Claudin5 protects the peripheral endothelial barrier in an organ and vessel-type-specific manner
Figures
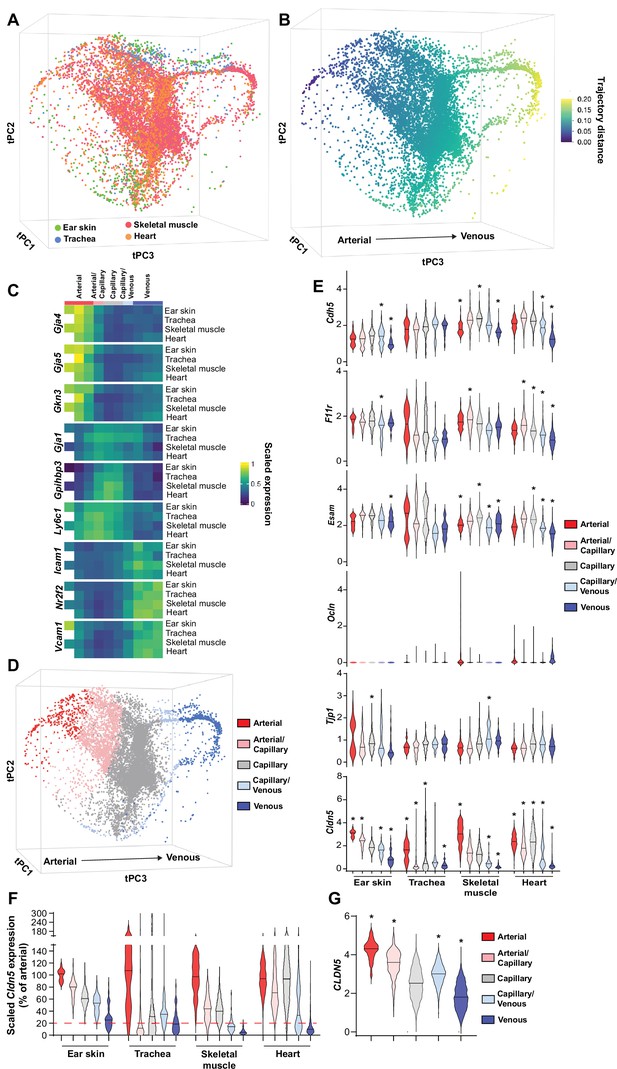
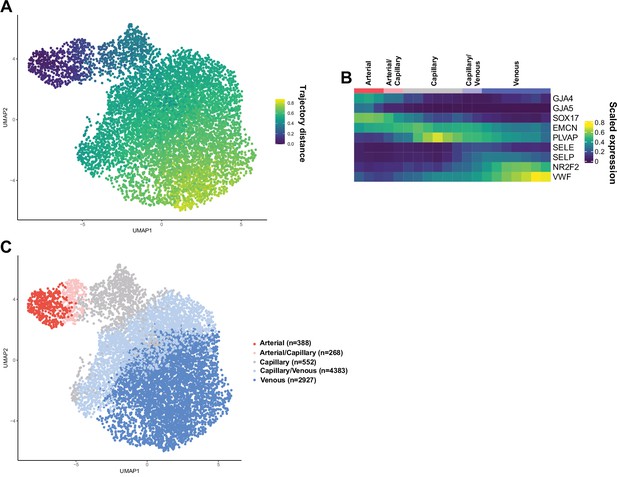
Patterning of the EC barrier at the single-cell level.
(A) Principal component analysis of the distances within 400 trajectories calculated with integrated data of murine datasets of ear skin, trachea, skeletal muscle, and heart blood endothelial cells (BECs). Colors illustrate the distribution of BECs (CD31+/CD45-/Lyve1-) for each organ. (B) Principal component analysis of trajectory distances colored by the distance along an isolated trajectory spanning from arterial to venous BEC. (C) Mean gene expression for each organ after equidistant binning of the isolated trajectory shown in B. Supervised vessel subset specifications (Top) based on the expression of previously established marker genes. (D) Principal component analysis of trajectory distances colored by the vessel subsets defined in C. (E) Violin plots of gene expression for BEC junctional components. Gene expression was normalized to account for differences in sample library size and has been imputed to account for dropouts in the data as described in Materials and methods. (F) Cldn5 expression in murine BEC datasets scaled per organ according to the mean expression in the arterial BECs of each organ. Red dashed line represents a fivefold reduction in expression compared to arterial BECs. (G) CLDN5 expression in human dermal BECs. n=534 ear skin, 559 trachea, 3498 skeletal muscle, 6423 heart and 8518 human BEC. * denotes statistical significance following differential gene expression analysis (Figure 1—source data 1–5).
-
Figure 1—source data 1
Spreadsheets detailing the results of the differential gene expression analysis conducted between mouse BEC subtypes in ear skin.
