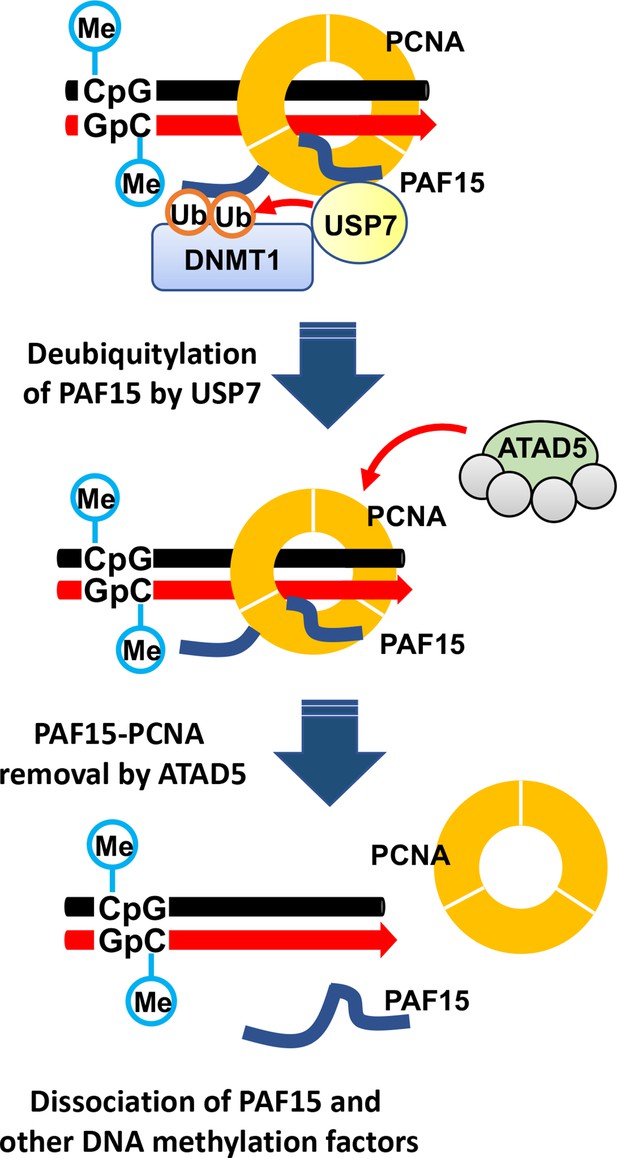
The termination of UHRF1-dependent PAF15 ubiquitin signaling is regulated by USP7 and ATAD5
Figures
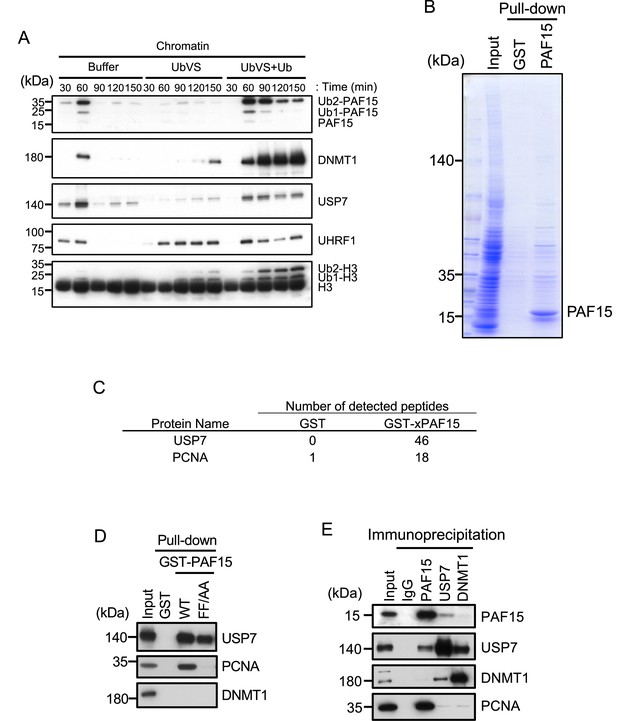
USP7 was identified as a PAF15 binding protein.
(A) Sperm chromatin was added to interphase egg extracts supplemented with either buffer (+Buffer), 20 µM UbVS (+UbVS), or 20 µM UbVS and 58 µM ubiquitin (+UbVS + Ub). Samples were analyzed by immunoblotting using the antibodies indicated. (B) Proteins pull-downed from interphase egg extracts by GST and GST-PAF15 were stained by Coomassie Brilliant Blue. (C) The samples from GST-PAF15 pull down were analyzed by nanoflow liquid chromatography-tandem mass spectrometry (nanoLC-MS/MS). Selected proteins were indicated in the table. (D) GST pull-down assay was performed by GST or GST-PAF15 wild-type (WT) or PIP mutant (FF/AA), and the samples were analyzed by immunoblotting using the antibodies indicated. (E) Immunoprecipitation was performed by PAF15, USP7, and DNMT1 antibodies-bound beads, and the samples were analyzed by immunoblotting using the antibodies indicated. Source data are provided as Figure 1—source data 1.
-
Figure 1—source data 1
Figure 1 Original blots.
