Hypoxia controls plasma membrane targeting of polarity proteins by dynamic turnover of PI4P and PI(4,5)P2
Figures
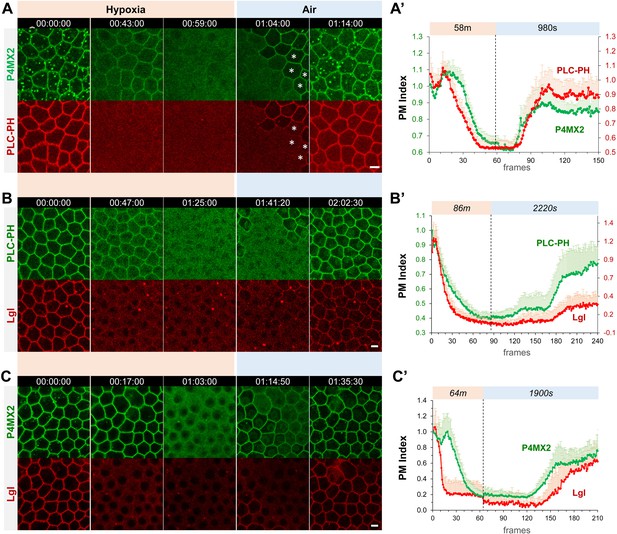
Hypoxia induces acute and reversible loss of P4M × 2::GFP and PLC-PH::RFP from the PM in Drosophila follicle cells.
(A–C) Representative frames showing follicle cells coexpressing P4M × 2::GFP and PLC-PH::RFP (A) or P4M × 2::GFP and Lgl::RFP (B) or PLC-PH::GFP and Lgl::RFP (C) undergoing hypoxia and reoxygenation. In A, at 1:04:00, asterisks (*) highlight cells that had already recovered P4M × 2::GFP but not PLC-PH::RFP. (A’–C’) Quantification of PM localizations of P4M × 2::GFP and PLC-PH::RFP (A’, n=18, 18, Figure 1—source data 1) or P4M × 2::GFP and Lgl::RFP (B’, n=20, 20, Figure 1—source data 2) or PLC-PH::GFP and Lgl::RFP (C’, n=20, 20, Figure 1—source data 3) during hypoxia and reoxygenation. PM Index: ratio of mean intensity of PM and cytosolic signals, normalized by the average value of the first three frames. Time stamp in hr:min:sec format. Scale bars: 5 µm.
-
Figure 1—source data 1
Hypoxia induces acute and reversible loss of PM PI4P and PIP2 in Drosophila follicle cells.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig1-data1-v2.xlsx
-
Figure 1—source data 2
Hypoxia induces acute and reversible loss of PM PIP2 and Lgl in Drosophila follicle cells.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig1-data2-v2.xlsx
-
Figure 1—source data 3
Hypoxia induces acute and reversible loss of PM PI4P and Lgl in Drosophila follicle cells.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig1-data3-v2.xlsx
Acute and reversible loss of PM localization of P4M × 2::GFP and PLC-PH::RFP under hypoxia in follicle cells.
Acute and reversible loss of PM localization of PLC-PH::GFP and Lgl::mCherry under hypoxia in follicle cells.
Acute and reversible loss of PM localization of P4M × 2::GFP and Lgl::mCherry under hypoxia in follicle cells.
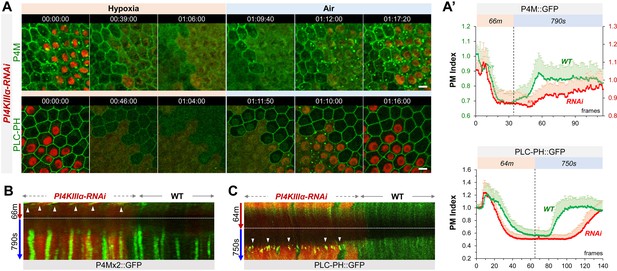
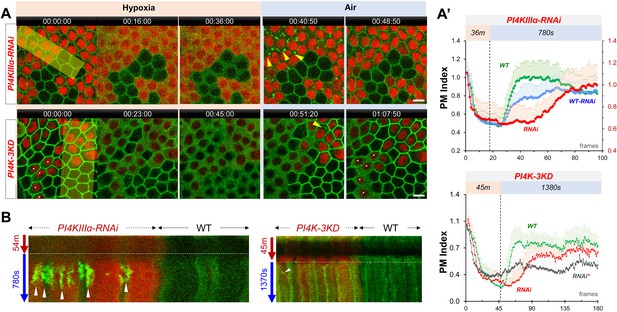
PI4KIIIα regulates PM PI4P and PIP2 homeostasis and dynamic turnover under hypoxia/reoxygenation.