- https://cdn.elifesciences.org/articles/78517/elife-78517-fig1-data1-v2.xlsx
-
Figure 1—source data 2
Spreadsheets detailing the results of the differential gene expression analysis conducted between mouse BEC subtypes in trachea.
- https://cdn.elifesciences.org/articles/78517/elife-78517-fig1-data2-v2.xlsx
-
Figure 1—source data 3
Spreadsheets detailing the results of the differential gene expression analysis conducted between mouse BEC subtypes in skeletal muscle.
- https://cdn.elifesciences.org/articles/78517/elife-78517-fig1-data3-v2.xlsx
-
Figure 1—source data 4
Spreadsheets detailing the results of the differential gene expression analysis conducted between mouse BEC subtypes in heart.
- https://cdn.elifesciences.org/articles/78517/elife-78517-fig1-data4-v2.xlsx
-
Figure 1—source data 5
Spreadsheet detailing the results of the differential gene expression analysis conducted between human dermal BEC subtypes.
- https://cdn.elifesciences.org/articles/78517/elife-78517-fig1-data5-v2.xlsx
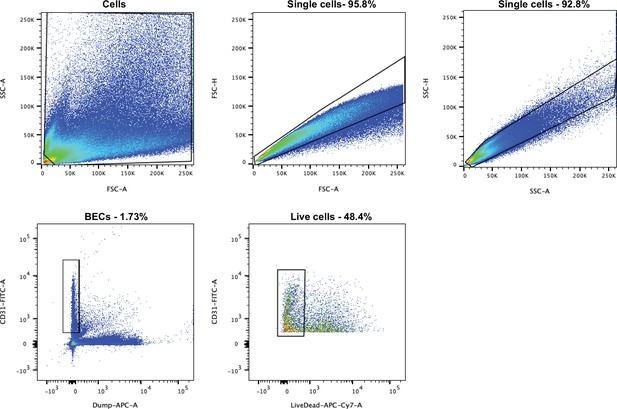
Gating strategy for the FACS isolation of single blood vessel BECs from the mouse ear skin.
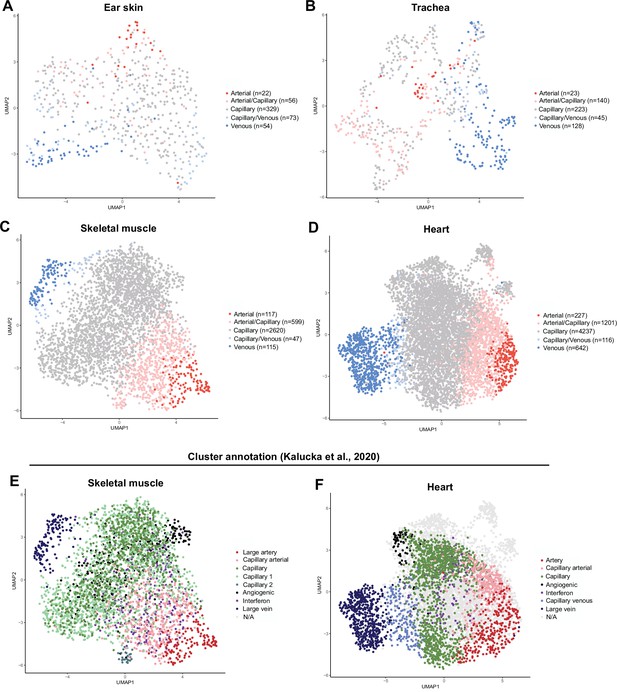
BEC subset allocation and clustering of individual mouse organs.
(A) Ear skin, (B) trachea, (C) skeletal muscle, and (D) heart mouse BECs colored by subset as defined by analysis of integrated data. The number of cells in each subtype are specified in each legend. (E) Skeletal muscle and (F) heart mouse BECs colored by clusters identified in the original publication of the data (Kalucka et al., 2020).