- https://cdn.elifesciences.org/articles/79013/elife-79013-fig1-data1-v2.zip
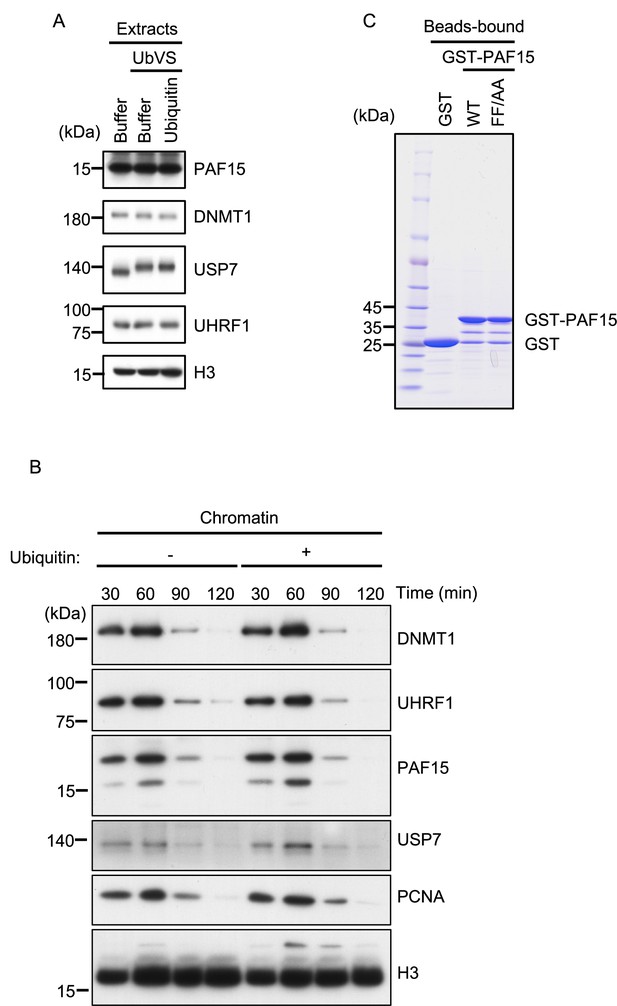
USP7 was identified as a PAF15 binding protein.
(A) Interphase egg extracts supplemented with UbVS or UbVS+Ub used in Figure 1A were analyzed by immunoblotting using the antibodies indicated. (B) Purified GST or GST-rPAF15 mutants used in Figure 1D were stained using CBB. (C) Sperm chromatin was added to interphase egg extracts supplemented with either buffer (−) and 58 µM ubiquitin (+). Samples were analyzed by immunoblotting using the antibodies indicated. Source data are provided as Figure 1—figure supplement 1—source data 1.
-
Figure 1—figure supplement 1—source data 1
Figure 1—figure supplement 1 Original blots.
- https://cdn.elifesciences.org/articles/79013/elife-79013-fig1-figsupp1-data1-v2.zip
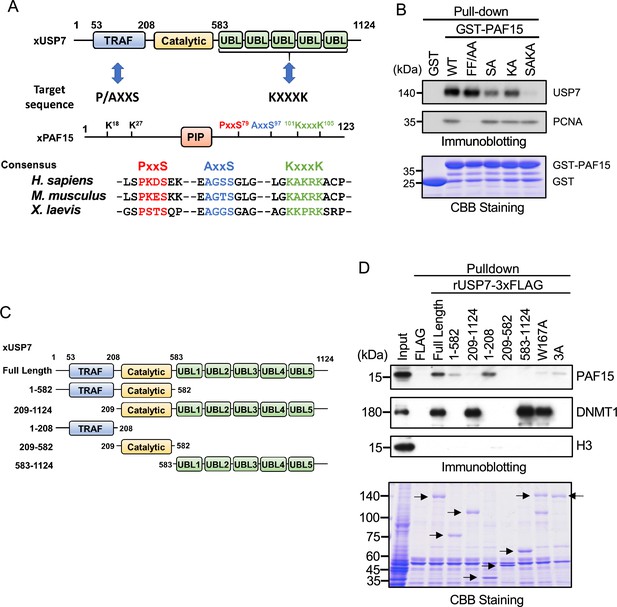
PAF15 associates with the TRAF and UBL1-2 domains of USP7.
(A) Schematic illustration of PAF15-USP7 binding experiment. USP7 recognizes P/A/ExxS or KxxxK motifs in its substrates via TRAF or ubiquitin-like (UBL) domains, respectively. PAF15 has three motifs, and alanine mutations were introduced at S79, S97, K101, and K105. (B) GST pull-down from the interphase egg extracts using GST-PAF15, P/AxxS mutant (S79A/S97A; SA), KxxxK mutant (K101A/K105A; KA), and triple mutant (SAKA). The samples were analyzed by immunoblotting using the antibodies indicated. Purified GST or GST-PAF15 mutants used in pull-down assay were stained using CBB. (C) Schematic illustration of rUSP7 truncation mutants employed in (D). (D) FLAG pull-down from interphase egg extracts using rUSP7-3xFLAG mutants presented in (C), W167A and 3 A point mutants. USP7 3 A: D780A/E781A/D786A. The samples were analyzed by immunoblotting using the antibodies indicated. Samples were also stained by CBB. Arrowheads indicate rUSP7 truncation mutants and point mutants. Source data are provided as Figure 2—source data 1.
-
Figure 2—source data 1
Figure 2 Original blots.
- https://cdn.elifesciences.org/articles/79013/elife-79013-fig2-data1-v2.zip
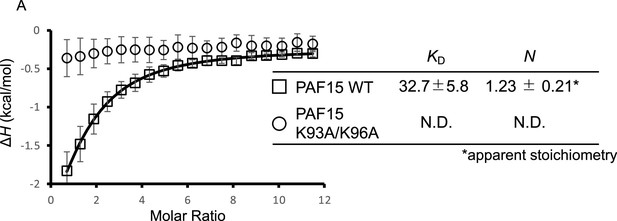
hUSP7561-1102 interacts with hPAF15 dependently on KxxxK motif.
(A) Superimposition of plots of enthalpy changes in the interaction between hUSP7561-1102 and full-length hPAF15 wild-type (WT) or K93A/K96A mutant by isothermal titration calorimetry (ITC) measurement. Data are presented as mean ± SEM from three biological replicates.
