Nuclear fascin regulates cancer cell survival
Figures
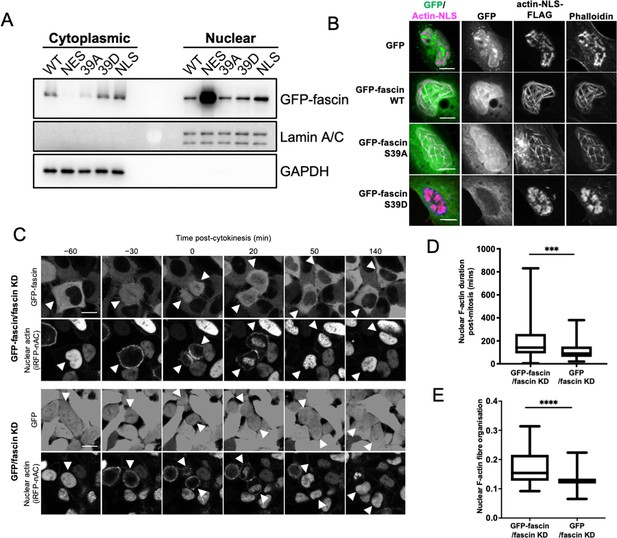
Nuclear fascin contributes to F-actin bundling.
(A) Representative western blot of fascin knockdown (KD) HeLa cells expressing specified GFP-fascin constructs subjected to biochemical fractionation. Nuclear and cytoplasmic compartments probed for GFP-fascin (80 kDa), Lamin A/C (69/62 kda) and GAPDH (36 kDa). Representative of three independent experiments. (B) Representative confocal images of nuclei of fascin KD HeLa cells co-expressing specified GFP-fascin constructs (green) and actin-NLS-FLAG construct, fixed and stained for FLAG (magenta) and F-actin (phalloidin). Scale bars are 10 µm. (C) Representative stills from time-lapse confocal movies of fascin KD HeLa cells co-expressing GFP or GFP-fascin (top panels) and iRFP-nAC nuclear F-actin probe (bottom panels) pre- and post-cytokinesis. Arrowheads point to dividing or daughter cells. Scale bars are 10 µm. (D) Quantification of duration of nuclear F-actin filaments in cells as in (C). (E) Organisation of nuclear F-actin in synchronised cells, 10 hr after release. For (D) and (E), N=89–100 cells/condition, pooled from three independent experiments. Graphs shows min/max and mean of dataset. ***=p < 0.001, ****=p < 0.0001.
-
Figure 1—source data 1
Figure 1A full western blots.
- https://cdn.elifesciences.org/articles/79283/elife-79283-fig1-data1-v1.tiff
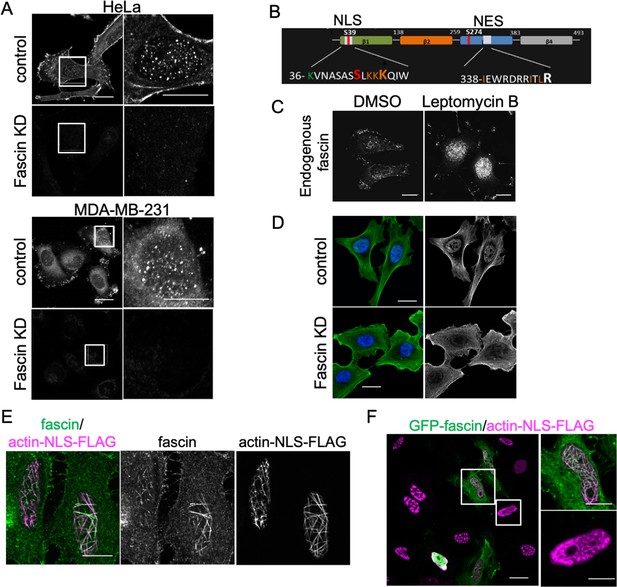
Fascin localises to the nucleus and colocalises with nuclear F-actin.
(A) Example images of WT and fascin knockdown (KD) HeLa (left panels) and MDA-MB-231 (right panels) cells fixed and stained for endogenous fascin. Inset panels show nuclear fascin localisation. Scale bars are 10 µm. (B) Schematic diagram of fascin domain structure indicating identified nuclear localisation signal (NLS) and nuclear export signal (NES) regions (colours denote mutated residues in mutant forms generated; Figure 1). (C) Example images of HeLa cells treated with leptomycin B fixed and stained for endogenous fascin. Scale bars are 10 µm. (D) Example images of WT and fascin KD HeLa cells fixed and stained using an antibody detecting endogenous nuclear actin (green) and DAPI (blue). Scale bars are 10 µm. (E) Example confocal images of HeLa cells expressing actin-NLS-FLAG fixed and stained for endogenous fascin (green) and FLAG (magenta). Scale bars are 10 µm. (F) Example confocal image of mixed population of fascin KD cells and those re-expressing GFP-fascin (green), transfected with actin-NLS-FLAG, fixed and stained for FLAG (magenta). Scale bars are 10 µm.