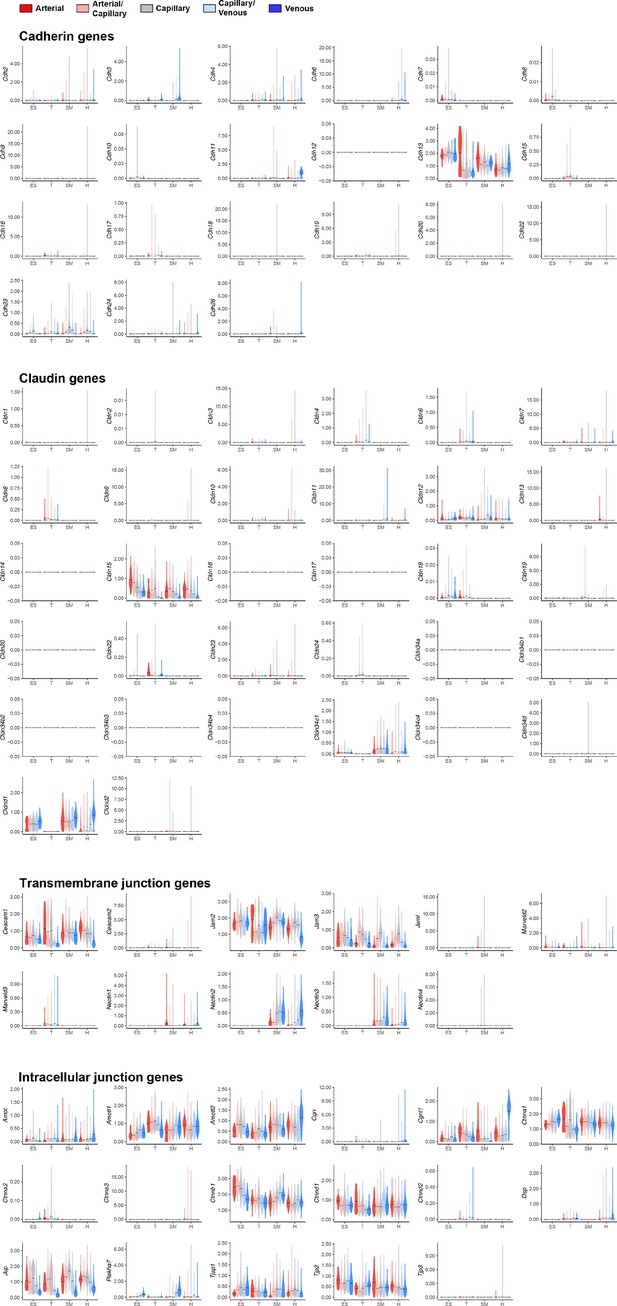
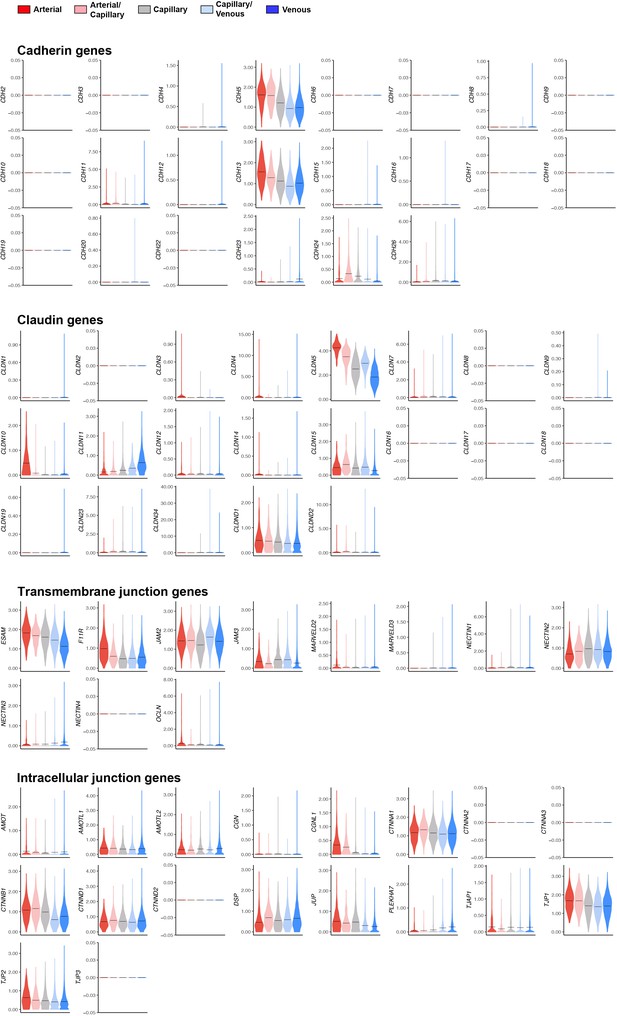
Expression of mouse endothelial junctional components.
Violin plots of gene expression for mouse endothelial junctional components.Gene expression was normalized to account for differences in sample library size and imputed to account for dropouts in the data as described in Materials and methods. ES, ear skin; T, trachea; SM, skeletal muscle; H, heart.
Analysis of human skin BECs.
(A) Uniform manifold approximation and projection (UMAP) of human dermal BECs showing the distance of an isolated trajectory calculated with tSpace. (B) Equidistant binning of the trajectory shown in A. with supervised annotation of the bins as vessel subsets. (C) UMAP colored by the vessel subsets specified in B.
Expression of human dermal endothelial junctional components.
Violin plots of gene expression for human dermal endothelial junctional components. Gene expression was normalized to account for differences in library size and imputed to account for dropouts in the data as described in Materials and methods.

Organotypic integrity of the EC barrier.
