Senataxin and RNase H2 act redundantly to suppress genome instability during class switch recombination
Figures
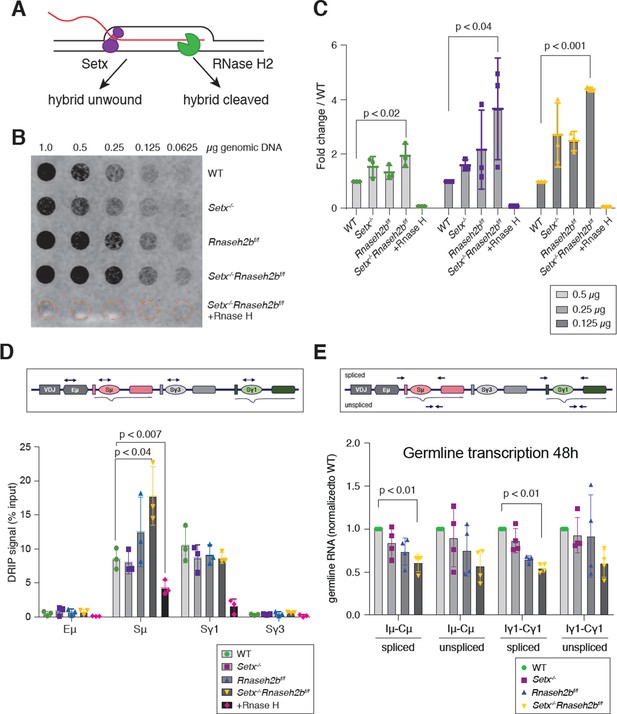
RNA-DNA hybrids are increased in Setx-/-Rnaseh2bf/f B cells during class switch recombination (CSR) to IgG1.
(A) Senataxin (SETX) and RNase H2 contribute to RNA:DNA hybrid removal; SETX helicase activity unwinds the nucleotide strands retaining both the RNA and DNA components while RNase H2 cleaves RNA, retaining only the DNA strand. (B) Dot blot analysis of R loop formation: twofold serial dilutions of genomic DNA starting at 1 µg were arrayed on a nitrocellulose membrane and probed using the S9.6 antibody; RNase H treatment of the Setx-/-Rnaseh2bf/f sample was used as the negative control. (C) Quantitation of 0.5 µg, 0.25 µg, and 0.125 µg dot blot from (B) using ImageJ; values were normalized to WT, set as 1. Figures are expressed as fold change relative to WT. Error bars are the standard deviation between different experiments; *p<0.05 comparing different genotypes using multiple t-test (n = 3 mice/genotype). (D) Diagram of the IgH locus showing the location of PCR products used for chromatin immunoprecipitation (ChIP) and DNA:RNA hybrid immunoprecipitation (DRIP) assays. DRIP assay was performed with S9.6 antibody using primary B cells after 72 hr of stimulation to IgG1 with LPS/IL-4/α-RP105; RNase H treatment of Setx-/-Rnaseh2bf/f sample was used as the negative control. Relative enrichment was calculated as ChIP/input, and the results were replicated in three independent experiments. Error bars show standard deviation; statistical analysis was performed using one-way ANOVA (n = 3 mice/genotype). (E) Diagram of the IgH locus showing the location of PCR products for germline transcription under IgG1 stimulation with LPS/IL-4/α-RP105. Real-time RT-PCR analysis for germline transcripts (Ix-Cx) at donor and acceptor switch regions in WT, Setx-/-, Rnaseh2bf/f, and Setx-/- Rnaseh2bf/f splenic B lymphocytes cultured for 48 hr with LPS/IL-4/α-RP105 stimulation. Expression is normalized to CD79b and is presented as relative to expression in WT cells, set as 1. Error bars show standard deviation; statistical analysis was performed using multiple t-test (n = 4 mice/genotype).
-
Figure 1—source data 1
Uncropped raw dot blot analysis of R loop formation.
Genomic DNA extracted from WT, Rnaseh2bf/f, Setx-/-, and Setx-/-Rnaseh2bf/f cells was digested with restriction enzyme cocktail and run on dot blot, RnaseH1-treated Setx-/-Rnaseh2bf/f as a negative control.
- https://cdn.elifesciences.org/articles/78917/elife-78917-fig1-data1-v1.zip
-
Figure 1—source data 2
Numerical data used to generate graphs in Figure 1C–E.
- https://cdn.elifesciences.org/articles/78917/elife-78917-fig1-data2-v1.xlsx
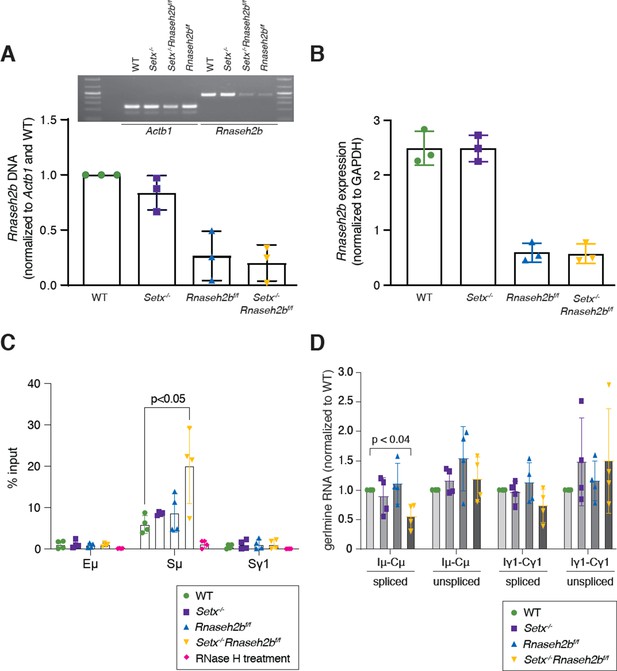
Rnaseh2b deletion efficiency by CD19-cre.