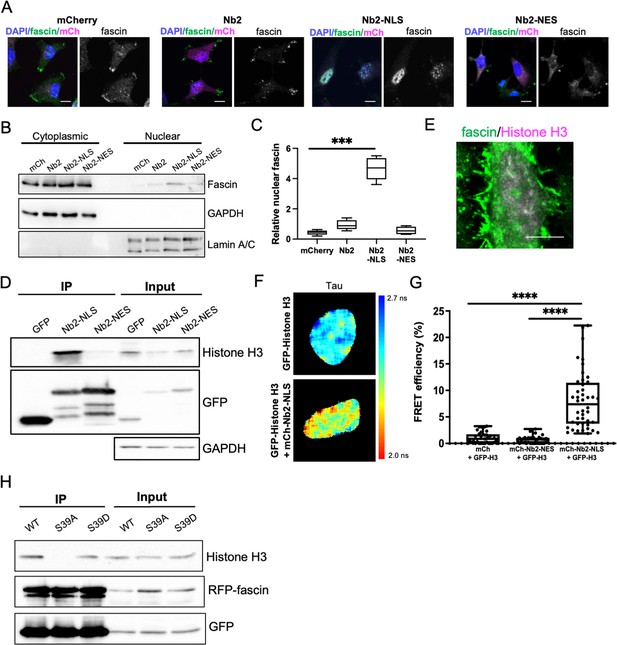
Nuclear fascin directly associates with Histone H3.
(A) Representative confocal images of fascin knockdown (KD) MDA-MB-231 cells expressing GFP-fascin (green) and specified mCherry-Nb2 constructs (magenta) fixed and stained with DAPI (blue). Scale bars are 10 µm. (B) Representative western blot of fascin KD MDA-MB-231 cells expressing GFP or GFP-fascin (80 kDa) subjected to biochemical fractionation. Nuclear and cytoplasmic compartments probed for GAPDH (36 kDa) and Lamin A/C (69/62 kDa) . (C) Quantification of data from (B) from four independent experiments. (D) Representative western blot of HeLa cells expressing GFP or GFP-Nb2-nuclear localisation signal (NLS)/nuclear export signal (NES), subjected to GFP-Trap and probed for Histone H3 (15 kDa), GFP (25 kDa and ~50 kDa Nbs). Input shown on right; GAPDH as loading control (36 kDa). Representative of four independent experiments. (E) Representative confocal images of HeLa nuclei fixed and stained for fascin (green) and Histone H3 (magenta). Scale bar is 10 µm. (F) Representative images of HeLa cells expressing GFP-Histone H3 (top panels) or GFP-Histone H3 and mCherry-Nb2-NLS (bottom panels). Donor and acceptor channels shown; lifetime shown in far-right panels. Scale bars are 10 µm. (G) Quantification of fluorescence resonance energy transfer (FRET) efficiency from data as in (F) plus mCherry and mCherry-Nb2-NES acceptor controls. N=35 cells, from four independent experiments. Graph shows min/max and mean of dataset; each point represents a single cell. (H) Representative western blot of HeLa cells expressing RFP-fascin WT, S39A or S39D (80 kDa), and GFP-Nb2-NLS (~50 kDa), subjected to GFP-Trap and probed for specified proteins (Histone H3 is 15 kDa). Input shown on right. Representative of four independent experiments.
-
Figure 2—source data 1
Figure 2B full western blots.
- https://cdn.elifesciences.org/articles/79283/elife-79283-fig2-data1-v1.tiff
-
Figure 2—source data 2
Table detailing proteins identified by mass spectrometry associated with Nb2-NLS or Nb2-NES immunoprecipitated from HeLa cells.
Peptide counts per identified protein shown for each protein/sample. Nuclear fascin (Nb2-NLS) enriched samples shown in tab 1. Full dataset shown in tab 2. Proteomics reporting details shown in tab 3.
- https://cdn.elifesciences.org/articles/79283/elife-79283-fig2-data2-v1.xlsx
-
Figure 2—source data 3
Figure 2D full western blots.