(A) Leakage patterning in Cldn5(BAC)-GFP mouse ear skin in response to intradermal histamine. Left, overlay of Cldn5(BAC)-GFP-positive and -negative vessels (visualised through circulating TRITC dextran). Arrowheads show sites of leakage. Right, stills of leakage in the vasculature shown on the left following intradermal histamine stimulation. (B) Leakage sites per vessel length in different vessel categories. +/+denotes capillary segments with full GFP expression,+/-denotes capillary segments with mixed GFP expression, -/- denotes capillary segments with no GFP expression. n=4, 2 or more acquisitions/mouse. (C) Proportion of Cldn5(BAC)-GFP-negative vessels susceptible or resistant to leakage. n=4, 2 or more acquisitions/mouse. (D–F) Leakage patterning in the ear skin (D), trachea (E) and back skin (F) in response to the systemic administration of histamine. Left, representative image. Dashed line shows progression of a blood vessel from arteriolar to venular. Right, representative fluorescent intensity line profile of Cldn5(BAC)-GFP and TRITC 2000 kDa dextran along the dashed line (Left). (G) Proportion of 2000 kDa FITC leakage area that occurs in vessels that are Cldn5(BAC)-GFP-positive (contain some positive cells) and Cldn5(BAC)-GFP-negative (contain no positive cells) in ear skin, back skin and trachea. n≥3 mice, 3 or more fields of view/mouse. (H) Fold change in 2000 kDa TRITC dextran extravasation from leakage permissive vessels in ear skin, back skin and trachea with and without systemic histamine stimulation. Dashed line represents unstimulated tissue. n=3 mice, 3 or more fields of view/mouse. (I) Fold change in tissue 2000 kDa FITC dextran following systemic histamine stimulation and formamide extraction of ear skin, skeletal muscle and heart. Dashed line represents unstimulated tissue. n=3 mice. Error bars; mean ± SD. Statistical significance: one-way ANOVA with Tukey’s post-hoc test (multiple comparisons; G–I).
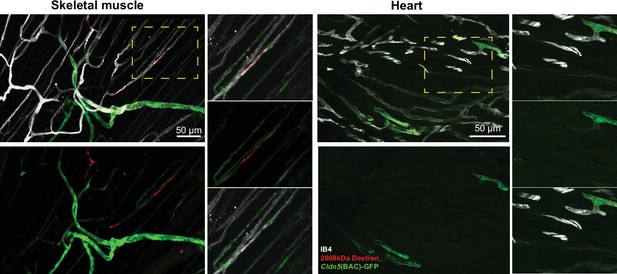
Histamine leakage in skeletal muscle and heart vasculature.
Leakage and Cldn5(BAC)-GFP expression patterning in skeletal muscle and heart in response to the systemic administration of histamine. Magnified images of dashed boxes are shown to the right of the main image.

Claudin5 exhibits organotypic protection of the EC barrier.
(A) Schematic illustration of systemic tamoxifen regime. (B) Representative western blot of Claudin5 protein expression in control and Cldn5 iECKO mice. (C) Quantification of Claudin5 protein expression in lung lysates of control and Cldn5 iECKO mice. n≥8 mice. (D) Cldn5 gene expression by qPCR on lung lysates of control and Cldn5 iECKO mice. n≥5 mice. (E) Claudin5 protein expression normalized to CD31 counter-staining in the ear skin of control and Cldn5 iECKO mice following systemic tamoxifen. Right, representative images of Claudin5 immunostaining in control and Cldn5 iECKO mice. n≥3 mice, 3 or more fields of view/mouse. (F) Blood vessel basal permeability to 10 kDa and 70 kDa dextran in ear skin, back skin, skeletal muscle, and heart of wildtype C57Bl/6 mice. Dashed lines represent background from control uninjected mice. n=3 mice. (G) Blood vessel basal permeability to 4 kDa, 10 kDa and 70 kDa dextran in ear skin, back skin, skeletal muscle, and heart of control and Cldn5 iECKO mice. Dashed lines represent control Cre-negative mice. n≥3 mice. (H–I) Leakage of 2000 kDa dextran in response to systemic histamine stimulation (10 mg/kg) in skeletal muscle (H) and ear skin (I). Top, quantification of tracer leakage area / vessel area normalized to control (Cre-negative) mice. Bottom, representative images. n≥7 mice, 3 or more fields of view/mouse. (J–K) Leakage of 2000 kDa dextran in response to systemic histamine stimulation (4 mg/kg) in back skin (J) and trachea (K). Top, quantification of tracer leakage area / vessel area normalized to control (Cre-negative) mice. Bottom, representative images. n≥8 mice, 3 or more fields of view/mouse. (L) Quantification of 2000 kDa dextran leakage in the ear skin of control and Cldn5 iECKO mice following Oxazolone-induced dermatitis. Right, representative images. n≥12 mice, 2 or more fields of view/mouse (M) Quantification of 2000 kDa dextran leakage in the back skin of control and Cldn5 iECKO mice following Oxazolone-induced dermatitis. n≥9 mice, 2 or more fields of view/mouse Error bars; mean ± SD. Statistical significance: two-tailed paired Student’s t test (C-E), H-M or one-way ANOVA with Tukey post-hoc test (multiple comparisons; F–G).