(A) Deletion of genomic DNA measured by PCR from splenic B cells. (B) Transcription at the Rnaseh2b locus measured by RT-qPCR across exon 6, downstream of the Cre-mediated deletion site. (C) DNA:RNA hybrid immunoprecipitation (DRIP) assay in resting B cells. DRIP assay was performed with S9.6 antibody using primary resting B cells; RNase H treatment of Setx-/-Rnaseh2bf/f sample was used as the negative control. Relative enrichment was calculated as chromatin immunoprecipitation (ChIP)/input, and the results were replicated in three independent experiments. Error bars show standard deviation; statistical analysis was performed using one-way ANOVA (n = 4 mice/genotype). (D) Real-time RT-PCR analysis for germline transcripts (Ix-Cx) at donor and acceptor switch regions in WT, Setx-/-, Rnaseh2bf/f, and Setx-/- Rnaseh2bf/f splenic B lymphocytes cultured for 72 hr with LPS/IL-4/α-RP105 stimulation. Expression is normalized to CD79b and is presented as relative to expression in WT cells, set as 1. Error bars show standard deviation; statistical analysis was performed using multiple t-test (n = 4 mice/genotype).
-
Figure 1—figure supplement 1—source data 1
Uncropped image of PCR analysis of Cre-mediated Rnaseh2b deletion.
PCR test of Rnaseh2b deletion efficiency on genomic DNA extracted from WT, Setx-/-, Rnaseh2bf/f, and Setx-/- Rnaseh2bf/f splenic B lymphocytes. Left and right represent two times results.
- https://cdn.elifesciences.org/articles/78917/elife-78917-fig1-figsupp1-data1-v1.zip
-
Figure 1—figure supplement 1—source data 2
Numerical data used to generate graphs in Figure 1—figure supplement 1A–D.
- https://cdn.elifesciences.org/articles/78917/elife-78917-fig1-figsupp1-data2-v1.xlsx

DNA:RNA hybrid formation at the Actb locus.
(A) Representative UCSC genome browser screenshot of DRIP-Seq signal in WT, Setx-/-, Rnaseh2bf/f, and Setx-/-Rnaseh2bf/f cells spanning 10 kb of the Actb locus (n = 2 mice/genotype). Locations of primers used in DRIP-qPCR shown in (B) are marked with black (gene body) and pink (3′UTR) triangles. (B) DRIP-qPCR along Actb locus in mouse primary B cells. Diagram of the IgH locus showing the location of PCR products used for DNA:RNA hybrid immunoprecipitation (DRIP) (top panel). DRIP-qPCR along Actb locus in mouse primary B cells (lower panel). DRIP assay was performed with S9.6 antibody using primary B cells after 72 hr of stimulation to IgG1 with LPS/IL-4/α-RP105; RNase H treatment of Setx-/-Rnaseh2bf/f sample was used as the negative control. Relative enrichment was calculated as chromatin immunoprecipitation (ChIP)/input, and the results were replicated in three independent experiments. Error bars show standard deviation; statistical analysis was performed using one-way ANOVA (n = 3 mice/genotype).
-
Figure 1—figure supplement 2—source data 1
Numerical data used to generate graph in Figure 1—figure supplement 2B.
- https://cdn.elifesciences.org/articles/78917/elife-78917-fig1-figsupp2-data1-v1.xlsx
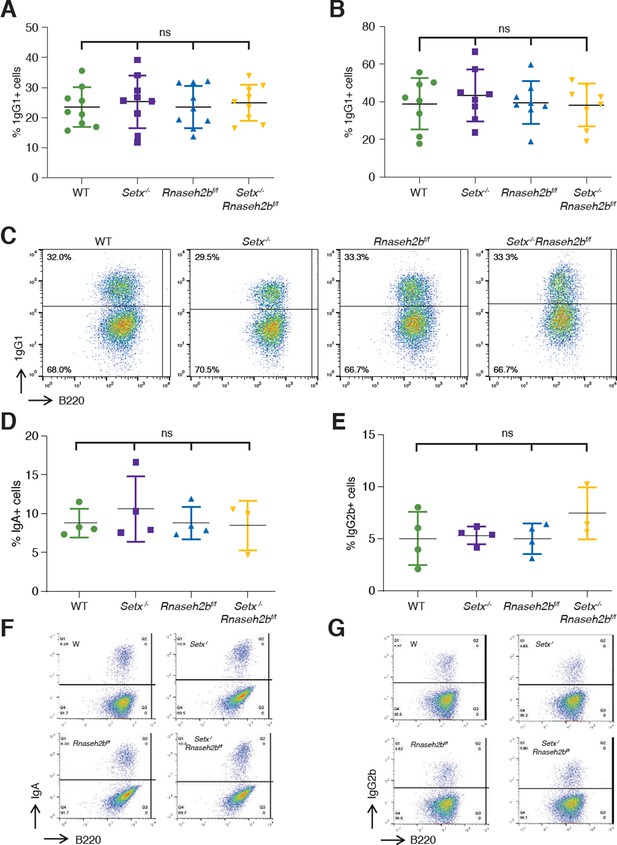
Class switch recombination (CSR) is not reduced in Setx-/-, Rnaseh2bf/f, or Setx-/-Rnaseh2bf/f B cells.