-
Figure 2—figure supplement 1—source data 1
- https://cdn.elifesciences.org/articles/79013/elife-79013-fig2-figsupp1-data1-v2.zip
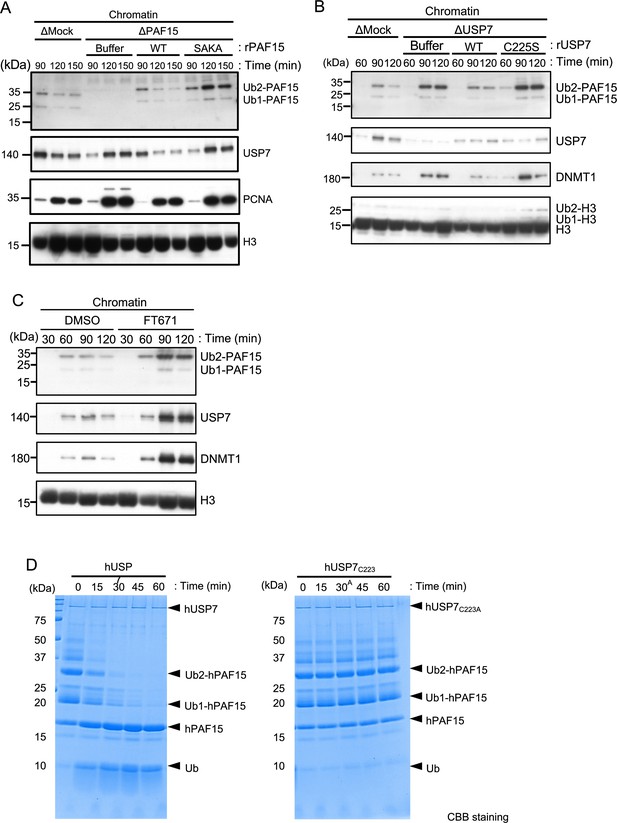
USP7 promotes PAF15 dissociation from chromatin.
(A) Sperm chromatin was added to Mock- or PAF15-depleted interphase extracts supplemented with either buffer (+Buffer), wild-type rPAF15-3xFLAG (+WT) or rPAF15 SAKA-3xFLAG (+SAKA). Chromatin fractions were isolated, and the samples were analyzed by immunoblotting using the antibodies indicated. (B) Sperm chromatin was added to Mock- or USP7-depleted interphase extracts supplemented with either buffer (+Buffer), wild-type rUSP7-3xFLAG (+WT) or catalytic mutant rUSP7 C225S-3xFLAG (+C225 S). Chromatin fractions were isolated, and the samples were analyzed by immunoblotting using the antibodies indicated. (C) Sperm chromatin was added to interphase extracts supplemented with either dimethyl sulfoxide (DMSO) (+DMSO) or FT671 (+FT671). Chromatin fractions were isolated, and the samples were analyzed by immunoblotting using the antibodies indicated. (D) Ubiquitylated hPAF15 was incubated with recombinant hUSP7 WT (left) or C223A catalytic mutant (right) at indicated times. The reaction products were analyzed by SDS-PAGE with CBB staining. Source data are provided as Figure 3—source data 1.
-
Figure 3—source data 1
Figure 3 Original blots.
- https://cdn.elifesciences.org/articles/79013/elife-79013-fig3-data1-v2.zip
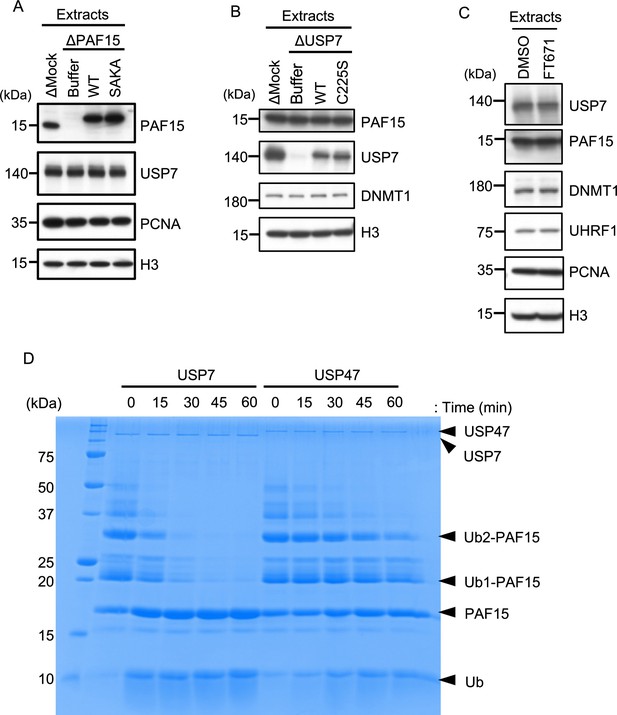
USP7 directly deubiquitylates PAF15 to promote PAF15 chromatin dissociation.
(A) rPAF15 WT-3xFLAG or SAKA-3xFLAG were added to PAF15-depleted extracts. The extracts used in Figure 3A were analyzed by immunoblotting using the antibodies indicated. (B) rUSP7 WT-3xFLAG or C225S-3xFLAG were added to USP7-depleted extracts. The extracts used in Figure 3B were analyzed by immunoblotting using the antibodies indicated. (C) Interphase egg extracts supplemented with dimethyl sulfoxide (DMSO) or the USP7 inhibitor FT671 used in Figure 3C were analyzed by immunoblotting using the antibodies indicated. (D) Ubiquitinated PAF15 was prepared by in vitro ubiquitylation using E1 (mouse UBA1), E2 (UBE2D3), E3 (UHRF1), ubiquitin, and C-teminal FLAG tagged PAF15. The reaction mixture was boiled, and the precipitant was removed by centrifugation. The supernatant containing ubiquitinated PAF15 was incubated with 50 nM recombinant USP7 or USP47. Incubations were stopped at the indicated time by adding SDS-PAGE loading buffer and analyzed by CBB staining. Source data are provided as Figure 3—figure supplement 1—source data 1.
-
Figure 3—figure supplement 1—source data 1
Figure 3—figure supplement 1 Original blots.
