Regional importation and asymmetric within-country spread of SARS-CoV-2 variants of concern in the Netherlands
Figures
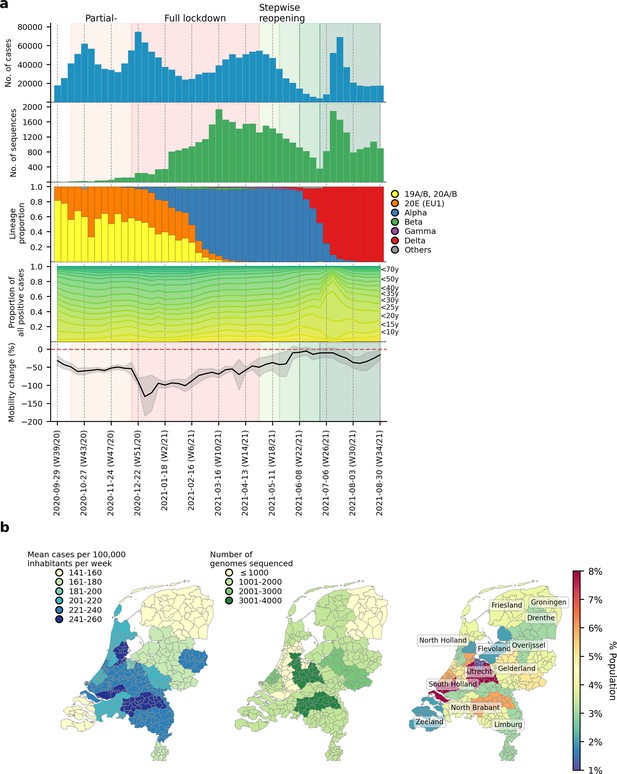
SARS-CoV-2 infections and VOCs circulating in the Netherlands from September 2020 to August 2021.
(A) Weekly number of laboratory-confirmed SARS-CoV-2 cases (1st panel from the top) and sequenced genomes (2nd panel). Lineage proportions of sequences colored by NextClade genotype designations (3rd panel). Breakdown of positive cases by age group from data provided by the Dutch National Institute for Public Health and Environment (4th panel). Aggregated weekly average percentage change in mobility to the baseline in the Netherlands from Google’s COVID-19 community mobility reports. Baseline mobility is the median value from a 5-week period between 3 January 2020 and 6 February 2020, prior to the COVID-19 pandemic in Europe (5th panel). (B) Mean number of laboratory-confirmed cases per 100,000 inhabitants (data from the Dutch National Institute for Public Health and Environment; left panel); total number of sequenced genomes in different Municipal and Regional Health Service (GGD) regions over the entire study period (middle panel); Percentage of Dutch population residing in each GGD region (right panel).
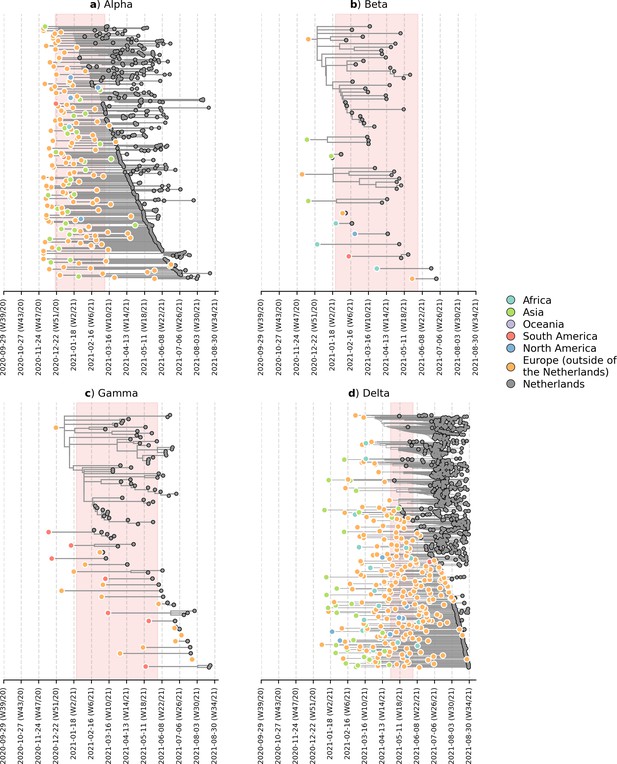
Likely overseas introduction of VOC lineages into the Netherlands at the continental level.