Percentage of cells undergoing CSR to IgG1 72 hr (A) and 96 hr (B) post-stimulation with LPS/IL-4/α-RP105. (C) Representative flow cytometry analyses of IgG1+ and B220 expression in response to LPS/IL-4/α-RP105. The percentage of IgG1+ B cells is indicated at top left. Percentage of cells undergoing CSR to IgA (D) or IgG2B (E) 72 hr post-stimulation. (F) Representative flow cytometry analyses of IgA+ and B220 expression in response to LPS/α-RP105/TGF-B/CD40L. (G) Representative flow cytometry analyses of IgG2B+ and B220 expression in response to LPS/α-RP105/TGF-B. Horizontal lines in dot plots indicate mean, and error bars show standard deviation. Statistical significance versus WT was determined by one-way ANOVA; each dot represents an independent mouse.
-
Figure 2—source data 1
Numerical data used to generate graphs in Figure 2A, B, D and E.
- https://cdn.elifesciences.org/articles/78917/elife-78917-fig2-data1-v1.xlsx
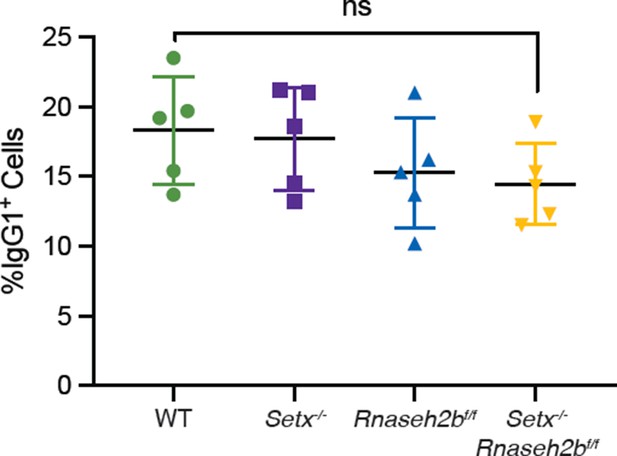
Percentage of cells undergoing class switch recombination (CSR) to IgG1 72 hr post-stimulation with LPS/IL-4 alone.
Horizontal lines in dot plots indicate mean, and error bars show standard deviation. Statistical significance versus WT was determined by one-way ANOVA; each dot represents an independent mouse.
-
Figure 2—figure supplement 1—source data 1
Numerical data used to generate graphs in Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/78917/elife-78917-fig2-figsupp1-data1-v1.xlsx
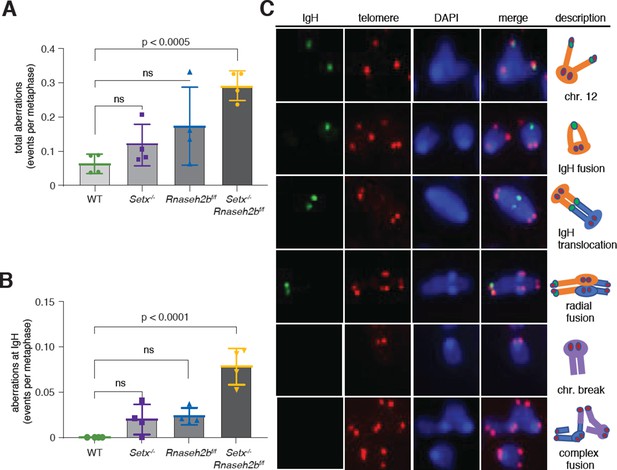
Increased IgH damage is observed in Setx-/-Rnaseh2bf/f B cells.
(A) Frequency of spontaneous DNA damage in WT, Setx-/-, Rnaseh2bf/f, and Setx-/-Rnaseh2bf/f cells. (B) Frequency of spontaneous DNA damage at IgH. (C) Representative images of the types of rearrangements produced. IgH-specific probe visualized in green, Telomere-specific probe visualized in red, DAPI is in blue. All cells were harvested 72 hr post-stimulation to IgG1 with LPS/IL-4/α-RP105. Error bars show standard deviation; statistical significance versus WT was determined by one-way ANOVA (n = 4 independent mice).
-
Figure 3—source data 1
Numerical data used to generate graphs in Figure 3A and B.
- https://cdn.elifesciences.org/articles/78917/elife-78917-fig3-data1-v1.xlsx
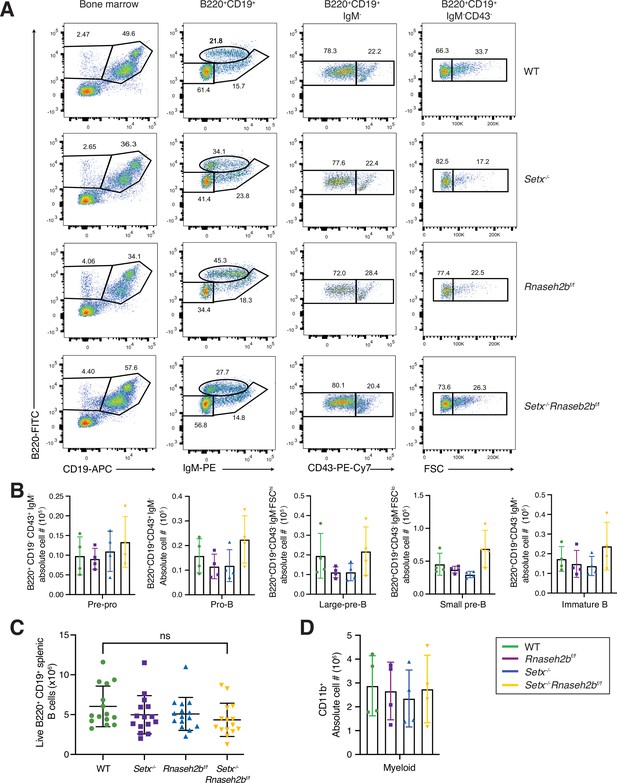
B cell development in the bone marrow is normal in cells lacking senataxin (Setx) and RNase H2B.