- https://cdn.elifesciences.org/articles/79013/elife-79013-fig3-figsupp1-data1-v2.zip
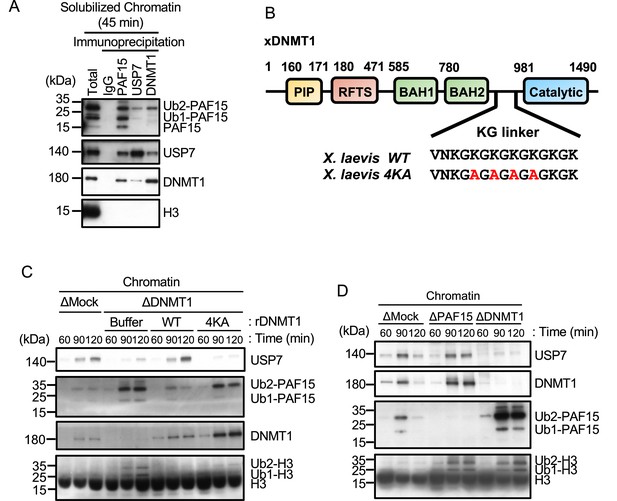
USP7 is recruited to chromatin through the interaction with DNMT1 for PAF15 deubiquitylation.
(A) Sperm chromatin was added to interphase extracts. Replicating chromatin was digested by micrococcal nuclease (MNase). Immunoprecipitation was performed by PAF15, USP7, and DNMT1 antibodies from the solubilized chromatin fraction, and the samples were analyzed by immunoblotting using the antibodies indicated. (B) Illustration of DNMT1 domain structure. The KG linker located between the bromo-adjacent homology (BAH) domain and catalytic domain contributes to interaction with USP7. rDNMT1 4KA mutant, using in (C), was introduced mutation at four lysines to alanine in the KG linker. (C) Sperm chromatin was added to Mock- or DNMT1-depleted interphase extracts supplemented with either buffer (+Buffer), wild-type rDNMT1-3xFLAG (+WT), or rDNMT1 4KA-3xFLAG (+4 KA). Chromatin fractions were isolated, and the samples were analyzed by immunoblotting using the antibodies indicated. (D) Sperm chromatin was added to Mock-, PAF15-, and DNMT1-depleted extracts. Chromatin fractions were isolated, and the samples were analyzed by immunoblotting using the antibodies indicated. Source data are provided as Figure 4—source data 1.
-
Figure 4—source data 1
Figure 4 Original blots.
- https://cdn.elifesciences.org/articles/79013/elife-79013-fig4-data1-v2.zip
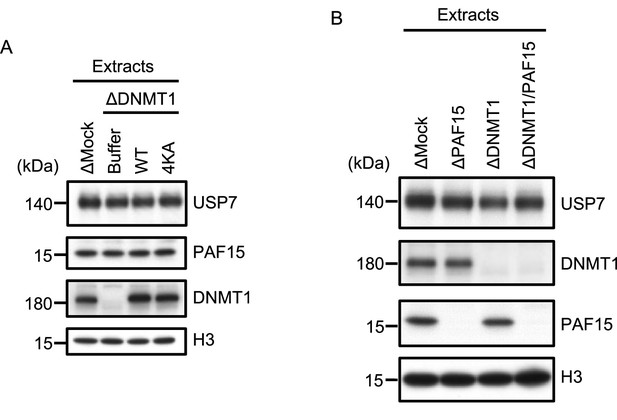
USP7 is recruited to chromatin through the interaction with DNMT1.
(A) rDNMT1WT-3xFLAG or 4KA-3xFLAG were added to DNMT1-depleted extracts. The extracts used in Figure 4C were analyzed by immunoblotting using the antibodies indicated. (B) Mock-, PAF15-, DNMT1-, or PAF15/DNMT1 co-depleted extracts used in Figure 4D were analyzed by immunoblotting using the antibodies indicated. Source data are provided as Figure 4—figure supplement 1—source data 1.
-
Figure 4—figure supplement 1—source data 1
Figure 4—figure supplement 1 Original blots.
- https://cdn.elifesciences.org/articles/79013/elife-79013-fig4-figsupp1-data1-v2.zip
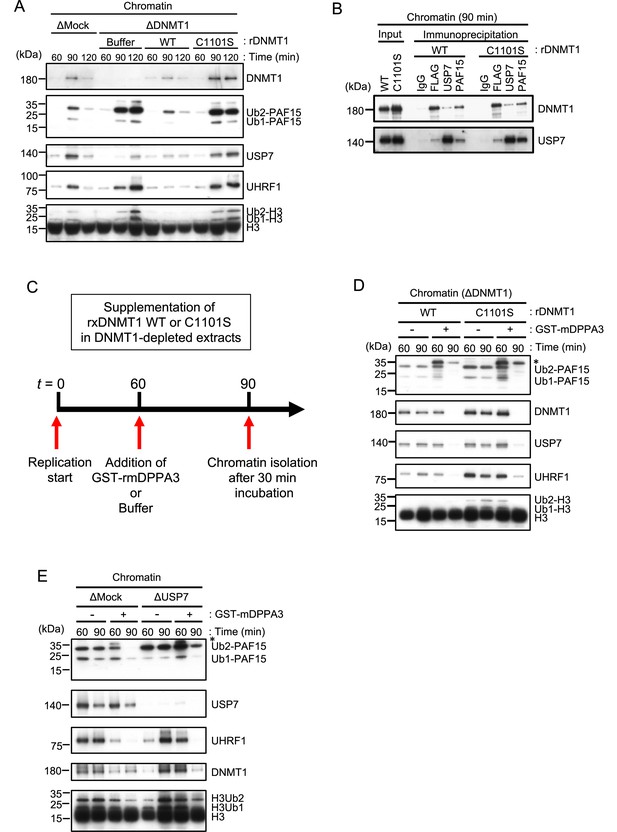
Unloading of PAF15 requires DNMT1-dependent DNA methylation.