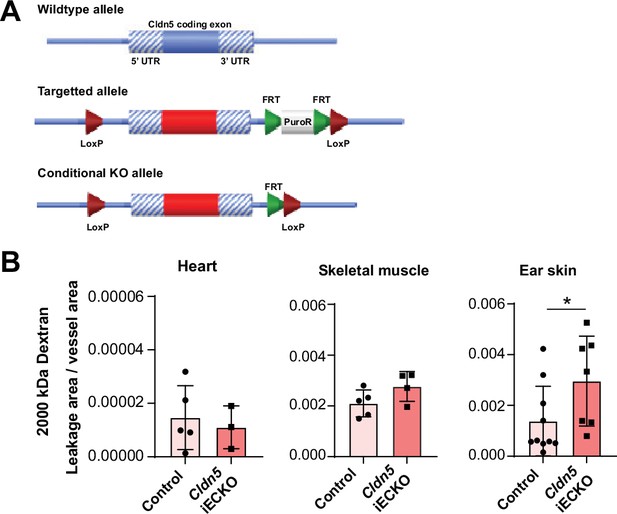
Cldn5 targeting and histamine-induced leakage quantification.
(A) Schematic diagram showing the targeting strategy of Cldn5 floxed mice. (B) Quantification of histamine-induced (10 mg/kg) leakage of 2000 kDa dextran from the heart, skeletal muscle and ear skin vasculature in control and Cldn5 iECKO mice. n≥3 mice, 3 or more fields of view/mouse. Error bars; mean ± SD. Statistical significance: two-tailed paired Student’s t test.
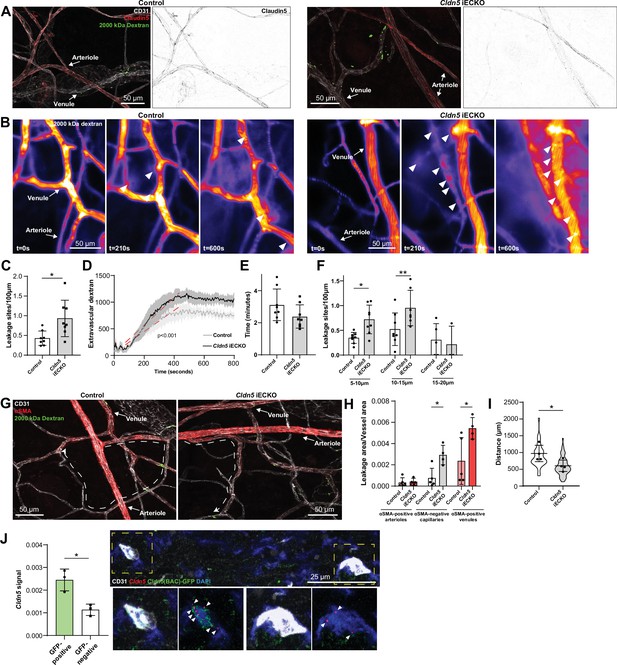
Loss of Claudin5 differentially affects vessel subtypes in the ear dermis.
(A) Representative images of histamine-induced 2000 kDa dextran leakage in the ear skin of control (left) and Cldn5 iECKO (right) mice. (B) Representative time-lapse images of 2000 kDa dextran leakage in response to intradermal histamine stimulation in the ear skin of control (left) and Cldn5 iECKO (right) mice. Arrowheads show sites of leakage. (C) Leakage sites per vessel length in response to intradermal histamine stimulation in the ear skin of control and Cldn5 iECKO mice. n≥7 mice, two or more acquisitions/mouse. (D) Quantification of extravascular 2000 kDa dextran over time in the ear skin of control and Cldn5 iECKO mice following intradermal histamine stimulation. Red dashed lines represent lines of best fit for the slope between leakage initiation and leakage termination. n≥7 mice, two or more acquisitions/mouse. (E) Lag period between intradermal histamine injection and initiation of leakage in the ear skin of control and Cldn5 iECKO mice. n≥7 mice, two or more acquisitions/mouse. (F) Leakage sites per length of post-arteriolar vessels of different diameter in response to intradermal histamine stimulation in the ear skin of control and Cldn5 iECKO mice. n≥7 mice, two or more acquisitions/mouse. n≥7 mice, two or more acquisitions/mouse. (G) Representative images of 2000 kDa dextran leakage in response to systemic histamine stimulation in the ear skin of control and Cldn5 iECKO mice counter-stained for αSMA. Dashed lines with arrows show distance from arteriolar/capillary transition to first site of leakage. (H) Leakage area/vessel area of 2000 kDa dextran in response to systemic histamine stimulation in αSMA-positive arterioles, αSMA-negative capillaries and αSMA-positive venules in the ear skin of control and Cldn5 iECKO mice. n≥4 mice, 3 or more fields of view/mouse. (I) Distance between arteriolar-capillary branch points and the first site of 2000 kDa dextran leakage in response to systemic histamine stimulation in the ear skin of control and Cldn5 iECKO mice. n≥4 mice, 3 or more fields of view/mouse. (J) Cldn5 mRNA expression in Cldn5(BAC)-GFP-positive and -negative vessels of the ear skin. Left, quantification of Cldn5 signal (Cldn5 mRNA particles/vessel area). Right, representative image. Dashed boxes are magnified below, arrowheads mark Cldn5 mRNA particles. n=3 mice, 4 or more fields of view/mouse. Error bars; mean ± SD. Statistical significance: two-tailed paired Student’s t test (C, E–J) and linear regression and ANCOVA (D).