(A) Flow cytometric gating of different developmental stages of B lymphopoiesis in the bone marrow (BM) of WT, Setx-/-Rnaseh2bf/f, and Setx-/-Rnaseh2bf/f mice (n = 4). Pre-pro-B cells are defined as B220+CD19- CD43+IgM-, pro-B cells are defined as B220+CD19+CD43+IgM-, large and small pre-B cells are defined as B220+ CD19+CD43−IgM−FSChi and B220+CD19+CD43−IgM−FSClo, respectively, and immature B cells are defined as B220+CD19+CD43−IgM+ FSC, forward scatter. (B) Absolute number of cells per mouse at different stages of B cell development in the BM of WT, Setx-/-, Rnaseh2bf/f, and Setx-/-Rnaseh2bf/f mice (n = 4). (C) Number of mature naïve B lymphocytes isolated from spleen. (D) Absolute number of myeloid compartment cells (CD11b + cells) in BM.
-
Figure 3—figure supplement 1—source data 1
Numerical data used to generate graphs in Figure 3—figure supplement 1B–D.
- https://cdn.elifesciences.org/articles/78917/elife-78917-fig3-figsupp1-data1-v1.xlsx

Ribonucleotide monophosphate (rNMP) incorporation and camptothecin (CPT) sensitivity in cells lacking senataxin (SETX) and RNase H2.
(A) Representative image of alkaline gel of genomic DNA (n = 3 independent mice/genotype). (B) Densitometry trace of representative alkaline gel shown in (A) with ImageJ software. (C) Representative image of native gel of genomic DNA (n = 3). (D) Densitometry trace of representative native gel shown in (A) with ImageJ software. (E) Frequency of DNA damage in WT, Setx-/-, Rnaseh2bf/f, and Setx-/-Rnaseh2bf/f cells after exposure to 5 nM CPT for 20 hr. (F) Frequency of DNA damage at IgH in CPT-treated cells. All cells were harvested 72 hours post-stimulation to IgG1 with LPS/IL-4/α-RP105. Statistical significance versus WT was determined by one-way ANOVA (n = 3 independent mice/genotype).
-
Figure 3—figure supplement 2—source data 1
Uncropped alkaline gel image from WT, Rnaseh2bf/f, Setx-/-, and Setx-/-Rnaseh2bf/f cells.
Genomic DNA from WT, Rnaseh2bf/f, Setx-/-, and Setx-/-Rnaseh2bf/f B cells was treated with NaOH for 3 hr before running the alkaline gel.
- https://cdn.elifesciences.org/articles/78917/elife-78917-fig3-figsupp2-data1-v1.zip
-
Figure 3—figure supplement 2—source data 2
Uncropped native gel image of WT, Rnaseh2bf/f, Setx-/-, and Setx-/-Rnaseh2bf/f B cells.
Genomic DNA from WT, Rnaseh2bf/f, Setx-/-, and Setx-/-Rnaseh2bf/f B cells was treated with NaCl (left) or NaOH (right) for 3 hr before running the TAE native gel.
- https://cdn.elifesciences.org/articles/78917/elife-78917-fig3-figsupp2-data2-v1.zip
-
Figure 3—figure supplement 2—source data 3
Numerical data used to generate graphs in Figure 3—figure supplement 2E and F.
- https://cdn.elifesciences.org/articles/78917/elife-78917-fig3-figsupp2-data3-v1.xlsx

B cell proliferation and cell cycle in response to stimulation.
(A) Flow cytometry analysis of cell proliferation with CFSE staining. CFSE-labeled primary B cells were stimulated with LPS/IL-4/α-RP105, and cell division was measured by CFSE dye dilution at 72 hr post-stimulation. (B) Representative flow cytometry analyses of IgG1 staining with CFSE staining in spleen primary B cells in response to LPS/IL-4/α-RP105. (C) The percentage of cells in each cell cycle. Error bars show the SD, and statistical analyses were performed using one-way ANOVA (n = 3 mice per genotype). (D) Representative cell cycle profiles for WT, Setx-/-, Rnaseh2bf/f, and Setx-/-Rnaseh2bf/f cells stimulated with LPS/IL-4/α-RP105.
-
Figure 3—figure supplement 3—source data 1
Numerical data used to generate graph in Figure 3—figure supplement 3C.
- https://cdn.elifesciences.org/articles/78917/elife-78917-fig3-figsupp3-data1-v1.xlsx
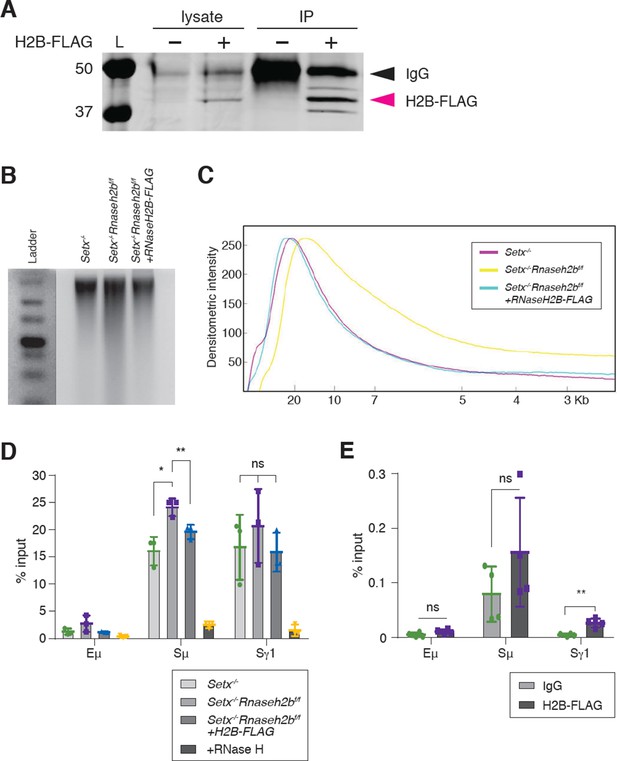
RNase H2 activity rescues DNA:RNA hybrid levels in stimulated B cells.