- https://cdn.elifesciences.org/articles/79283/elife-79283-fig2-data3-v1.tiff
-
Figure 2—source data 4
Figure 2H full western blots.
- https://cdn.elifesciences.org/articles/79283/elife-79283-fig2-data4-v1.tiff
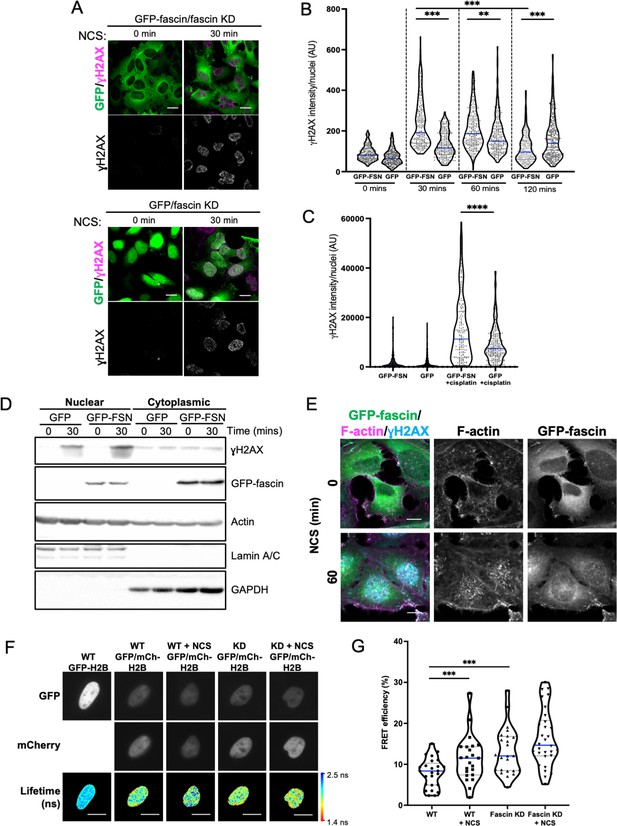
Nuclear fascin promotes efficient DNA damage response.
(A) Representative confocal images of fascin knockdown (KD) HeLa cells expressing GFP or GFP-fascin (green) fixed and stained for ɣH2AX (magenta) before (0 min) or after (30 min) treatment with 0.5 µg/ml neocarzinostatin (NCS). Scale bars are 10 µm. (B) Quantification of ɣH2AX levels in cells from data as in (A). N=300–350 cells/condition, pooled from three independent experiments. (C) Quantification of ɣH2AX levels in cells treated with 5 µM cisplatin for 18 hr. N=240–300 cells/condition, pooled from three independent experiments. (D) Representative western blot of fascin KD HeLa cells expressing GFP (25 kDa) or GFP-fascin (FSN; 80 kDa) subjected to biochemical fractionation. Nuclear and cytoplasmic compartments probed for ɣH2AX (~15 kDa), Actin (42 kDa) and Lamic A/C (69/62 kDa). Representative of three independent experiments. (E) Representative confocal images of fascin KD HeLa cells expressing GFP-fascin (green) fixed and stained for ɣH2AX (cyan) and phalloidin (magenta) before (0 min) or after (60 min) treatment with NCS. Scale bars are 10 µm. (F) Representative images of WT or fascin KD HeLa cells expressing GFP-Histone H2B (left panels) or GFP-Histone H2B and mCherry-Histone H2B, with or without 30 min NCS treatment. Donor and acceptor channels shown; lifetime shown in bottom panels. Scale bars are 10 µm. (G) Quantification of fluorescence resonance energy transfer (FRET) efficiency from data as in (F). N=35 cells, pooled from three independent experiments. All graphs show min/max and mean of dataset; for figures in (B), (C), and (G), data is shown as violin plot with each point representing a single cell and mean shown in blue. **=p < 0.01, ***=p < 0.001, ****=p < 0.0001.
-
Figure 3—source data 1
Figure 3D full western blots.