(A) Representative frames showing follicle cells expressing P4M::GFP and PLC-PH::GFP undergoing hypoxia and reoxygenation. PI4KIIIα-RNAi cells are labeled by RFP. (A’) PM localization of P4M::GFP (n=24, 23, Figure 2—source data 1) and PLC-PH::GFP (n=24, 24, Figure 2—source data 2) quantified in boundaries between wild type (WT) cells and between PI4KIIIα-RNAi (RNAi) cell. (B) Kymographs showing the persistent P4M::GFP puncta in both wild type and RNAi cells after hypoxia. White arrowheads point to puncta in RNAi cells at onset of hypoxia. Kymograph made by the maximum projection of 250 pixel wide line reslice of the time-lapse movie. (C) Kymograph showing the transient PLC-PH::GFP puncta (white arrowhead) in RNAi cells only. Kymograph made by the maximum projection of 300pixel wide line reslice of the time-lapse movie. Time stamp in hr:min:sec format. Scale bars: 5 µm.
-
Figure 2—source data 1
PI4KIIIα regulates PM PI4P homeostasis and dynamic turnover under hypoxia/reoxygenation.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig2-data1-v2.xlsx
-
Figure 2—source data 2
PI4KIIIα regulates PM PIP2 homeostasis and dynamic turnover under hypoxia/reoxygenation.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig2-data2-v2.xlsx
PM P4M::GFP in PI4KIIIα-RNAi cells show accelerated loss under hypoxia and delayed recovery under reoxygenation.
PM PLC-PH::GFP in PI4KIIIα-RNAi cells show accelerated loss under hypoxia and delayed recovery under reoxygenation.
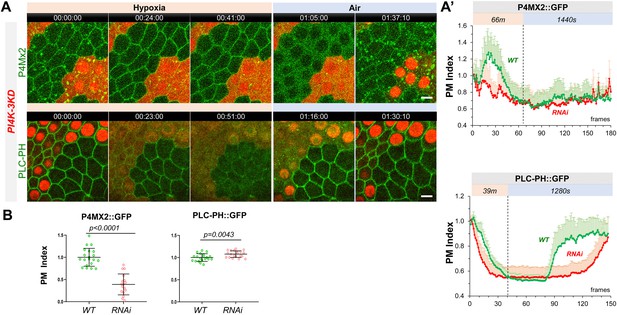
PM PI4P and PIP2 show accelerated loss under hypoxia and delayed recovery under reoxygenation in PI4K-3KD RNAi cells.
(A) Representative frames showing follicle cells expressing P4M × 2::GFP or PLC-PH::GFP undergoing hypoxia and reoxygenation. PI4K-3KD cells are labeled by RFP. (A’) PM localization of P4M × 2::GFP (n=10, 5, Figure 3—source data 1) and PLC-PH::GFP (n=14, 15, Figure 3—source data 2) quantified in boundaries between wild type (WT) cells and between PI4K-3KD (RNAi) cell during hypoxia and regoxygenation. (B) Strong reduction of PM P4M × 2::GFP (n=20, 17, Figure 3—source data 3) but not PLC-PH::GFP (n=20, 20, Figure 3—source data 4) in PI4K-3KD follicle cells. Time stamp in hr:min:sec format. Scale bars: 5µm.
-
Figure 3—source data 1
PI4P showed accelerated loss under hypoxia and delayed recovery under reoxygenation in PI4K-3KD RNAi cells.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig3-data1-v2.xlsx
-
Figure 3—source data 2
PIP2 showed accelerated loss under hypoxia and delayed recovery under reoxygenation in PI4K-3KD RNAi cells.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig3-data2-v2.xlsx
-
Figure 3—source data 3
Reduction of PM PI4P in PI4K-3KD RNAi cells.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig3-data3-v2.xlsx
-
Figure 3—source data 4
PM PI4P unchanged in PI4K-3KD RNAi cells.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig3-data4-v2.xlsx
-
Figure 3—source data 5
Dynamic turnover of PM PI4P under hypoxia and reoxygenation in PI4K-2KD cells.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig3-data5-v2.xlsx
-
Figure 3—source data 6
Dynamic turnover of PM PIP2 under hypoxia and reoxygenation in PI4K-2KD cells.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig3-data6-v2.xlsx
-
Figure 3—source data 7
Dynamic turnover of PM Lgl under hypoxia and reoxygenation in PI4K-2KD cells.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig3-data7-v2.xlsx
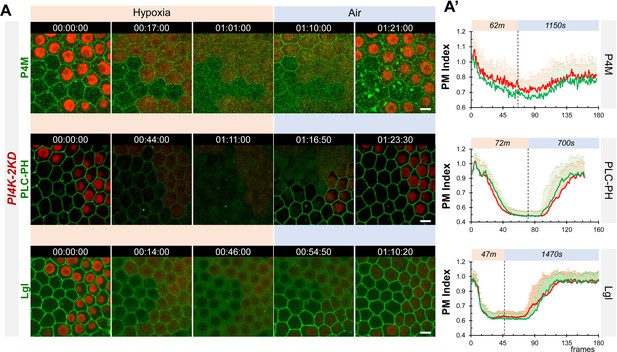
Dynamic turnover of PM P4M::GFP, PLC-PH::GFP and Lgl::GFP under hypoxia and reoxygenation in PI4K-2KD cells is similar to wild type cells.