(A) Total cell lysates were extracted from Setx-/-Rnaseh2bf/f cells stimulated for 96 hr with LPS/IL-4/α-RP105. Cells were infected with empty vector or retrovirus, expressing FLAG-RNaseH2B, then subjected to immunoprecipitation and immunoblotting with indicated antibodies. (B) Representative image of alkaline gel from Setx-/-, Setx-/-Rnaseh2bf/f EV, and Setx-/-Rnaseh2bf/f+FLAG-RNaseH2B cells (n = 3 independent mice/genotype). (C) Densitometry trace of representative alkaline gel in (B). (D) DNA:RNA hybrid immunoprecipitation (DRIP) assay was performed with S9.6 antibody on Setx-/-, Setx-/-Rnaseh2bf/f, and Setx-/-Rnaseh2bf/f+FLAG-RNaseH2B-expressing cells stimulated with LPS/IL-4/α-RP105 for 96 hr. RNase H treatment of Setx-/-Rnaseh2bf/f sample was a negative control. Relative enrichment was calculated as chromatin immunoprecipitation (ChIP)/input, and the results were replicated in three independent experiments. Error bars show standard deviation; statistical analysis was performed using one-way ANOVA. (E) ChIP analysis for FLAG-RNaseH2B occupancy in Eμ, Sμ, and Sγ regions of primary B cells in response to LPS/IL-4/α-RP105 stimulation. Relative enrichment was calculated as ChIP/input. Error bars show standard deviation. Statistical analysis was performed using Student’s t-test (n = 3 mice/genotype).
-
Figure 4—source data 1
Uncropped western blot for RNaseH2B-FLAG expression in B cells under retroviral infection.
Immunoprecipitation and immunoblotting test for RNaseH2B-FLAG protein expression under retroviral infection in Setx-/-Rnaseh2bf/f B cells.
- https://cdn.elifesciences.org/articles/78917/elife-78917-fig4-data1-v1.zip
-
Figure 4—source data 2
Uncropped alkaline gel from retrovirally infected cells.
Uncropped image of alkaline gel from Setx-/-, Setx-/-Rnaseh2bf/f EV, and Setx-/-Rnaseh2bf/f+FLAG-RNaseH2B cells; left and right represent two times results.
- https://cdn.elifesciences.org/articles/78917/elife-78917-fig4-data2-v1.zip
-
Figure 4—source data 3
Numerical data used to generate graphs in Figure 4D and E.
- https://cdn.elifesciences.org/articles/78917/elife-78917-fig4-data3-v1.xlsx

Independent repeats of FLAG-RNaseH2B chromatin immunoprecipitation (ChIP).
Separate graphs for the four independent repeats of the FLAG-RNaseH2B ChIP shown in in Figure 4E.
-
Figure 4—figure supplement 1—source data 1
Numerical data used to generate graphs in Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/78917/elife-78917-fig4-figsupp1-data1-v1.xlsx
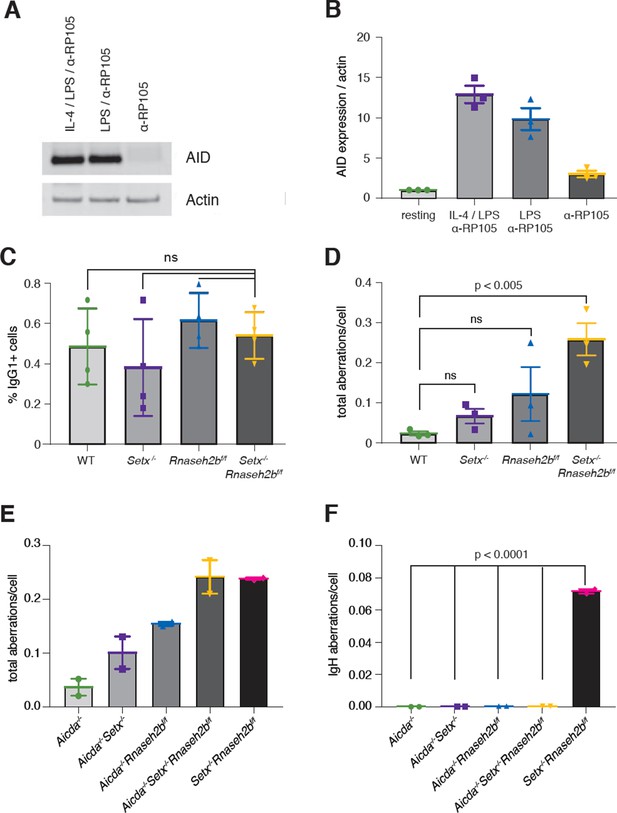
Activation-induced cytidine deaminase (AID) activity is required for persistent IgH breaks in Setx-/-Rnaseh2bf/f B cells.