- https://cdn.elifesciences.org/articles/79283/elife-79283-fig3-data1-v1.tiff
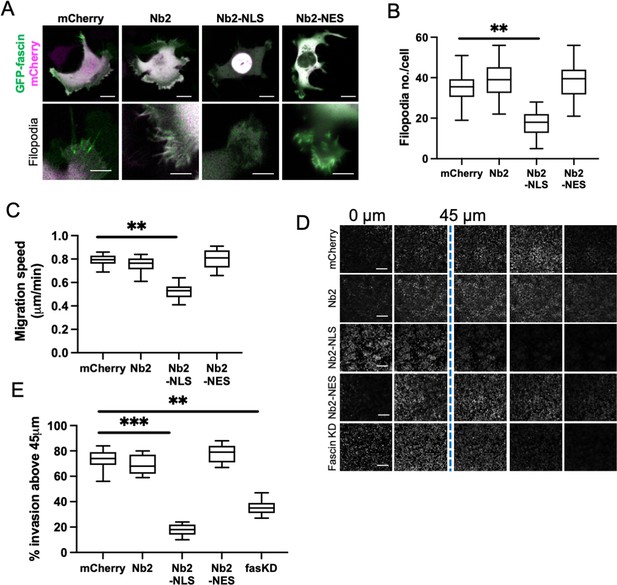
Sustained nuclear fascin reduces cell invasion.
(A) Representative confocal images of fascin knockdown (KD) MDA-MB-231 cells expressing GFP-fascin (green) and Nb2 constructs (magenta). Zoom region of filopodia shown below each. Scale bars are 10 µm. (B) Quantification of filopodia number/cell from data as in (A) from 30 cells, pooled from three independent experiments. (C) Quantification of 2D migration speed of cells as in (A) from 16 hr time-lapse movies. (D) Representative images of confocal Z-stacks from inverted invasion assays from cells as in (A), fixed after 48 hr invasion and stained for DAPI (shown). Fascin KD cells additionally shown (bottom row). (E) Quantification of invasion from data as in (D). Three wells imaged per experiment (3 fields of view/well), pooled from three independent experiments. All graphs show min/max and mean of dataset. **=p < 0.01, ***=p < 0.001, ****=p < 0.0001.
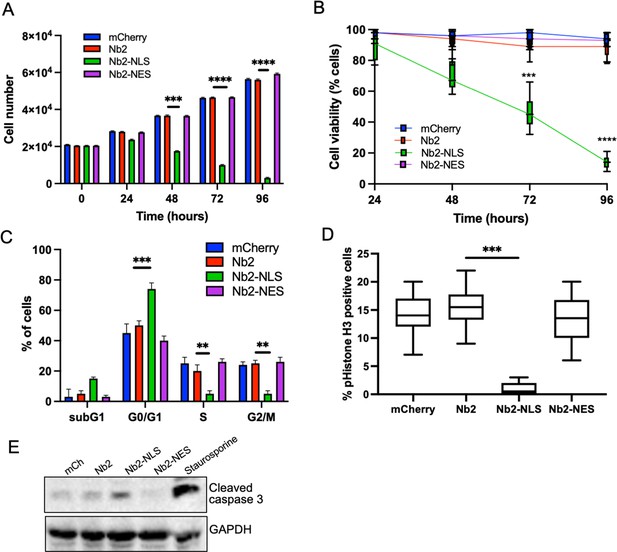
Forced and sustained nuclear fascin reduces cancer cell viability.
(A) Quantification of proliferation of MDA-MB-231 cells expressing specified mCherry-Nb2 constructs over 96 hr. Cell nuclei counted from three replicate wells per condition per experiment. Mean ± SEM are shown. Representative of three independent experiments. (B) Quantification of cell viability using MTT assays of MDA-MB-231 cells expressing specified mCherry-Nb2 constructs over 96 hr. Three replicate wells per condition per experiment. Min/max and mean are shown. Representative of three independent experiments. (C) Quantification of cell cycle stage of MDA-MB-231 cells expressing specified mCherry-Nb2 constructs after 48 hr using FACS analysis of PI-stained cells. Three samples per condition were analysed. Mean ± SEM are shown. Representative of three independent experiments. (D) Quantification of pS10-Histone H3 staining from confocal images of MDA-MB-231 cells expressing specified mCherry-Nb2 constructs after 48 hr. N=90 cells analysed per condition, pooled from three independent experiments. Graph shows min/max and mean of dataset. ***=p < 0.001, ****=p < 0.0001. (E) Representative western blot of MDA-MB-231 cells expressing specified mCherry-Nb2 constructs after 72 hr probed for cleaved caspase 3 (~32 kDa) and GAPDH (36 kDa). Staurosporine (1 µm) treated cells (24 hr) were used as a positive control.
-
Figure 5—source data 1
Figure 5E full western blots.