(A) Sperm chromatin was added to Mock- or DNMT1-depleted interphase extracts supplemented with either buffer (+Buffer), wild-type rDNMT1-3xFLAG (+WT), or catalytic mutant rDNMT1 C1101S-3xFLAG (+C1101 S). Chromatin fractions were isolated, and the samples were analyzed by immunoblotting using the antibodies indicated. (B) Sperm chromatin was added to DNMT1-depleted interphase extracts supplemented wild-type rDNMT1-3xFLAG (+WT) or catalytic mutant rDNMT1 C1101S-3xFLAG (+C1101 S). Replicating chromatin was digested by micrococcal nuclease (MNase). Immunoprecipitation was performed by PAF15, USP7 antibodies-bound beads, and FLAG affinity beads in the solubilized chromatin fraction solution, and the samples were analyzed by immunoblotting using the antibodies indicated. (C) A schema of an experiment described in D. (D) Sperm chromatin was added to DNMT1-depleted interphase extracts supplemented wild-type rDNMT1-3xFLAG (+WT) or catalytic mutant rDNMT1 C1101S-3xFLAG (+C1101 S). After 60 min, the extracts were supplemented with either buffer (−) or GST-mDPPA3 61–150 (+). Chromatin fractions were isolated, and the samples were analyzed by immunoblotting using the antibodies indicated. The asterisk indicates a non-specific band. (E) Sperm chromatin was added to USP7-depleted interphase extracts. After 90 min, the extracts were supplemented with either buffer (−) or GST-mDPPA3 61–150 (+). Chromatin fractions were isolated, and the samples were analyzed by immunoblotting using the antibodies indicated. The asterisk indicates a non-specific band. Source data are provided as Figure 5—source data 1.
-
Figure 5—source data 1
Figure 5 Orignal blots.
- https://cdn.elifesciences.org/articles/79013/elife-79013-fig5-data1-v2.zip
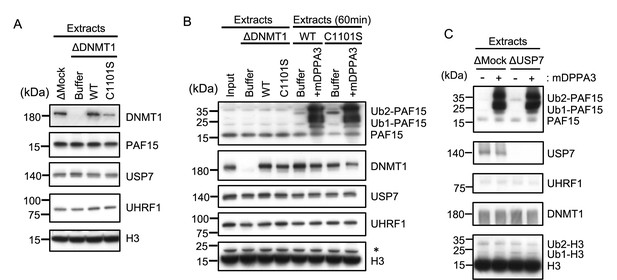
Unloading of PAF15 requires DNMT1-dependent DNA methylation.
(A) rDNMT1 WT-3xFLAG or C1101S-3xFLAG were added to DNMT1-depleted extracts. The extracts used in Figure 5A were analyzed by immunoblotting using the antibodies indicated. (B) rDNMT1 WT-3xFLAG or C1101S-3xFLAG were added to DNMT1-depleted extracts used in Figure 5D. After 60 min, these extracts were supplemented with GST-mDPPA3. The extracts were analyzed by immunoblotting using the antibodies indicated. The asterisk indicates a non-specific band. (C) Replicating Mock- or USP7-depleted extracts at 90 min were supplemented with either buffer (−) or GST-mDPPA3 (+). The extracts were used in Figure 5E and analyzed by immunoblotting using the antibodies indicated. Source data are provided as Figure 5—figure supplement 1—source data 1.
-
Figure 5—figure supplement 1—source data 1
Figure 5—figure supplement 1 Original blots.
- https://cdn.elifesciences.org/articles/79013/elife-79013-fig5-figsupp1-data1-v2.zip
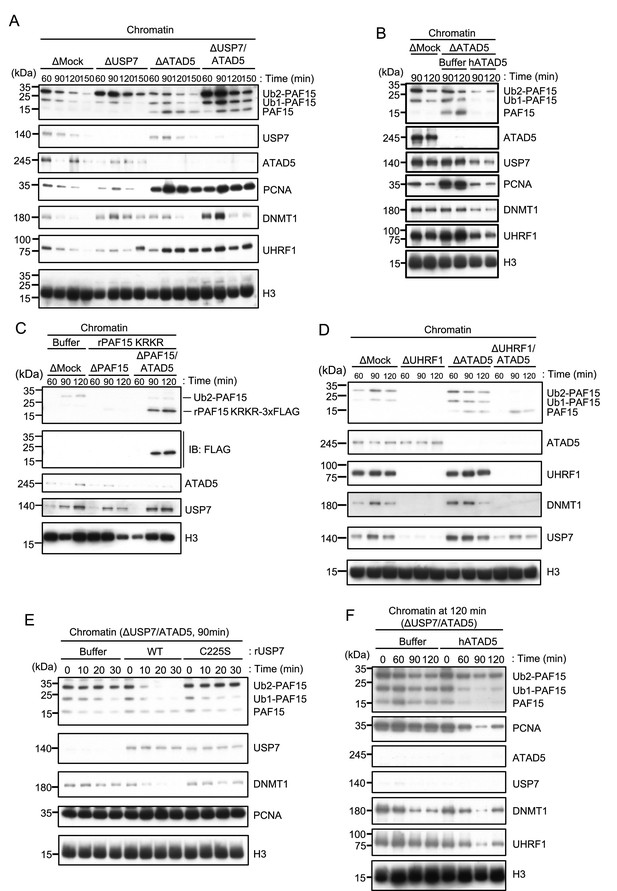
ATAD5 unloads PAF15Ub0 from chromatin.