(A) Representative frames showing follicle cells expressing P4M::GFP, PLC-PH::GFP or Lgl::GFP undergoing hypoxia and reoxygenation. PI4K-2KD RNAi cells are labeled by RFP. (A’) PM localization of P4M::GFP (n=10,10, Figure 3—source data 5), PLC-PH::GFP (n=14,19, Figure 3—source data 6) and Lgl::GFP (n=15,17, Figure 3—source data 7) quantified in boundaries between wild type (WT) cells and between PI4KIIIα-RNAi (RNAi) cells. Scale bars: 5µm.
PM P4M × 2::GFP in PI4K-3KD-RNAi cells show accelerated loss under hypoxia and delayed recovery under reoxygenation.
PM PLC-PH::GFP in PI4K-3KD-RNAi cells show accelerated loss under hypoxia and delayed recovery under reoxygenation.
PI4Ks regulate the PM localization of Lgl::GFP under hypoxia and reoxygenation.
(A) Representative frames showing Lgl::GFP PM localization during hypoxia and reoxygenation. RNAi cells are labeled by RFP. Yellow arrowheads: transient Lgl::GFP puncta (only few highlighted). *: RNAi cells that failed to recover Lgl::GFP to PM. (A’) (TOP) PM localization of Lgl::GFP quantified in boundaries between wild type (WT) cells (n=23), between PI4KIIIα-RNAi (RNAi) cells (n=24) and between WT and RNAi (WT-RNAi) cells (n=24). (Figure 4—source data 1). (BOTTOM). PM localization of Lgl::GFP quantified in boundaries between wild-type (WT) cells (n=20), between PI4K-3KD (RNAi) cells (n=10) and between cells failed recovery (RNAi*) (n=10). (Figure 4—source data 2). (B) Kymograph highlights the transient Lgl::GFP puncta (arrowheads). Each kymograph was made by reslicing the movie with the maximum projection of a 150 or 250-pixel wide line (yellow bands in A). Time stamp in hr:min:sec format. Scale bars: 5 µm.
-
Figure 4—source data 1
Dynamic turnover of PM Lgl under hypoxia and reoxygenation in PI4K-IIIα-RNAi cells.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Dynamic turnover of PM Lgl under hypoxia and reoxygenation in PI4K-3KD cells.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig4-data2-v2.xlsx
PM Lgl::GFP in PI4KIIIα-RNAi cells show accelerated loss under hypoxia and delayed recovery under reoxygenation.
PM Lgl::GFP in PI4K-3KD-RNAi cells show accelerated loss under hypoxia and delayed recovery under reoxygenation.
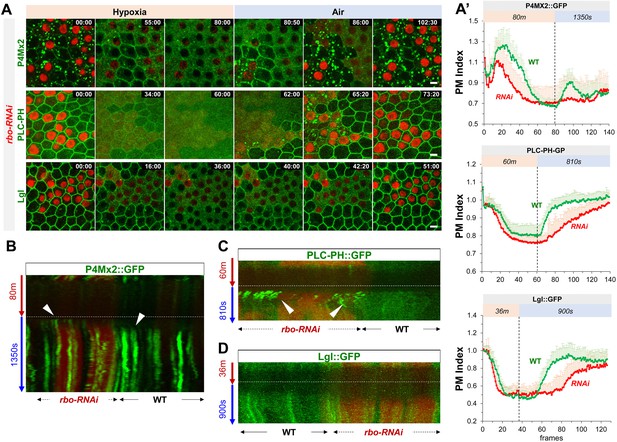
Rbo regulates the homeostasis and dynamic turnover of PI4P, PIP2 and Lgl under hypoxia/reoxygenation.
(A) Representative frames follicle cells expressing P4M × 2::GFP or PLC-PH::GFP or Lgl::GFP undergoing hypoxia and reoxygenation. rbo-RNAi cells are labeled by RFP. Time stamp in min:sec format. (A’) PM localization of P4M::GFP (n=10, 10, Figure 5—source data 1), PLC-PH::GFP (n=20, 20, Figure 5—source data 2) and Lgl::GFP (n=20, 20, Figure 5—source data 3) in A quantified in wild type (WT) and rbo-RNAi (RNAi) cells. (B) Kymograph highlights the earlier onset of P4M::GFP puncta in post-hypoxia RNAi cells. White arrowheads point to the onset of puncta in post-hypoxia RNAi and WT cells. (C) Kymograph highlights the transient PLC-PH::GFP puncta (white arrowheads) seen only in post-hypoxia RNAi cells. (D) Kymograph showing the absence of Lgl::GFP puncta in post-hypoxia RNAi cells. Scale bars: 5µm.