(A) AID protein levels in WT B cells 72 hr post-stimulation to indicated isotypes (IgG1, LPS/IL-4/α-RP105; IgG3 with LPS/α-RP105; and α-RP105 alone). Actin served as a loading control. (B) Quantification of AID protein expression relative to Actin for three independent experiments, with AID expression in resting cells set as 1. Error bars show standard deviation. (C) Percent of cells undergoing class switch recombination (CSR) to IgG1 in B cells in response to α-RP105 stimulation. Error bars show standard deviation; statistical significance between each genotype was determined by one-way ANOVA (n = 3 mice/genotype). (D) Frequency of total spontaneous DNA damage under anti-RP105 treatment in vitro. Error bars show the standard deviation; statistical significance versus WT was determined by one-way ANOVA (n = 3 mice/genotype). (E) Frequency of total spontaneous DNA damage 72 hr post-stimulation with LPS/IL-4/α-RP105 in Aicda-/-, Aicda-/-Setx-/-, Aicda-/-Rnaseh2bf/f, Aicda-/-Setx-/-Rnaseh2bf/f, and Setx-/-Rnaseh2bf/f cells (n = 2 independent mice/genotype). (F) Frequency of spontaneous IgH damage in Aicda-/-, Aicda-/-Setx-/-, Aicda-/-Rnaseh2bf/f, Aicda-/-Setx-/-Rnaseh2bf/f, and Setx-/-Rnaseh2bf/f cells 72 hr after stimulation with LPS/IL-4/α-RP105. Error bars show the standard deviation; statistical significance versus WT was determined by one-way ANOVA.
-
Figure 5—source data 1
Uncropped Western blots of activation-induced cytidine deaminase (AID) and actin protein expression.
AID protein expression in WT B cells 72 hr post-stimulation with different reagents. Actin served as a loading control.
- https://cdn.elifesciences.org/articles/78917/elife-78917-fig5-data1-v1.zip
-
Figure 5—source data 2
Numerical data used to generate graphs in Figure 5B–F.
- https://cdn.elifesciences.org/articles/78917/elife-78917-fig5-data2-v1.xlsx

Aicda expression, enrichment, and recruitment in cells lacking senataxin (SETX) and RNase H2.
(A) Activation-induced cytidine deaminase (AID) mRNA quantitation by RT-qPCR in resting cells, LPS/IL-4/α-RP105, LPS/α-RP105, and α-RP105-only stimulated cells. Error bars show the standard deviation from three independent experiments. (B) AID protein expression in WT, Setx-/-, Rnaseh2bf/f, Setx-/-Rnaseh2bf/f, and Aicda-/- cells 72 hr post-stimulation to IgG1. Western blots were probed with an anti-AID antibody and anti-actin as loading control. Aicda-/- cells were used as a negative control. (C) Quantification of AID protein expression versus WT related to B; error bars show standard deviation; statistical analysis was performed using multiple t-test (n = 3 mice/genotype). (D) Chromatin immunoprecipitation (ChIP) analysis for AID occupancy in Sμ and Sγ regions of primary B cells in response to LPS/IL-4/α-RP105 stimulation at 60 hr post-stimulation. Relative enrichment was calculated as ChIP/input. Error bars show standard deviation (n = 3 mice/genotype); statistical analysis versus Aicda-/- control was performed using Student’s t-test, *p-value<0.05 compared to Aicda-/-. (E) ChIP analysis for Pol ll (Ser5) occupancy in Sμ and Sγ regions of primary B cells in response to LPS/IL-4/α-RP105 stimulation. Relative enrichment was calculated as fold change relative to WT set to 1; error bars show standard deviation (n = 5 mice/genotype).
-
Figure 5—figure supplement 1—source data 1
Activation-induced cytidine deaminase (AID) expression test.
Whole-cell lysis from WT, Setx-/-, Rnaseh2bf/f, Setx-/-Rnaseh2bf/f, and Aicda-/- cells 72 hr post-stimulation to IgG1 were run on SDS-PAGE. Western blots were probed with an anti-AID antibody and anti-actin as loading control. Aicda-/- cells were used as a negative control.
- https://cdn.elifesciences.org/articles/78917/elife-78917-fig5-figsupp1-data1-v1.zip
-
Figure 5—figure supplement 1—source data 2
Numerical data used to generate graphs in Figure 5—figure supplement 1A, C–E.
- https://cdn.elifesciences.org/articles/78917/elife-78917-fig5-figsupp1-data2-v1.xlsx
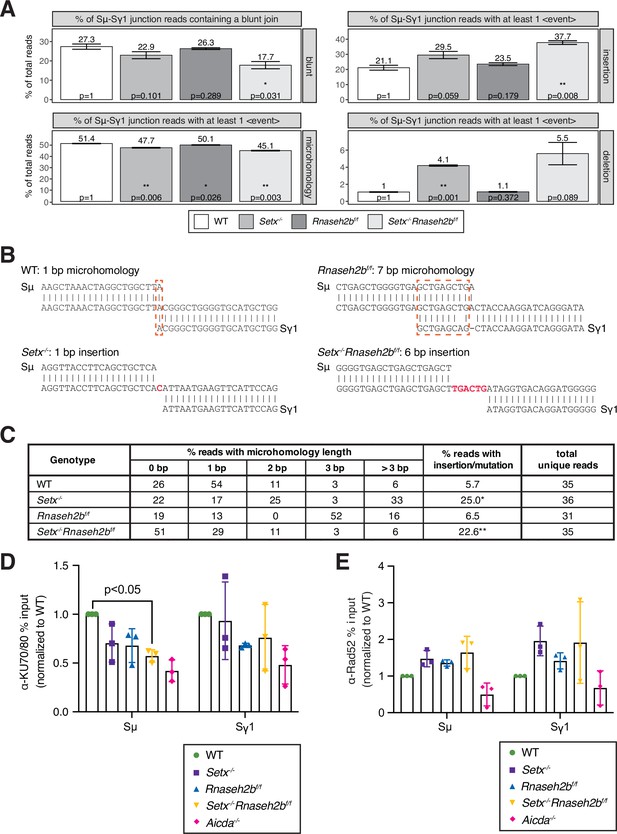
Altered switch junctions in in Setx-/-, Rnaseh2bf/f, and Setx-/-Rnaseh2bf/f B cells.