(A) Sperm chromatin was added to Mock-, USP7-, ATAD5-, and USP7/ATAD5-depleted interphase extracts and isolated at indicated time points. Chromatin bound proteins were analyzed by immunoblotting. (B) Sperm chromatin was added to Mock-, ATAD5-depleted interphase extracts supplemented with either buffer (+Buffer) or recombinant hATAD5-RFCs (+ATAD5). Chromatin fractions were isolated, and the samples were analyzed by immunoblotting using the antibodies indicated. (C) Recombinant PAF15 K18R/K27R-3xFLAG was supplemented to PAF15- and PAF15/ATAD5-depleted extracts, and chromatin fractions were isolated. Chromatin bound proteins were confirmed by immunoblotting. (D) Sperm chromatin was added to Mock-, UHRF1-, ATAD5-, and UHRF1/ATAD5-depleted extracts and isolated at indicated time point. Chromatin bound proteins were analyzed by immunoblotting. (E) Sperm chromatin was added to USP7/ATAD5-depleted extracts and isolated at 90 min. The chromatin was supplemented with either buffer (+Buffer), USP7 WT-3xFLAG (+WT), or USP7 C225S-3xFLAG (+C225 S) and re-isolated at indicated time points. Chromatin bound proteins were analyzed by immunoblotting. (F) Sperm chromatin was added to Mock- and USP7/ATAD5-depleted extracts. After replication at 90 min, the extracts were supplemented with either buffer (+Buffer) or recombinant hATAD5-RFCs (+ATAD5). Chromatin fractions were isolated, and the samples were analyzed by immunoblotting using the antibodies indicated. Source data are provided as Figure 6—source data 1.
-
Figure 6—source data 1
Figure 6 Original blots.
- https://cdn.elifesciences.org/articles/79013/elife-79013-fig6-data1-v2.zip
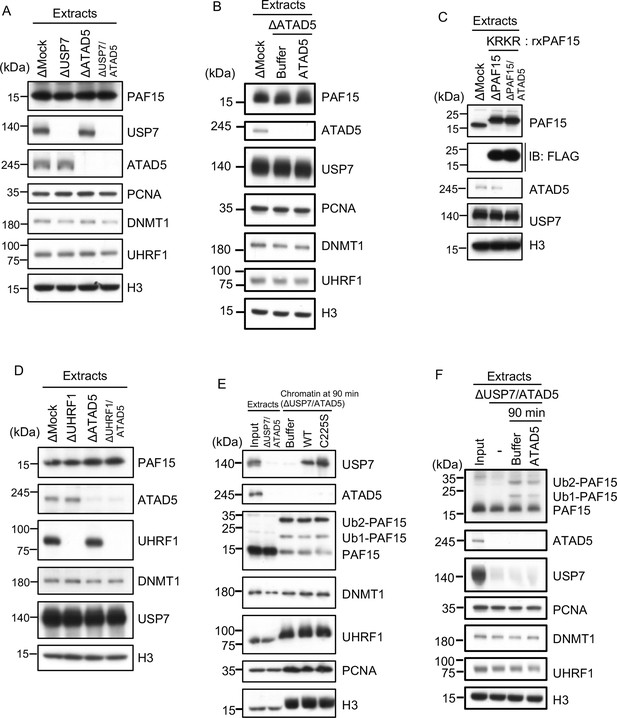
ATAD5 unloads PAF15Ub0 from chromatin.
(A) Mock-, USP7-, ATAD5-, or USP7/ATAD5 co-depleted extracts were used in Figure 6A and analyzed by immunoblotting using the antibodies indicated. (B) rhATAD5-RFCs was added to ATAD5-depleted extracts. The extracts were used in Figure 6B and analyzed by immunoblotting using the antibodies indicated. (C) rPAF15 KRKR-3xFLAG was added to PAF15- or PAF15/ATAD5 co-depleted extracts. The extracts were used in Figure 6C and analyzed by immunoblotting using the antibodies indicated. (D) Mock-, UHRF1-, ATAD5-, or UHRF1/ATAD5 co-depleted extracts were used in Figure 6D and analyzed by immunoblotting using the antibodies indicated. (E) rUSP7WT-3xFLAG or C225S-3xFLAG were added to USP7/ATAD5 co-depleted extracts. The extracts were used in Figure 6E and analyzed by immunoblotting using the antibodies indicated. (F) Replicating USP7/ATAD5 co-depleted extracts at 90 min were supplemented with either buffer or rhATAD5-RFCs. The extracts were used in Figure 6F and analyzed by immunoblotting using the antibodies indicated. Source data are provided as Figure 6—figure supplement 1—source data 1.
-
Figure 6—figure supplement 1—source data 1
Figure 6—figure supplement 1 Original blots.
- https://cdn.elifesciences.org/articles/79013/elife-79013-fig6-figsupp1-data1-v2.zip
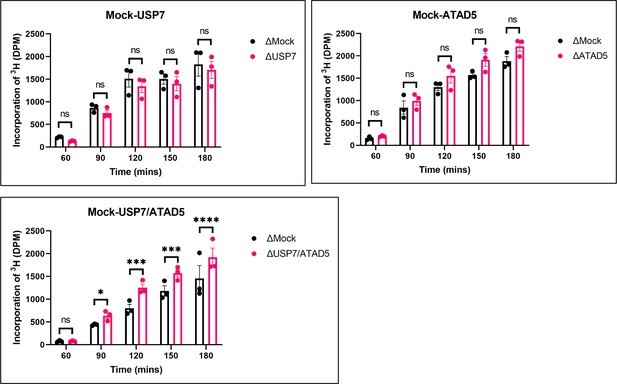
PAF15 dissociation negatively regulates aberrant increase of DNA methylation.
Sperm chromatin and radiolabeled S-(methyl-3H)-adenosyl-L-methionine were added to either Mock- and USP7-, ATAD5-, or USP7/ATAD5 co-depleted extracts. Purified DNA samples were analyzed to determine the efficiency of DNA methylation. Data are presented as mean ± SEM from three biological replicates. Multiple comparisons were performed by two-way repeated measure ANOVA (RM ANOVA) followed by Sidak’s multiple comparison test. ns; not significant, ∗p<0.05, ∗∗∗p<0.001, and ∗∗∗∗p<0.0001. Source data are provided as Figure 7—source data 1.