-
Figure 5—source data 1
Dynamic turnover of PM PI4P under hypoxia and reoxygenation in rbo-RNAi cells.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig5-data1-v2.xlsx
-
Figure 5—source data 2
Dynamic turnover of PM PIP2 under hypoxia and reoxygenation in rbo-RNAi cells.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig5-data2-v2.xlsx
-
Figure 5—source data 3
Dynamic turnover of PM Lgl under hypoxia and reoxygenation in rbo-RNAi cells.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig5-data3-v2.xlsx
-
Figure 5—source data 4
PM localization of Rbo is resistant to hypoxia.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig5-data4-v2.xlsx
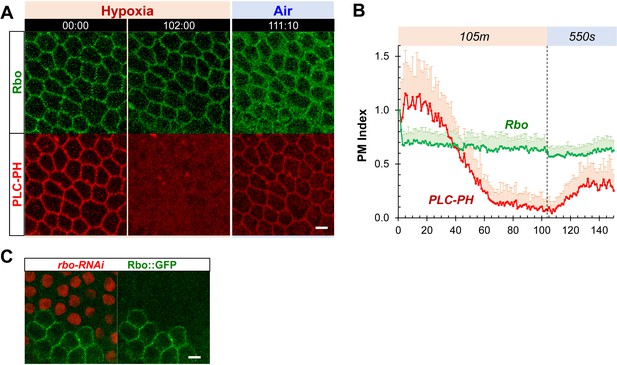
PM localization of Rbo is resistant to hypoxia.
(A) PM localization of Rbo::GFP persisted under hypoxia, regardless of the transient loss of PM PIP2. (B) Quantification of PM Rbo::GFP and PLC-PH::RFP under hypoxia (105 min) and reoxygenation (550 sec). n=22. (C) Lack of Rbo::GFP expression in rbo-RNAi follicle cells (labeled by the expression of nuclear RFP) Scale bars: 5 µm.
PM localization of rbo::GFP is resistant to hypoxia.
PM P4M × 2::GFP in rbo-RNAi cells show accelerated loss under hypoxia and delayed recovery under reoxygenation.
PM PLC-PH::GFP in rbo-RNAi cells show accelerated loss under hypoxia and delayed recovery under reoxygenation.
PM Lgl::GFP in Rbo-RNAi cells show accelerated loss under hypoxia and delayed recovery under reoxygenation.
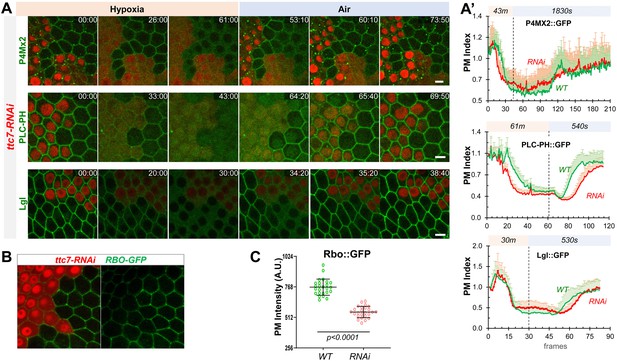
YPP1/TTC7 regulates the homeostasis and dynamic turnover of PI4P, PIP2 and Lgl in cells undergoing hypoxia and reoxygenation.
(A) Representative frames follicle cells expressing P4M × 2::GFP or PLC-PH::GFP or Lgl::GFP undergoing hypoxia and reoxygenation. ttc7-RNAi cells are labeled by RFP. Time stamp in min:sec format. (A’) PM localization changes of P4M × 2::GFP (n=10, 10, Figure 6—source data 1), PLC-PH::GFP (n=20, 20, Figure 6—source data 2) and Lgl::GFP (n=20, 20, Figure 6—source data 3) in A quantified in wild type (WT) and ttc7-RNAi (RNAi) cells.(B and C) Reduction of PM RBO::GFP in ttc7-1-RNAi cells (n=24, 24, Figure 6—source data 4). Scale bars: 5µm.
-
Figure 6—source data 1
Dynamic turnover of PM PI4P under hypoxia and reoxygenation in ttc7-RNAi cells.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig6-data1-v2.xlsx
-
Figure 6—source data 2
Dynamic turnover of PM PIP2 under hypoxia and reoxygenation in ttc7-RNAi cells.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig6-data2-v2.xlsx
-
Figure 6—source data 3
Dynamic turnover of PM Lgl under hypoxia and reoxygenation in ttc7-RNAi cells.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig6-data3-v2.xlsx
-
Figure 6—source data 4
Reduction of Rbo::GFP in ttc7-RNAi cells.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig6-data4-v2.xlsx
PM P4M × 2::GFP in ttc7-RNAi cells show loss under hypoxia and delayed recovery under reoxygenation.
PM PLC-PH::GFP in ttc7-RNAi cells show accelerated loss under hypoxia and delayed recovery under reoxygenation.
PM Lgl::GFP in ttc7-RNAi cells show accelerated loss under hypoxia and delayed recovery under reoxygenation.