For each VOC lineage, a time-scaled maximum likelihood phylogeny using the Dutch and their nearest overseas neighboring sequences was inferred. Discrete trait analyses were performed to infer the likely continental region of ancestral states. Subtrees or singletons with ancestral nodes attributed to an overseas origin but subtend only Dutch sequences are drawn. Shaded plot area denotes the timespan when a targeted flight restriction was imposed on the country where the VOC lineage first emerged (i.e. (A) Alpha, United Kingdom.; (B) Beta, South Africa; (C) Gamma, Brazil; (D) Delta, India).
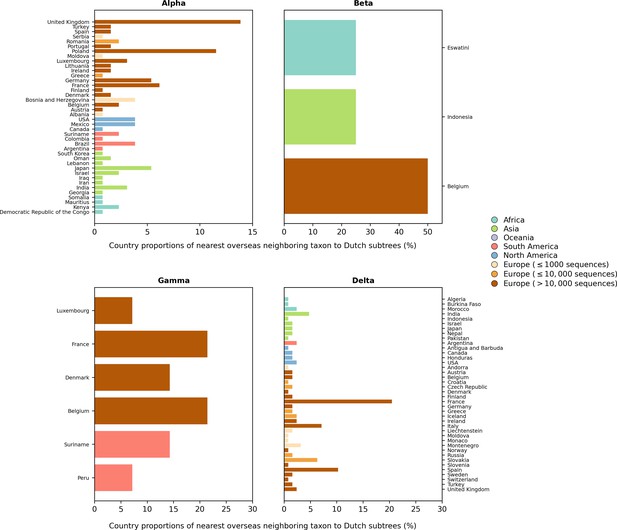
Distribution of countries of the nearest overseas neighboring taxon to Dutch subtrees.
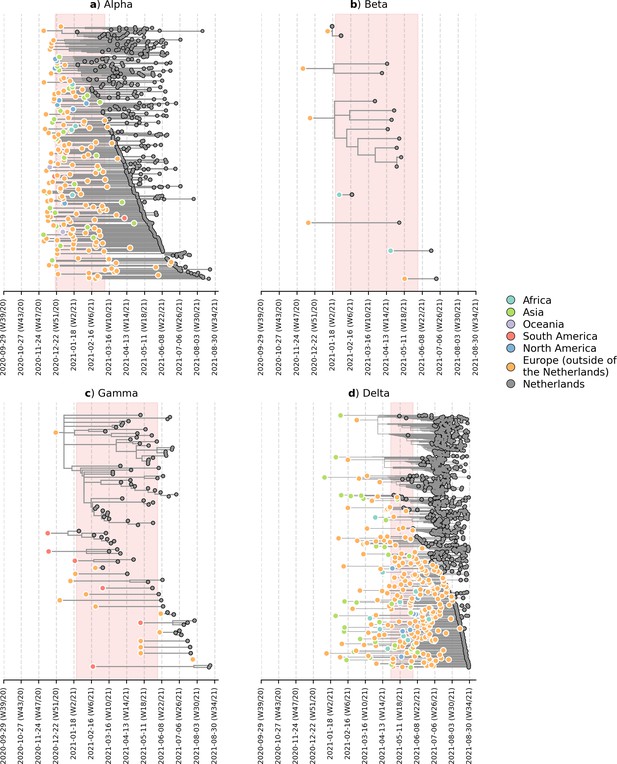
Likely overseas introduction of VOC lineages into the Netherlands at the continental level (based on phylogenetic analyses using an independently subsampled set of sequence data from that used in Figure 2).
For each VOC lineage, a time-scaled maximum likelihood phylogeny using the Dutch and their nearest overseas neighboring sequences was inferred. Discrete trait analyses were performed to infer the likely continental region of ancestral states. Subtrees or singletons with ancestral nodes attributed to an overseas origin but subtend only Dutch sequences are drawn. Shaded plot area denotes the timespan when a targeted flight restriction was imposed on the country where the VOC lineage first emerged (i.e. (A) Alpha, United Kingdom.; (B) Beta, South Africa; (C) Gamma, Brazil; (D) Delta, India).