(A) Linear amplification-mediated high-throughput genome-wide translocation sequencing (LAM-HTGTS) analysis showing the percentage of sequenced junction events harboring blunt joins, microhomology (MH) use, insertions, or deletions at junction sites. LAM-HTGTS uses two technical replicates from genomic DNA isolated from cells 72 hr after LPS/IL-4/α-RP105 stimulation. p-Values are calculated using Student’s t-test. Sequencing reads may harbor more than one event class (insertion, deletion, mutation, MH) with the resulting junctions having a more complex result; a junction exhibiting MH may also have a deletion or mismatches in flanking DNA. Thus, junction types were quantified on whether they contained at least one event of the class listed in the side bar (deletions, insertions, MH). (B) Representative nucleotide sequences surrounding representative Sμ-Sγ junctions from WT, Setx-/-, Rnaseh2bf/f, and Setx-/-Rnaseh2bf/f B cells from Sanger sequencing of cloned junctions. Overlap was determined by identifying the longest region at the switch junction of perfect uninterrupted donor/acceptor identity. Sμ and Sγ1 germline sequences are shown above and below each junction sequence, respectively. Regions of MH at junctions are boxed with a dashed red line, and insertions are in red bold text. Genomic DNA from sequencing experiments was isolated from two independent mice for each genotype. (C) Table with absolute numbers of uniquely mapping cloned switch junctions harboring MH and insertions in WT, Setx-/-, Rnaseh2bf/f, and Setx-/-Rnaseh2bf/f B cells 72 hr after stimulation with LPS/IL-4/α-RP105. p-Values were calculated using the chi-square goodness-of-fit test: *WT vs. Setx-/- 6.19 × 10–7, ** WT vs. Setx-/-Rnaseh2bf/f: 1.25 × 10–5. (D) Chromatin immunoprecipitation (ChIP) analysis for KU70/KU80 occupancy in Sμ and Sγ regions of primary B cells in response to LPS/IL-4/α-RP105 stimulation. Relative enrichment was calculated as fold change relative to WT set to 1; error bars show standard deviation (n = 3 mice/genotype), statistical analysis versus WT was performed using Student’s t-test. (E) ChIP analysis for RAD52 occupancy in Sμ and Sγ regions of primary B cells in response to LPS/IL-4/α-RP105 stimulation. Relative enrichment was calculated as fold change relative to WT set to 1; error bars show standard deviation (n = 3 mice/genotype).
-
Figure 6—source data 1
Numerical data used to generate graphs in Figure 6D and E.
- https://cdn.elifesciences.org/articles/78917/elife-78917-fig6-data1-v1.xlsx

Features of Sμ-Sγ1 junction sites.
(A) Average number of reads from two runs of linear amplification-mediated high-throughput genome-wide translocation sequencing (LAM-HTGTS) spanning IgM-IgG1 junctions from WT, Setx-/-, Rnaseh2bf/f, and Setx-/-Rnaseh2bf/f cells 72 hr post-stimulation with LPs/IL4/α-RP105. (B) Average read lengths for analyzed junctions spanning IgM-IgG1. (C) Percentage of reads with different sizes of deletions, insertions, or microhomology events at junction sites from WT, Setx-/-, Rnaseh2bf/f, and Setx-/-Rnaseh2bf/f cells. with 1 bp, 2 bp, 3 bp, and 4 or more bp of each event type are shown. (D) Frequency of mutations for each genotype within IgM-IgG1 junction reads. Graphs from top to bottom show the total mutations per kilobase (kb), the number of C to T transition mutations per kb, and the number of G to A transition mutations per kb. p-Values are calculated using Student’s t-test. Sequencing reads may harbor more than one event class (insertion, deletion, mutation, microhomology [MH]) with the resulting junctions having a more complex result; a junction exhibiting MH may also have a deletion or mismatches in flanking DNA. Thus, junction types were quantified on whether they contained at least one event of the class listed in the header (deletions, insertions, MH).
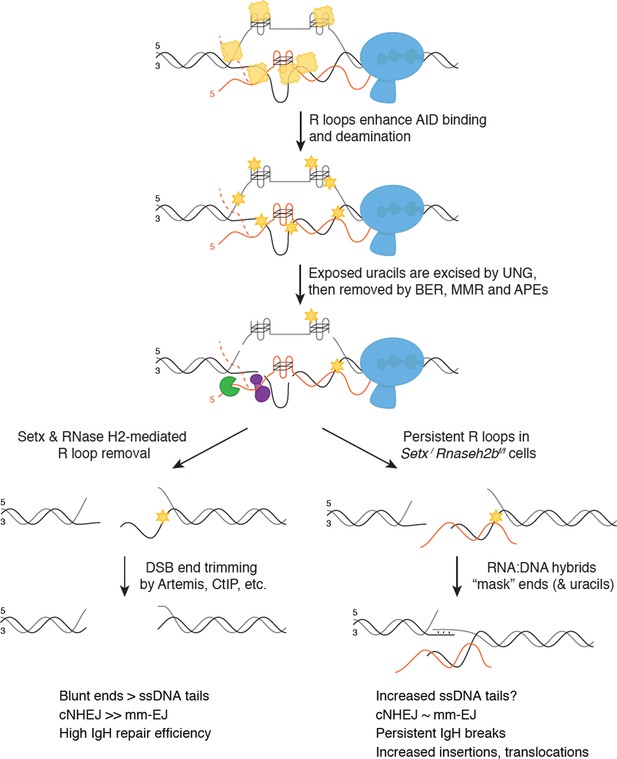
Model for senataxin (SETX) and RNase H2 in promoting efficient class switch recombination (CSR).