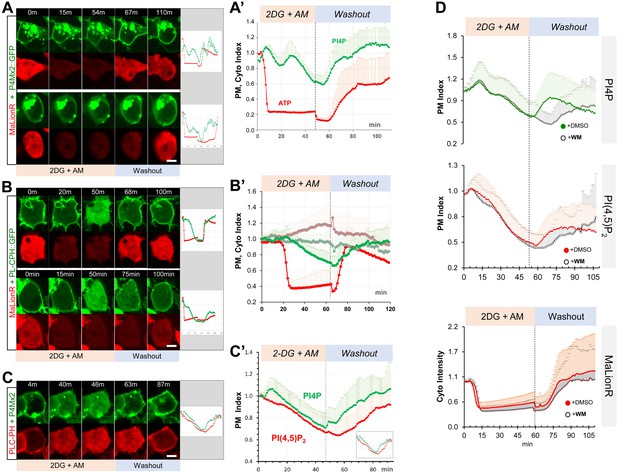
ATP inhibition induces acute and reversible loss of P4M × 2::GFP and PLC-PH::RFP from the PM in HEK293 cells.
(A, B) Representative cells showing the PM localization of P4M × 2::GFP (A) or PLC-PH::GFP (B) and MaLionR ATP sensor during ATP inhibition and subsequent washout with low-glucose medium.(A) Top cell: P4M × 2::GFP recovery on PM was immediately followed by ATP sensor brightness increase (n=9). Bottom cell: measurable ATP increase lagged well behind the PM recovery of P4M × 2::GFP (n=6). Intracellular P4M × 2::GFP puncta were overexposed but excluded from quantification. (A’) Normalized quantification of PM localization of P4M × 2::GFP and MaLionR intensity (n=15, 19 cells, respectively). Figure 7—source data 1. (B) Top cell: synchronous PLC-PH::GFP recovery and ATP senor brightness increase (n=13). Bottom cell: PLC-PH::GFP PM recovery slightly preceded the detectable ATP increase (n=12). (B’) Normalized quantification of PM localization of PLC-PH::GFP (green) and MaLionR intensity (red) in ATP inhibition cells (solid dots, n=13) or serum-free medium treated cells (blank diamonds, n=12 cells) Figure 7—source data 2. (C) A representative cell showing the PM localization of P4M × 2::GFP and PLC-PH::RFP during ATP inhibition and subsequent washout. (C’) Quantification of PM P4M × 2::GFP and PLC-PH::GFP (n=13 cells) (Figure 7—source data 3). (D) Normalized PM localization index of P4M × 2::GFP, PLC-PH::RFP and cyto index of MaLionR sensor in cells treated with ATP inhibition followed by washout with buffer containing DMSO or Wortmannin (WM, 20 µm). (n=48, all samples). Figure 7—source data 4; 5. Scale bars: 10 µm.
-
Figure 7—source data 1
Correlation between ATP inhibition PM PI4P turnovery in HEK293 cells.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig7-data1-v2.xlsx
-
Figure 7—source data 2
Correlation between ATP inhibition PM PIP2 turnovery in HEK293 cells.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig7-data2-v2.xlsx
-
Figure 7—source data 3
ATP inhibition induces acute and reversible loss of PM PI4P and PIP2 in HEK293 cells.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig7-data3-v2.xlsx
-
Figure 7—source data 4
Wortmannin inhibits PM PI4P recovery post ATP inhibition.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig7-data4-v2.xlsx
-
Figure 7—source data 5
Post-inhibition recovery of ATP was not delayed by wortmannin treatment.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig7-data5-v2.xlsx
-
Figure 7—source data 6
Acute and reversible reduction of intracellular ATP in Drosophila follicle cells undery hypoxia.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig7-data6-v2.xlsx
-
Figure 7—source data 7
Acute and reversible reduction of intracellular ATP in HEK203 cells undery hypoxia.
- https://cdn.elifesciences.org/articles/79582/elife-79582-fig7-data7-v2.xlsx
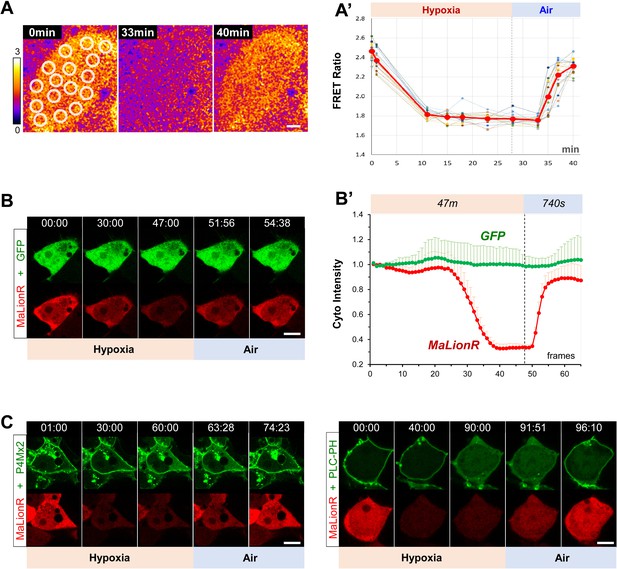
Real-time monitoring of intracellular ATP changes in follicular cells and HEK293 cells undergoing hypoxia and reoxygenation.