-
Figure 7—source data 1
Figure 7 DNA methylation.
- https://cdn.elifesciences.org/articles/79013/elife-79013-fig7-data1-v2.zip
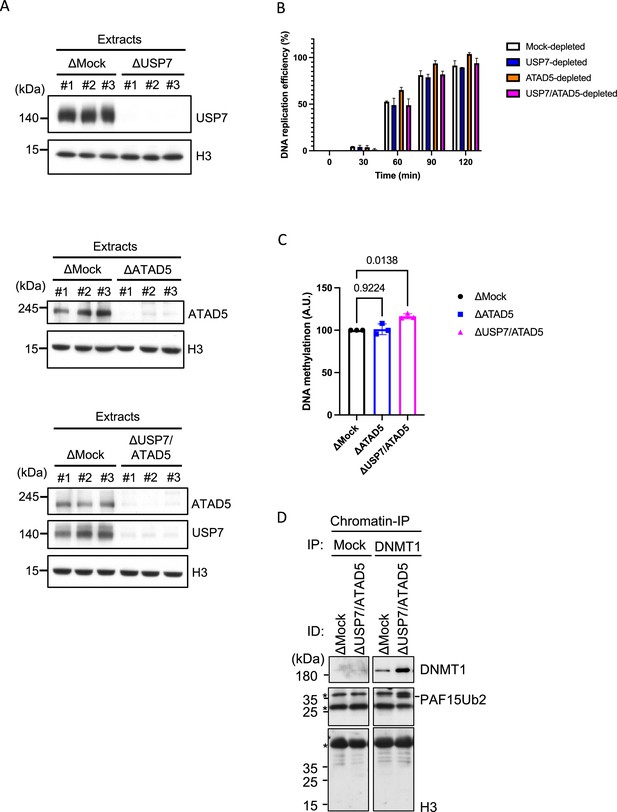
DNA replication and methylation analysis in USP7 or ATAD5-depleted extracts.
(A) Mock-, USP7-, ATAD5-, or USP7/ATAD5 co-depleted extracts used in Figure 7 were analyzed by immunoblotting using the antibodies indicated. (B) Sperm chromatin and radiolabeled (α-32P) dCTP were added to Mock-, USP7-, ATAD5-, or USP7/ATAD5 co-depleted extracts. Purified DNA samples were analyzed to determine the efficiency of DNA replication. Data are presented as mean ± SEM from three biological replicates. (C) Sperm chromatin and radiolabeled S-(methyl-3H)-adenosyl-L-methionine were added to either Mock-, ATAD5-, or USP7/ATAD5 co-depleted extracts. Purified DNA samples were analyzed to determine the efficiency of DNA methylation. Data are presented as mean ± SEM from three biological replicates. Multiple comparisons were performed by two-way repeated measure ANOVA (RM ANOVA) followed by Sidak’s multiple comparison test. DNA methylation (A.U) was calculated relative to the value of mock-depleted control reactions. (D) Sperm chromatin was added to Mock- or USP7/ATAD5-depleted extracts. Replicating chromatin was isolated and digested by micrococcal nuclease (MNase). Immunoprecipitation was performed by DNMT1 antibodies-bound beads, and the samples were analyzed by immunoblotting using the antibodies indicated. Source data are provided as Figure 7—figure supplement 1—source data 1.
-
Figure 7—figure supplement 1—source data 1
Figure 7—figure supplement 1 Original blots, DNA replication, and methylation data.
- https://cdn.elifesciences.org/articles/79013/elife-79013-fig7-figsupp1-data1-v2.zip
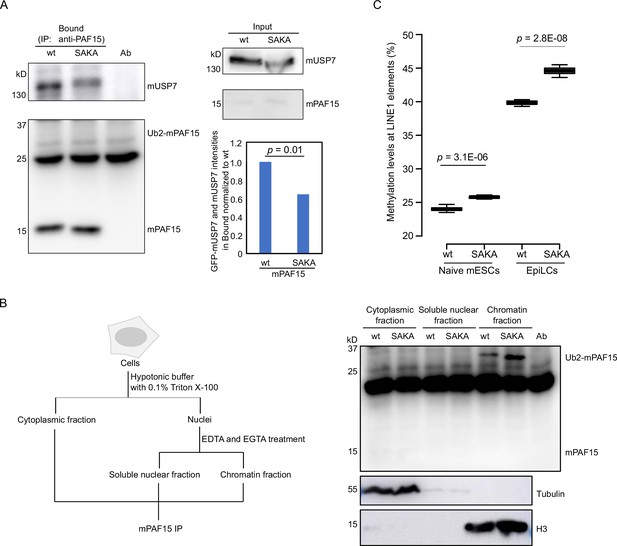
mUSP7 interacts with mPAF15 to assure a complete DNA methylation maintenance.
(A) Immunoprecipitation (IP) of endogenous mPAF15 from whole cell lysates of wt and mPAF15 SAKA mouse embryonic stem cells (mESCs) using an anti-PAF15 antibody. Bound fractions were subjected to immunoblotting with anti-USP7 and PAF15 antibodies. The bar plot shows the quantifications of the relative GFP-mUSP7 and mUSP7 co-precipitated with mPAF15. The error bar stands for SD from three biological replicates. A paired t-test with two tails was done, and p value was indicated. (B) Scheme of the cell fractionation experiment described in Figure 7B (left). IP of endogenous mPAF15 from cytosolic, soluble nuclear, and chromatin fractions using an anti-PAF15 antibody. Bound fractions were subjected to immunoblotting with PAF15 antibody. Immunoblotting with anti-Tubulin and anti-H3 antibodies is used to indicate the cytosolic and chromatin fractions, respectively. (C) Boxplot shows the DNA methylation levels of LINE-1 elements in both wt and mPAF15 SAKA naïve and epiblast-like cells (EpiLCs). Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles; outliers are represented by dots. Data sets from four biological replicates were tested for significance with an unpaired t-test with one tail was performed, and p values are indicated.