4-hydroxytamoxifen-mediated Claudin5 loss, histamine receptor expression and vessel-specific leakage in Cldn5 iECKO mice.
(A) Schematic illustration of topical 4-hydroxytamoxifen regime. (B) Claudin5 protein expression in the ear skin of control and Cldn5 iECKO mice following topical tamoxifen treatment. Left, quantification. Right, representative image of Claudin5 expression in Cldn5 fl/fl; Rosa26lox-STOP-lox-YFP; Cdh5CreERT2 mice following tamoxifen treatment. (C) Leakage of 2000 kDa dextran in the ear skin of topically tamoxifen treated control or Cldn5 iECKO mice in response to histamine (10 mg/kg). Left, quantification. Right, representative image of leakage and Claudin5. (D) Violin plots for gene expression of histamine receptors Hrh1 and Hrh2. Gene expression was normalized to account for differences in sample library size and imputed to account for dropouts in the data as described in Methods. (E) Image showing the expression of Cldn5(BAC)-GFP and αSMA in the mouse ear dermis. Dashed box is shown magnified to right. (F) Quantification of Claudin5 expression in αSMA-positive arterioles, αSMA-negative capillaries and αSMA-positive venules in control and Cldn5 iECKO mice. Error bars; mean ± SD. Statistical significance: two-tailed paired Student’s t test.
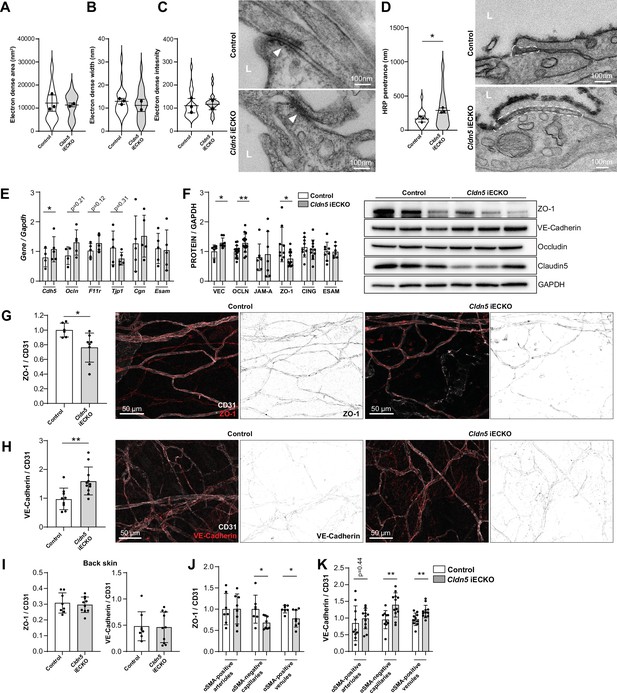
Claudin5 regulates junction protein expression.
(A–C) Area (A) width (B) and intensity (C) of electron dense regions in the ear skin of control and Cldn5 iECKO mice after visualisation by TEM. Right, representative TEM images of junctions in the ear skin of control and Cldn5 iECKO mice. Junctions can be seen within electron dense regions (arrowheads). L, lumen. n≥2 mice, 6 or more fields of view/mouse. (D) Distance of HRP penetrance into EC junctions in the ear skin of control and Cldn5 iECKO mice following systemic histamine stimulation. Right, representative TEM images of HRP penetrance (visualized by electron dense 3,3′-Diaminobenzidine (DAB) reaction precipitate) into EC junctions in the ear skin of control and Cldn5 iECKO mice following systemic histamine stimulation. Dashed regions show areas of disrupted junction into which HRP has penetrated. Note that the typical electron dense area is lacking due to absence of uranyl acetate staining. L, lumen. n≥2 mice, 6 or more fields of view/mouse. (E) Gene expression of AJ- and TJ-associated genes in lung lysates of control and Cldn5 iECKO mice. n≥4 mice. (F) Expression of AJ- and TJ- associated proteins in lung lysates of control and Cldn5 iECKO mice. Right, representative western blots of AJ- and TJ- associated proteins in lung lysates of control and Cldn5 iECKO mice. n≥4 mice. (G) Expression of ZO-1 in ear skin blood vessels of control and Cldn5 iECKO mice. Left, quantification of ZO-1. Right, representative images of ZO-1 in the ear skin of control and Cldn5 iECKO mice. n≥6 mice, 3 or more fields of view/mouse. (H) Expression of VE-Cadherin in ear skin blood vessels of control and Cldn5 iECKO mice. Left, quantification of VE-Cadherin. Right, representative images of VE-Cadherin in the ear skin of control and Cldn5 iECKO mice. n≥9 mice, 3 or more fields of view/mouse. (I) Quantification of ZO-1 (left) and VE-Cadherin (right) in back skin blood vessels of control and Cldn5 iECKO mice. n≥8 mice, 2 or more fields of view/mouse. (J) Quantification of ZO-1 in different vessel subtypes in the ear skin of control and Cldn5 iECKO mice. n≥3 mice, 3 or more fields of view/mouse. (K) Quantification of VE-Cadherin in different vessel subtypes in the ear skin of control and Cldn5 iECKO mice. n≥3 mice, 3 or more fields of view/mouse. Error bars; mean ± SD. Statistical significance: two-tailed paired Student’s t test.