(A) Heat map of the (uncalibrated) FRET ratio of AT[NL] (an ATeam FRET-based ATP sensor) in follicular cells of a dissected ovary undergoing hypoxia and reoxygenation ex vivo. (A’) Quantification of FRET ratios in A. Dashed lines are the mean FRET ratios measured at different spots (white circles in A) over the time. Thick red line: average of the all measurements. (Figure 7—source data 6). (B) A representative HEK293 cell expressing GFP and MaLionR ATP sensor undergoing hypoxia and reoxygenation. (B’) Quantification of GFP and MaLionR intensities in HEK293 cells undergoing hypoxia and reoxygenation (n=12). GFP served as a negative control that did not show intensity changes under hypoxia and reoxygenation. (Figure 7—source data 7) (C) Representative HEK293 cells expressing MaLionR and P4M × 2 GFP or PLC-PH::GFP undergoing hypoxia and reoxygenation. Time stamp in min:sec format. Scale bars: 10 µm.
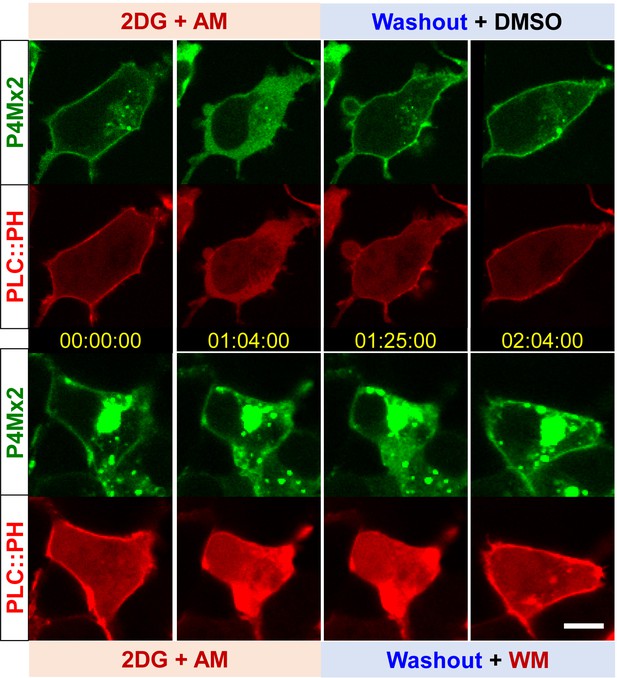
Wortmannin inhibits PM PI4P recovery after ATP inhibition.
Representative cells showing the PM localization of P4M × 2:GFP and PLC-PH::RFP during ATP inhibition and washout with DMSO or wortmannin (WM). Scale bar: 10 µm.
HEK293 cells expressing GFP and MaLionR ATP sensor undergoing hypoxia and reoxygenation.
HEK293 cells expressing P4M × 2::GFP and MaLionR undergoing acute and transient ATP inhibition.
HEK293 cells expressing PLC-PH::GFP and MaLionR undergoing acute and transient ATP inhibition.
HEK293 cells expressing P4M × 2::GFP and PLC-PH::RFP undergoing acute and transient ATP inhibition.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (D. melanogaster) | ubi-P4M::GFP | This paper | See “Materials and methods” | |
| Genetic reagent (D. melanogaster) | ubi-P4M × 2::GFP | This paper | See “Materials and methods” | |
| Genetic reagent (D. melanogaster) | ubi-PLC-PH::GFP | This paper | See “Materials and methods” | |
| Genetic reagent (D. melanogaster) | ubi-PLC-PH::RFP | This paper | See “Materials and methods” | |
| Genetic reagent(D. melanogaster) | y[1] M{RFP[3xP3.PB] GFP[E.3xP3]=vas int.Dm}ZH-2A w[*]; PBac{y[+]-attP-9A}VK00022 | Bloomington Drosophila Stock Center | BDSC:24868; RRID:BDSC_24868 | |
| Genetic reagent (D. melanogaster) | y[1] w[*] P{y[+t7.7]=nos-phiC31\int.NLS}X; PBac{y[+]-attP-3B}VK00040 | Bloomington Drosophila Stock Center | BDSC:35568; RRID:BDSC_35568 | |
| Genetic reagent (D. melanogaster) | PI4K-2KD | This paper | See “Materials and methods” | |
| Genetic reagent (D. melanogaster) | PI4K-3KD | This paper | See “Materials and methods” | |
| Genetic reagent (D. melanogaster) | UAS-AT1.03NL1 | DGRC#117,011 Tsuyama et al., 2017 | DGRC#117,011 | |
| Genetic reagent (D. melanogaster) | Cy2-Gal4 | Gift from David Bilder Queenan et al., 1997 | ||
| Genetic reagent (D. melanogaster) | rbo::GFP | Gift from Kendal Broadie Huang et al., 2004 | ||
| Genetic reagent (D. melanogaster) | lgl::mCherry | Dong et al., 2015 | ||
| Genetic reagent (D. melanogaster) | lgl::GFP | Dong et al., 2015 | ||
| Genetic reagent (D. melanogaster) | UAS-PI4KIIIα-RNAi | Bloomington Drosophila Stock Center | BDSC:35256; FLYB:FBst0035256; RRID:BDSC_35256 | FlyBase symbol: P{TRiP.GL00144}attP2 |