When B cells are stimulated to undergo class switching, PolII-mediated transcription opens duplex DNA at recombining S regions. R loops formed during transcription promote activation-induced cytidine deaminase (AID) binding to ssDNA on the non-template strand first. SETX and RNase H2 then cooperate to remove switch region R loops, exposing ssDNA on the template strand for AID to bind. Extensive AID activity and uracil removal on both strands results in the formation of double-stranded breaks (DSBs) with limited single-strand DNA (ssDNA) overhangs at break ends (‘blunted’ ends), which are predominantly repaired by classical non-homologous end joining (cNHEJ). When SETX and RNase H2B are absent, R loops forming at S region are not efficiently removed and error-prone EJ is increased. This persistent R loop/RNA:DNA hybrid may affect CSR in two ways. One possible mechanism for increased error-prone EJ is that persistent R loops reduce the extent of ssDNA available for AID binding specifically on the template strand, increasing ssDNA tail length at DSB ends. Alternatively, persistent RNA:DNA hybrids may alter DNA repair protein recruitment to DSB ends, impeding end processing and/or ligation. Both possibilities reduce NHEJ efficiency, but do not affect overall CSR levels as the majority of breaks with long ssDNA tails are repaired by error-prone EJ. However, a subset of breaks are not repaired, leading to persistent DSBs that manifest as chromosome breaks and translocations in mitotic spreads.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus musculus) | Rnaseh2bf/f | PMID:2802351 | RRID:MGI:5911393 | Dr. Axel Roers (University of Technology Dresden) |
| Genetic reagent (M. musculus) | Setx-/- | PMID:23593030 | RRID:MGI:5697060 | Dr. Martin F Lavin (Queensland Institute of Medical Research) |
| Genetic reagent (M. musculus) | Cd19cre | PMID:9092650 | RRID:MGI:5614310 | Dr. Klaus Rajewsky (University of Cologne) |
| Genetic reagent (M. musculus) | Aicda-/- | PMID:11007474 | RRID:MGI:2156156 | |
| Recombinant DNA reagent | Migr1-RNaseH2B-FLAG | This paper | N-terminally FLAG tagged mouse Rnaseh2b | |
| Antibody | Anti-S9.6 (mouse monoclonal) | Chedin lab | DRIP (1:500) Dot blot (1:1000) | |
| Antibody | IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 680 (goat anti-mouse polyclonal) | Thermo Fisher | Cat# A21057 | Dot blot (1:5000) |
| Antibody | APC CD19 (rat anti-mouse monoclonal) | BD Pharmingen | Cat# 561738 | Flow cytometry (1:100) |
| Antibody | PE-Cy7-CD43 (rat anti-mouse monoclonal) | BD Pharmingen | Cat# 562866 | Flow cytometry (1:20) |
| Antibody | PE -IgM (rat anti-mouse monoclonal) | BD Pharmingen | Cat# 562033 | Flow cytometry (1:100) |
| Antibody | BV421-CD11b (rat anti-mouse monoclonal) | BD Pharmingen | Cat# 562605 | Flow cytometry (1:100) |
| Antibody | PerCP- CD45R/B220 (rat anti-mouse monoclonal) | BD Pharmingen | Cat# 553093 | Flow cytometry (1:100) |
| Antibody | Anti-AID (mouse monoclonal) | Thermo Fisher | Cat# 39-2500 | CHIP (1:200) WB (1:500) |
| Antibody | Anti-KU70/KU80 (mouse monoclonal) | Invitrogen | Cat# MA1-21818 | CHIP (1:100) |
| Antibody | Anti-Rad52 (rabbit polyclonal) | ABclonal | Cat# A3077 | CHIP (1:500) |
| Antibody | Anti-RNA polymerase II (mouse monoclonal) | Abcam | Cat# AB5408 | CHIP (1:500) |
| Antibody | IgG- Isotype Control (mouse polyclonal) | Abcam | Cat# AB37355 | CHIP (1:500) |
| Antibody | Anti-CD180 (rat anti-mouse monoclonal) | BD Pharmingen | Cat# 552128 | CSR (1:2000) |
| Peptide, recombinant protein | Recombinant murine IL-4 | PeproTech | Cat# 214-14 | CSR (1:2000) |
| Peptide, recombinant protein | Lipopolysaccharides | MilliporeSigma | Cat# L2630 | CSR (1:2000) |
| Commercial assay or kit | Beckman Coulter AMPURE XP | Beckman Coulter | Cat# A63881 | Size selection |
| Commercial assay or kit | SPHERO AccuCount Blank Particles | Spherotech | Cat# ACBP-50-10 | Flow cytometry |
Additional files
-
Supplementary file 1
Summary of individual fluorescent in situ hybridization (FISH) results for experiments shown in Figures 3 and 5.
- https://cdn.elifesciences.org/articles/78917/elife-78917-supp1-v1.xlsx
-
Supplementary file 2
Primers used in qPCR and junction analysis.
Source data files
- https://cdn.elifesciences.org/articles/78917/elife-78917-supp2-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/78917/elife-78917-mdarchecklist1-v1.pdf