Endothelial junction protein expression in Cldn5 iECKO mice.
(A) Representative western blots of ESAM, Cingulin, and JAM-A expression in control and Cldn5 iECKO mouse lung lysates. (B) Scatter graphs showing the correlation between Claudin5 expression levels and other EC junction proteins in mouse lung lysates. Both control (red dots) and Cldn5 iECKO (black dots) mice are represented. Line of best fit following linear regression analysis is shown.
Videos
Histamine-mediated leakage in Cldn5(BAC)-GFP mice.
Extravasation of circulating 2000 kDa TRITC Dextran (pseudocolor) after intradermal injection of histamine in the ear dermis of Cldn5(BAC)-GFP mice. The first 30 frames show a still at t=0 to show Cldn5(BAC)-GFP expression (green) overlayed with 2000 kDa Dextran (grey).
Histamine-mediated leakage in control and Cldn5 iECKO mice.
Extravasation of circulating 2000 kDa FITC Dextran (pseudocolor) in control (left) and Cldn5 iECKO (right) mice after intradermal injection of histamine in the ear dermis.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus musculus) | C57BL/6 J | Taconic | B6-F/M | |
| Genetic reagent (Mus musculus) | Cldn5(BAC)-GFP | Laviña et al., 2018 | N/A | |
| Genetic reagent (Mus musculus) | Cldn5 iECKO | This paper | N/A | See Figure 3—figure supplement 1 and Methods-Animals |
| Genetic reagent (Mus musculus) | Cldn5fl/fl; Rosa26lox-STOP-lox-YFP; Cdh5CreERT2 | This paper | N/A | See Methods-Animals |
| Antibody | Mouse monoclonal anti-GAPDH | Millipore | MAB374 | 1:1,000 |
| Antibody | Rat monoclonal anti-CD31 | BD Biosciences | 553370 | 1:100 |
| Antibody | Goat polyclonal anti-CD31 | R&D Systems | AF3628 | 1:100 |
| Antibody | Goat polyclonal anti-VE-Cadherin | R&D Systems | AF1002 | 1:100, 1:1000 |
| Antibody | Chicken polyclonal anti-GFP | Abcam | Ab13970 | 1:100 |
| Antibody | Rabbit polyclonal anti-Claudin5 | ThermoFischer Scientific | 341600 | 1:100, 1:1000 |
| Antibody | Rabbit polyclonal anti-ZO-1 | ThermoFischer Scientific | 617300 | 1:100, 1:1000 |
| Antibody | Rabbit polyclonal anti-Occludin | ThermoFischer Scientific | 711500 | 1:1,000 |
| Antibody | Rabbit monoclonal anti-JAM-A | Martìn-Padura et al., 1998 | N/A | 1:1000 |
| Antibody | Rabbit polyclonal anti-Cingulin | Cardellini et al., 1996 | N/A | 1:1000 |
| Antibody | Goat polyclonal anti-ESAM | R&D Systems | AF2827 | 1:1000 |
| Antibody | Goat polyclonal anti-collagen IV | Merck Millipore | AB789 | 1:100 |
| Antibody | Mouse monoclonal anti-αSMA FITC | Sigma Aldrich | F3777 | 1:100 |
| Antibody | Mouse monoclonal anti- αSMA Cy3 | Sigma Aldrich | C6198 | 1:100 |
| Antibody | Rat monoclonal Anti-CD16/32 | ThermoFischer Scientific | 14-0161-85 | 1:100 |
| Antibody | Rat monoclonal Anti-CD31 FITC | BD Biosciences | 553372 | 1:50 |
| Antibody | Rat monoclonal Anti-CD45 APC | BioLegend | 103112 | 1:50 |
| Antibody | Rat monoclonal Anti-Lyve1 eFluor 660 | ThermoFischer Scientific | 50–0443082 | 1:50 |
| Antibody | Donkey polyclonal anti-rat alexa 488 | ThermoFischer Scientific | A21208 | 1:400 |
| Antibody | Donkey polyclonal anti-rat alexa 594 | ThermoFischer Scientific | A21209 | 1:400 |
| Antibody | Donkey polyclonal anti-rabbit alexa 488 | ThermoFischer Scientific | A21206 | 1:400 |
| Antibody | Donkey polyclonal anti-rabbit alexa 568 | ThermoFischer Scientific | A10042 | 1:400 |
| Antibody | Donkey polyclonal anti-goat alexa 647 | ImmunoResearch Laboratories | 705-605-147 | 1:400 |
| Antibody | Donkey polyclonal anti-chicken alexa 488 | ImmunoResearch Laboratories | 703-545-155 | 1:400 |