| Genetic reagent (D. melanogaster) | UAS-Rbo-RNAi | Vienna Drosophila Resource Center | VDRC:v47751; FLYB:Bst0467525 | FlyBase symbol: P{GD14013}v47751 |
| Genetic reagent (D. melanogaster) | UAS-ttc7-RNAi | Vienna Drosophila Resource Center | VDRC:v35881; FLYB: FBst0461391 | FlyBase symbol: P{GD13893}v35881 |
| Genetic reagent (D. melanogaster) | UAS-Plc21C-RNAi | Bloomington Drosophila Stock Center | BDSC:33719; FBst0033719 RRID:BDSC_33719 | |
| Genetic reagent (D. melanogaster) | UAS-pis-RNAi | Bloomington Drosophila Stock Center | BDSC:55602; FBst0055602; RRID:BDSC_55602 | |
| Genetic reagent (D. melanogaster) | UAS-PI4KIIIα-RNAi | Bloomington Drosophila Stock Center | BDSC:38242; FBst0038242; RRID:BDSC_38242 | |
| Genetic reagent (D. melanogaster) | UAS-synj-RNAi | Bloomington Drosophila Stock Center | BDSC:44420; FBst0044420; RRID:BDSC_44420 | |
| Genetic reagent (D. melanogaster) | UAS-rdgβ-RNAi | Bloomington Drosophila Stock Center | BDSC:44523; FBst0044523; RRID:BDSC_44523 | |
| Genetic reagent (D. melanogaster) | UAS-INPP5E-RNAi | Bloomington Drosophila Stock Center | BDSC:41701; FBst0041701; RRID:BDSC_41701 | |
| Genetic reagent (D. melanogaster) | UAS-CG5026-RNAi | Bloomington Drosophila Stock Center | BDSC:42759; FBst0042759; RRID:BDSC_42759 | |
| Genetic reagent (D. melanogaster) | UAS-Pten-RNAi | Bloomington Drosophila Stock Center | BDSC:33643; FBst0033643; RRID:BDSC_33643 | |
| Genetic reagent (D. melanogaster) | UAS-Pi3k21B-RNAi | Bloomington Drosophila Stock Center | BDSC:36810; FBst0036810; RRID:BDSC_36810 | |
| Genetic reagent (D. melanogaster) | UAS-fwd-RNAi | Bloomington Drosophila Stock Center | BDSC:35257; FBst0035257; RRID:BDSC_35257 | |
| Genetic reagent (D. melanogaster) | UAS-mtm-RNAi | Bloomington Drosophila Stock Center | BDSC:31552; FBst0031552; RRID:BDSC_31552 | |
| Genetic reagent (D. melanogaster) | UAS-PIP4K-RNAi | Bloomington Drosophila Stock Center | BDSC:35660; FBst0035660; RRID:BDSC_35660 | |
| Genetic reagent (D. melanogaster) | UAS-PI4KIIIα-RNAi | Bloomington Drosophila Stock Center | BDSC:35643; FBst0035643; RRID:BDSC_35643 | |
| Genetic reagent (D. melanogaster) | UAS-INPP5E-RNAi | Bloomington Drosophila Stock Center | BDSC:34037; FBst0034037; RRID:BDSC_34037 | |
| Genetic reagent (D. melanogaster) | UAS-CG3632-RNAi | Bloomington Drosophila Stock Center | BDSC:38341; FBst0038341; RRID:BDSC_38341 | |
| Genetic reagent (D. melanogaster) | UAS-mtm-RNAi | Bloomington Drosophila Stock Center | BDSC:38339; FBst0038339; RRID:BDSC_38339 | |
| Genetic reagent (D. melanogaster) | UAS-PIP4K-RNAi | Bloomington Drosophila Stock Center | BDSC:35338; FBst0035338; RRID:BDSC_35338 | |
| Genetic reagent (D. melanogaster) | UAS-INPP5E-RNAi | Bloomington Drosophila Stock Center | BDSC:34037; FBst0034037; RRID:BDSC_34037 | |
| Genetic reagent (D. melanogaster) | UAS-CG3632-RNAi | Bloomington Drosophila Stock Center | BDSC:38341; FBst0038341; RRID:BDSC_38341 | |
| Genetic reagent (D. melanogaster) | UAS-mtm-RNAi | Bloomington Drosophila Stock Center | BDSC:38339; FBst0038339; RRID:BDSC_38339 | |
| Genetic reagent (D. melanogaster) | UAS-PIP4K-RNAi | Bloomington Drosophila Stock Center | BDSC:35338; FBst0035338; RRID:BDSC_35338 | |
| Genetic reagent (D. melanogaster) | UAS-FIG4-RNAi | Bloomington Drosophila Stock Center | BDSC:38291; FBst0038291; RRID:BDSC_38291 | |