-
Figure 8—source data 1
Figure 8 Original blots and DNA methylation data.
- https://cdn.elifesciences.org/articles/79013/elife-79013-fig8-data1-v2.zip
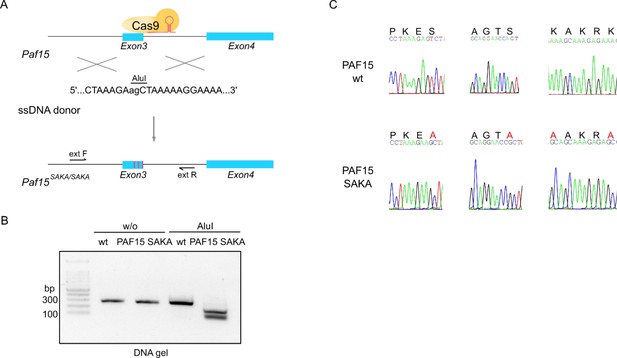
Generation and characterization of mouse embryonic stem cell (mESC) lines carrying the SAKA mutants by genome editing.
(A) Schematic representation of the CRISPR/Cas9 gene editing strategy. The restriction enzyme recognition site for restriction fragment length polymorphism (RFLP) screening is shown. (B) Genotyping using RFLP analysis of PAF15 SAKA mutant line and digestion by the AluI enzyme. (C) DNA sequencing traces confirming the successful mutations of the SAKA in Paf15.
-
Figure 8—figure supplement 1—source data 1
Figure 8—figure supplement 1 Original gel and sequencing data.
- https://cdn.elifesciences.org/articles/79013/elife-79013-fig8-figsupp1-data1-v2.zip
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (insect cells) | Sf9 | This paper | Cell line maintained in M. Nakanishi Lab | |
| Cell line (mouse cells) | mESC J1 line | This paper | Cell line maintained in H. LeonhardtLab | |
| Antibody | Anti-Xenopus PAF15 (Rabbit polyclonal) | PMID:32145273 | PMID:32145273 | WB (1:500) Nakanishi Lab |
| Antibody | Anti-Xenopus DNMT1 (Rabbit polyclonal) | PMID:24013172 | PMID:24013172 | WB (1:500) Nakanishi Lab |
| Antibody | Anti-USP7 (Rabbit polyclonal) | Thermo Fisher Scientific | Cat# A300-033A, RRID:AB_203276 | WB (1:1000) |
| Antibody | Anti-Xenopus UHRF1 (Rabbit polyclonal) | PMID:24013172 | WB (1:500) Nakanishi Lab | |
| Antibody | Anti-histone H3 (Rabbit polyclonal) | abcam | Cat# ab1791, RRID:AB_302613 | WB (1:3000) |
| Antibody | Anti-PCNA (mouse monoclonall) | Santa Cruz Biotechnology | Cat# sc-56, RRID:AB_628110 | WB (1:1000) |
| Antibody | Anti-Xenopus ATAD5 (rabbit polyclonal) | This study | WB (1:1000) | |
| Antibody | Anti-tubulin (mouse monoclonal) | Sigma-Aldrich | Cat# T9026, RRID:AB_477593 | WB (1:2000) |
| Antibody | Anti-PAF15 (mouse monoclonal) | Santa Cruz Biotechnology | Cat# sc-390515 | WB (1:500) |
| Antibody | HRP-anti-mouse IgG (rabbit polyclonal) | Sigma-Aldrich | Cat# A9044, RRID:AB_258431 | WB (1:5000) |
| Recombinant DNA reagent | pGEX4T-3-xPAF15 | PMID:32145273 | PMID:32145273 | Nakanishi Lab |
| Recombinant DNA reagent | pVL1392-xUSP7-3xFLAG | This study | Expression and purification of xUSP7 in insect cells | |
| Recombinant DNA reagent | pVL1392-xDNMT1-3xFLAG | PMID:29053958 | Nakanishi Lab | |
| Recombinant DNA reagent | pGEX-4T-3-mDPPA3 | PMID:33235224 | Nakanishi Lab | |
| Recombinant DNA reagent | pGEX-6P-1-hUSP7 (561–1102) | This study | Expression and purification of hUSP7 fragment in bacteria cells | |
| Recombinant DNA reagent | pGEX-4T-1-SUMO-hPAF15-FLAG | This study | Expression and purification of full-length hPAF15 in bacteria cells | |
| Recombinant DNA reagent | pCSII-EF-mini-AzamiGreen ATAD5 | This study | Expression and purification of full-length hATAD5 in human 293T cells | |
| Recombinant DNA reagent | pCSII-EF-RFC2, 3, 4 and 5 | This study | Expression and purification of full-length hRFC complex in human 293T cells | |
| Peptide, recombinant protein | Ubiquitin | R&D systems (Boston biochem) | U-100H | 58 μM |
| Peptide, recombinant protein | Recombinant Human Ubiquitin Vinyl Sulfone Protein | R&D systems (Boston biochem) | U-202 | 20 μM |
| Commercial assay or kit | EZ DNA Methylation-Gold Kit | Zymo | D5005 | |
| Chemical compound and drug | FT-671 | MedChem Express | HY-107985 | 100 μM |
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/79013/elife-79013-mdarchecklist1-v2.docx
-
Supplementary file 1
MS-based quantification of GST interacting protein.
- https://cdn.elifesciences.org/articles/79013/elife-79013-supp1-v2.xlsx
-
Supplementary file 2
MS-based quantification of xPAF15 interacting proteins.
- https://cdn.elifesciences.org/articles/79013/elife-79013-supp2-v2.xlsx
-
Supplementary file 3
Oligonucleotides used in this study.
- https://cdn.elifesciences.org/articles/79013/elife-79013-supp3-v2.xlsx