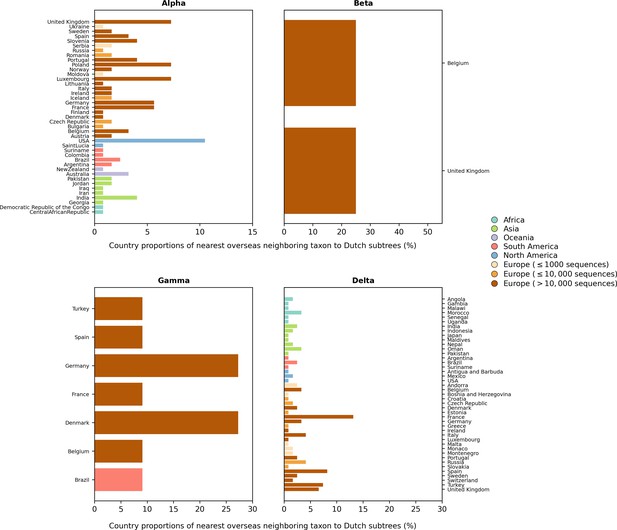
Distribution of countries of the nearest overseas neighboring taxon to Dutch subtrees.
(based on phylogenetic analyses using an independently subsampled set of sequence data from that used in Figure 2 and Figure 2—figure supplement 1).
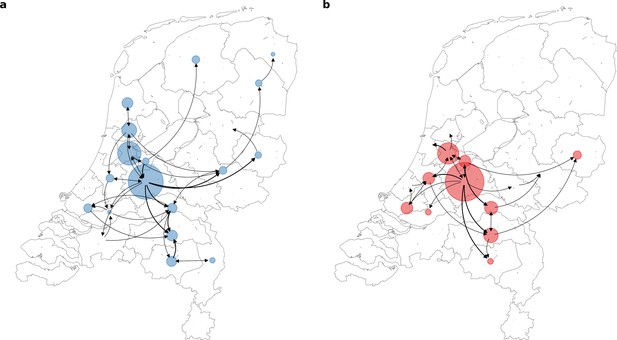
Source-sink dynamics the Alpha and Delta variants of concern (VOC) within the Netherlands during the first four weeks after their respective detection.
(A) Alpha (between 2 and 29 December 2020); (B) Delta (between 20 April and 18 May 2021).
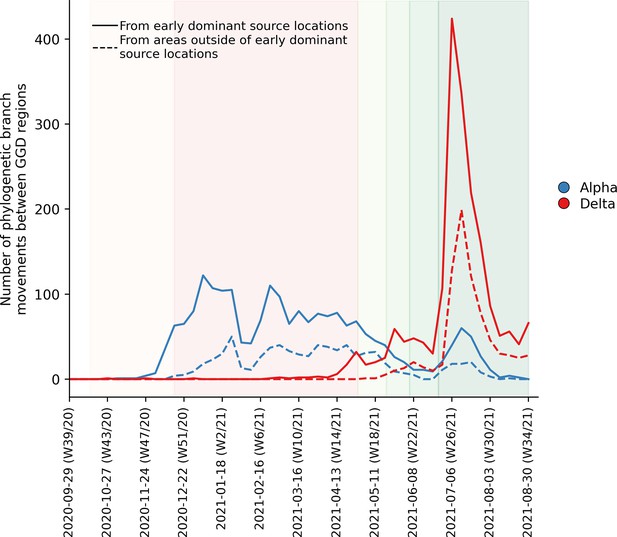
Estimated number of phylogenetic branch movements and growth rate estimations of Alpha and Delta VOCs in the Netherlands.
Solid line shows the number of branch movements from early dominant source regions including North and South Holland, Utrecht and Brabant. Dashed line shows number of branch movements from areas outside of these early dominant source locations.
Additional files
-
Supplementary file 1
Accession codes of all Dutch sequencing data that have been deposited in the GISAID database (https://www.gisaid.org).
- https://cdn.elifesciences.org/articles/78770/elife-78770-supp1-v1.txt
-
Supplementary file 2
Acknowledgement table of authors of global genome sequence data deposited in the GISAID database that were used in this study.
- https://cdn.elifesciences.org/articles/78770/elife-78770-supp2-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/78770/elife-78770-mdarchecklist1-v1.pdf
-
Reporting standard 1
SRQR Checklist.
- https://cdn.elifesciences.org/articles/78770/elife-78770-repstand1-v1.docx