- https://cdn.elifesciences.org/articles/79283/elife-79283-fig5-data1-v1.tiff
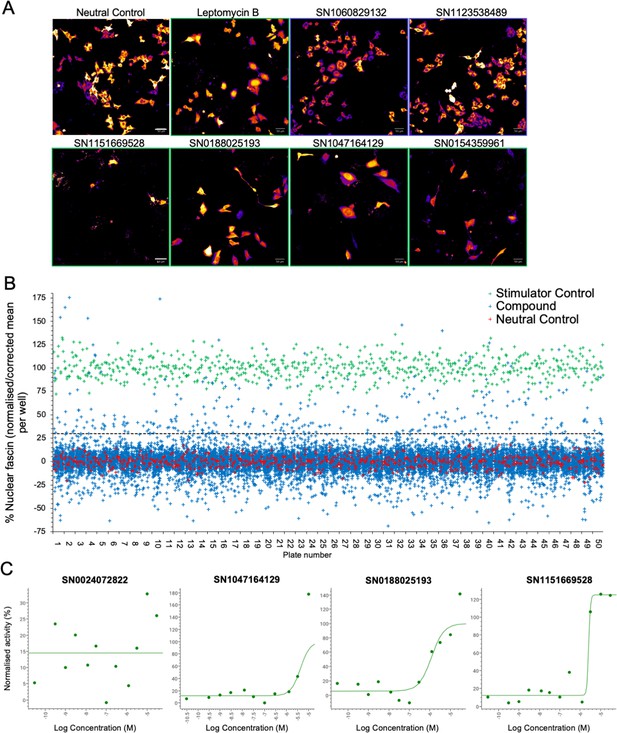
Identification of pathways controlling nuclear fascin.
(A) Example images from high-content screen showing mScarlet-fascin expressed in fascin knockdown (KD) cells (images shown in fire LUT for clarity) treated with specified compounds from small molecule library. Images framed in blue denote no increase in nuclear fascin; those in green denote high nuclear fascin. Scale bars are 50 µm. (B) Plot showing quantification of the mean % nuclear fascin/cell/well from each well of the entire screen (50×384 well plates). Data are normalised to neutral/stimulator controls and corrected for well position. Compound library data shown in blue, neutral controls in red and stimulator controls in green. (C) Representative compound response curves from selected compounds with normalised mean % nuclear fascin/cell/well plotted against compound concentration.
-
Figure 6—source data 1
Table detailing nuclear fascin and nuclear actin bundling levels in mScarlet-fascin/fascin knockdown (KD) HeLa cells treated with 12 increasing doses of the indicated compounds from the small molecule library.
- https://cdn.elifesciences.org/articles/79283/elife-79283-fig6-data1-v1.xlsx
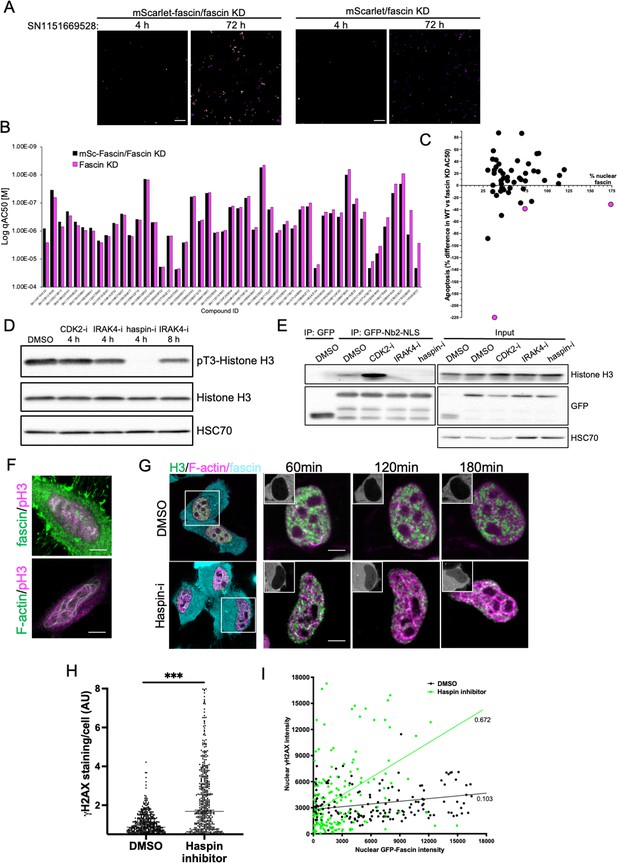
Haspin inhibition promotes sustained nuclear fascin and fascin-dependent apoptosis.