| Antibody | Sheep polyclonal anti-mouse HRP | Cytiva | NA931 | 1:10,000 |
| Antibody | Sheep polyclonal anti-rabbit HRP | Cytiva | NA934 | 1:10,000 |
| Sequence-based reagent | Cldn5 probe | ACD Bio | 491611-C2 | |
| Sequence-based reagent | 3-plex negative control probes | ACD Bio | 320871 | |
| Sequence-based reagent | 3-plex positive control probes | ACD Bio | 320811 | |
| Sequence-based reagent | GAPDH | ThermoFischer Scientific | Mm99999915_g1 | |
| Sequence-based reagent | Cldn5 | ThermoFischer Scientific | Mm00727012_s1 | |
| Sequence-based reagent | Cdh5 | ThermoFischer Scientific | Mm00486938_m1 | |
| Sequence-based reagent | Tjp1 | ThermoFischer Scientific | Mm01320638_m1 | |
| Sequence-based reagent | Ocln | ThermoFischer Scientific | Mm00500912_m1 | |
| Sequence-based reagent | F11r | ThermoFischer Scientific | Mm00554113_m1 | |
| Sequence-based reagent | Cgn | ThermoFischer Scientific | Mm01263534_m1 | |
| Sequence-based reagent | Esam | ThermoFischer Scientific | Mm00518378_m1 | |
| Peptide, recombinant protein | Collagenase IV | Worthington | LS004183 | |
| Peptide, recombinant protein | DNase I | Worthington | LS006333 | |
| Peptide, recombinant protein | HRP | SigmaAldrich | 77332 | |
| Commercial assay or kit | RNAscope Fluorescent Multiplex Assay | ACD Bio | 322340, 320851 | |
| Commercial assay or kit | RNeasy Plus kit | Qiagen | 74034 | |
| Commercial assay or kit | iScript Adv cDNA Kit for RT-qPCR | Bio-Rad | 1725038 | |
| Chemical compound, drug | Tamoxifen | SigmaAldrich | T5648 | |
| Chemical compound, drug | 4-hydroxytamoxifen | SigmaAldrich | H7904 | |
| Chemical compound, drug | Oxazolone | SigmaAldrich | E0753 | |
| Chemical compound, drug | Histamine | SigmaAldrich | H7125 | |
| Other | Live/Dead near IR cell stain | ThermoFischer Scientific | L10119 | See Materials and methods-Ear dermal single cell isolation |
| Other | Phosphatase inhibitor cocktail | Roche | 04906837001 | See Materials and methods-Western blot analysis |
| Other | LDS sample buffer | Invitrogen | NP0007 | See Materials and methods-Western blot analysis |
| Other | Sample reducing agent | Invitrogen | NP0009 | See Materials and methods-Western blot analysis |
| Other | MOPS SDS running buffer | Invitrogen | NP0001 | See Materials and methods-Western blot analysis |
| Other | PVDF membrane | Thermofischer Scientific | 88518 | See Materials and methods-Western blot analysis |
| Other | NuPAGE transfer buffer | Novex | NP006 | See Materials and methods-Western blot analysis |
| Other | RNAlater | ThermoFischer Scientific | AM7024 | See Materials and methods-quantitative PCR |
| Other | 2000 kDa FITC Dextran | SigmaAldrich | FD2000S | See Materials and methods-permeability analysis |
| Other | 2000 kDa TRITC Dextran Lysine Fixable | ThermoFischer Scientific | D7139 | See Materials and methods-permeability analysis |
| Other | 10 kDa TRITC Dextran | ThermoFischer Scientific | D1817 | See Materials and methods-permeability analysis |
| Other | 4 kDa TRITC Dextran | Tdb labs | TD4 | See Materials and methods-permeability analysis |
| Other | 10 kDa FITC Dextran | Tdb labs | FD10 | See Materials and methods-permeability analysis |
| Other | 70 kDa TRITC Dextran | Tdb labs | TD70 | See Materials and methods-permeability analysis |
| Other | 2000 kDa FITC Dextran lysine fixable | Tdb labs | FLD2000 | See Materials and methods-permeability analysis |
| Other | Anti-Isolectin GS-IB4 | Molecular Probes | I32450 | See Materials and methods-Immunohistochemistry |





