 and IL-6 family complexes.
(A) Ribbon representation of the IL-27 signaling complex, containing gp130 (red), p28 (blue), Epstein-Barr Virus-Induced 3 (Ebi3; purple), and IL-27Rα (green). Sites 1, 2, and 3 are noted with a numbered circle. (B) Ribbon representation of the IL-23 signaling complex (IL-12 family), containing IL-23R (red), IL-23p19 (blue), p40 (purple), and IL-12Rβ1 (green) (PDB 6WDQ). Sites 1 and 3 are noted as in (A), with the unoccupied site 2 marked with a gray ‘X’ and the distinct IL-12/IL-23 site 2 marked with a circled number 2. (C) Ribbon representation of a model of the IL-12 signaling complex (IL-12 family), containing IL-12Rβ2 (red), IL-12p35 (blue), p40 (purple), and IL-12Rβ1 (green) (Baek et al., 2021; Glassman et al., 2021a). Sites 1 and 3 are noted as in (A), with the unoccupied site 2 marked with a gray ‘X’ and the distinct IL-12/IL-23 site 2 marked with a circled number 2. (D) Ribbon representation of the IL-6 signaling complex (IL-6 family), containing gp130 (red and green), IL-6 (blue), and IL-6Rα (purple) (PDB 1P9M). Secondary copies of each protein in the complex are colored in white. Sites 1, 2, and 3 are noted as in (A). (E) Ribbon representation of the viral IL-6 signaling complex (IL-6 family), containing gp130 (red and green) and (blue) (PDB 1I1R). Secondary copies of each protein in the complex are colored in white. Sites 2 and 3 are noted as in (A), with the unoccupied site 1 marked with a gray ‘X’.
Tables
CryoEM data collection, refinement, and validation statistics.
| IL-27 Complex (PDB 7U7N/EMD-26382) | |
|---|---|
| Data collection and processing | |
| Magnification | 105,000 |
| Voltage (keV) | 300 |
| Electron exposure (e-/Å2) | 60 |
| Defocus range (µm) | –0.8 to –2.0 |
| Pixel size (Å) | 0.839 |
| Symmetry imposed | C1 |
| Initial particle images | 6,387,370 |
| Final particle images | 548,147 |
| Map resolution FSC threshold (Å) | 0.143 |
| Map resolution (Å) | 3.47 |
| Refinement | |
| Initial model used (PDB) | AlphaFold |
| Model resolution FSC threshold (Å) | 0.143 |
| Model resolution (Å) | 1.9 |
| Map sharpening B-factor (Å2) | 189.6 |
| Model Composition | |
| Non-hydrogen atoms | 7,284 |
| Protein residues | 884 |
| Ligands | 16 |
| B-factors (Å2) | |
| Protein | 102.97 |
| Ligand | 114.62 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.610 |
| Validation | |
| MolProbity score | 1.60 |
| Clashscore | 8.81 |
| EMringer score | 2.33 |
| Rotamer outliers (%) | 0.89 |
| Ramachandran plot | |
| Favored (%) | 97.37 |
| Allowed (%) | 2.63 |
| Outliers (%) | 0.00 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | Human embryonic kidney cells | GIBCO | Expi293 | |
| Recombinant DNA reagent | pD649-IL-27Rα (plasmid) | This paper | See: Methods - Cloning and protein expression | |
| Recombinant DNA reagent | pD649-Ebi3 (plasmid) | This paper | See: Methods - Cloning and protein expression | |
| Recombinant DNA reagent | pD649-p28-gp130 (plasmid) | This paper | See: Methods - Cloning and protein expression | |
| Software, algorithm | Data collection software | SerialEM | SerialEM | |
| Software, algorithm | Data processing software | Structura Biotechnology Inc. | cryoSPARC | |
| Software, algorithm | Data sharpening software | Sanchez-Garcia et al., 2021 | DeepEMhancer | |
| Software, algorithm | Initial modeling software | Jumper et al., 2021 | AlphaFold | |
| Software, algorithm | Graphics software | Pettersen et al., 2021 | UCSF ChimeraX | |
| Software, algorithm | Modeling and refinement software | Adams et al., 2010 | Phenix | |
| Software, algorithm | Modeling and refinement software | Emsley and Cowtan, 2004 | Coot | |
| Software, algorithm | Model validation software | Barad et al., 2015 | EMRinger | |
| Software, algorithm | Model validation software | Chen et al., 2010 | MolProbity |