(A) Example images from live cell time-lapse apoptosis screen in fascin knockdown (KD) HeLa cells expressing mScarlet-fascin or mScarlet, showing caspase reporter activity (images shown in fire LUT for clarity) after treatment with compound SN1151669528 (1.25 µM) for 4 or 72 hr. Scale bars are 150 µm. (B) Plot showing AC50 of specified compounds based on apoptosis screen data. (C) Correlation plot from apoptosis screen data showing % difference in apoptosis AC50 between fascin KD HeLa cells expressing mScarlet-fascin (WT) or mScarlet (KD), plotted as a function of mean % nuclear fascin/cell/well from original screen (as in Figure 6B), after treatment with hit compounds. Magenta data points denote hits selected with significantly lower death in KD cells vs. WT. (D) Representative western blot of HeLa cells treated with target inhibitors (1.25 µM) probed for pT3/total Histone H3 (15 kDa) and HSC70 (70 kDa). Representative of three independent experiments. (E) Representative western blots of HeLa cells expressing GFP or GFP-Nb2-NLS treated with specified compounds (1.25 µM, 4 hr) followed by GFP-Trap and probing for Histone H3 (15 kDa), GFP (80 kDa for GFP-FSN and 25 kDa for GFP) and HSC70 (70 kDa). Representative of three independent experiments. (F) Example confocal images of HeLa cells expressing iRFP-nAC nuclear actin probe, stained for fascin and pT3-Histone H3 (magenta). Scale bars are 10 µm. (G) Representative images of HeLa cells co-expressing GFP-Histone H3 (green), iRFP-nAC (magenta), and mScarlet-fascin (cyan) treated with DMSO or haspin inhibitor followed by time-lapse imaging. First panels show whole cells at time 0, subsequent panels show single nuclei with mScarlet-fascin as inset panels at specified time points. Scale bars are 10 µm. (H) ɣH2AX levels in cells treated with DMSO or 1.25 µM haspin inhibitor for 4 hr. Each point represents a single cell; representative of three independent experiments. ***=p < 0.001. (I) Correlation plot between nuclear GFP-fascin and ɣH2AX intensity from data as in (H). Values represent linear regression slope. NLS, nuclear localisation signal.
-
Figure 7—source data 1
Table detailing apoptosis levels in mScarlet-fascin/fascin knockdown (KD) (WT) or mScarlet/fascin KD (KD) HeLa cells treated with 12 increasing doses of the indicated compounds from the small molecule library for 72 hr.
- https://cdn.elifesciences.org/articles/79283/elife-79283-fig7-data1-v1.xlsx
-
Figure 7—source data 2
Table detailing nuclear fascin and nuclear actin bundling levels in mScarlet-fascin/fascin knockdown (KD) HeLa cells treated with 12 increasing doses of the indicated compounds from the small molecule library.
- https://cdn.elifesciences.org/articles/79283/elife-79283-fig7-data2-v1.xlsx
-
Figure 7—source data 3
Figure 7D full western blots.
- https://cdn.elifesciences.org/articles/79283/elife-79283-fig7-data3-v1.tiff
-
Figure 7—source data 4
Figure 7E full western blots.
- https://cdn.elifesciences.org/articles/79283/elife-79283-fig7-data4-v1.tiff
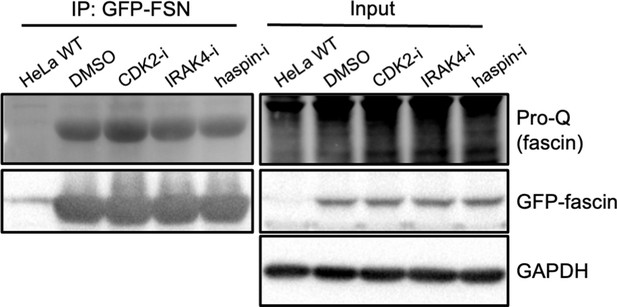
Haspin and IRAK4 do not promote fascin phosphorylation.
Representative western blots of fascin knockdown (KD) HeLa cells expressing GFP-fascin treated with specified inhibitors subjected to GFP-Trap then probed for total phosphorylated protein (top lanes, ~80 kDa in GFP-FSN lane) and GFP-FSN (~80 kDa). Input shown on right. GAPDH (36 kDa) provided as loading for input. Representative of three independent experiments.
-
Figure 7—figure supplement 1—source data 1
Figure Supplement 2 full western blots.
- https://cdn.elifesciences.org/articles/79283/elife-79283-fig7-figsupp1-data1-v1.tiff