| Genetic reagent (D. melanogaster) | UAS-CG5026-RNAi | Bloomington Drosophila Stock Center | BDSC:38309; FBst0038309; RRID:BDSC_38309 | |
| Genetic reagent (D. melanogaster) | UAS-PI4KIIα-RNAi | Bloomington Drosophila Stock Center | BDSC:35278; FBst0035278; RRID:BDSC_35278 | |
| Genetic reagent (D. melanogaster) | UAS-norpA-RNAi | Bloomington Drosophila Stock Center | BDSC:31113; FBst0031113; RRID:BDSC_31113 | |
| Genetic reagent (D. melanogaster) | UAS-sktl-RNAi | Bloomington Drosophila Stock Center | BDSC:27715; FBst0027715; RRID:BDSC_27715 | |
| Genetic reagent (D. melanogaster) | UAS-fwd-RNAi | Bloomington Drosophila Stock Center | BDSC:29396; FBst0029396; RRID:BDSC_29396 | |
| Genetic reagent (D. melanogaster) | UAS-CG6707-RNAi | Bloomington Drosophila Stock Center | BDSC:28316; FBst0028316; RRID:BDSC_28316 | |
| Genetic reagent (D. melanogaster) | UAS-synj-RNAi | Bloomington Drosophila Stock Center | BDSC:34378; FBst0034378; RRID:BDSC_34378 | |
| Genetic reagent (D. melanogaster) | UAS-pis-RNAi | Bloomington Drosophila Stock Center | BDSC:29383; FBst0029383; RRID:BDSC_29383 | |
| Genetic reagent (D. melanogaster) | UAS-sl-RNAi | Bloomington Drosophila Stock Center | BDSC:32385; FBst0032385; RRID:BDSC_32385 | |
| Genetic reagent (D. melanogaster) | UAS-CG42271-RNAi | Bloomington Drosophila Stock Center | BDSC:29411; FBst0029411; RRID:BDSC_29411 | |
| Genetic reagent (D. melanogaster) | UAS-Plc21C-RNAi | Bloomington Drosophila Stock Center | BDSC:31270; FBst0031270; RRID:BDSC_31270 | |
| Genetic reagent (D. melanogaster) | UAS-Pi3K92E-RNAi | Bloomington Drosophila Stock Center | BDSC:35798; FBst0035798; RRID:BDSC_35798 | |
| Genetic reagent (D. melanogaster) | UAS-Pi3K59F-RNAi | Bloomington Drosophila Stock Center | BDSC:33384; FBst0033384; RRID:BDSC_33384 | |
| Genetic reagent (D. melanogaster) | UAS-sl-RNAi | Bloomington Drosophila Stock Center | BDSC:35604; FBst0035604; RRID:BDSC_35604 | |
| Genetic reagent (D. melanogaster) | UAS-CG9784-RNAi | Bloomington Drosophila Stock Center | BDSC:34723; FBst0034723; RRID:BDSC_34723 | |
| Genetic reagent (D. melanogaster) | UAS-Pi3K68D-RNAi | Bloomington Drosophila Stock Center | BDSC:34621; FBst0034621; RRID:BDSC_34621 | |
| Genetic reagent (D. melanogaster) | UAS-Ocrl-RNAi | Bloomington Drosophila Stock Center | BDSC:34722; FBst0034722; RRID:BDSC_34722 | |
| Genetic reagent (D. melanogaster) | UAS-sktl-RNAi | Bloomington Drosophila Stock Center | BDSC:35198; FBst0035198; RRID:BDSC_35198 | |
| Genetic reagent (D. melanogaster) | UAS-Pten-RNAi | Bloomington Drosophila Stock Center | BDSC:25841; FBst0025841; RRID:BDSC_25841 | |
| Genetic reagent (D. melanogaster) | UAS-CG33981,fab1-RNAi | Bloomington Drosophila Stock Center | BDSC:35793; FBst0035793; RRID:BDSC_35793 | |
| Genetic reagent (D. melanogaster) | UAS-Plc21C-RNAi | Bloomington Drosophila Stock Center | BDSC:32438; FBst0032438; RRID:BDSC_32438 | |
| Genetic reagent (D. melanogaster) | UAS-rdgB-RNAi | Bloomington Drosophila Stock Center | BDSC:28796; FBst0028796; RRID:BDSC_28796 | |
| Genetic reagent (D. melanogaster) | UAS-CG3530-RNAi | Bloomington Drosophila Stock Center | BDSC:25864; FBst0025864; RRID:BDSC_25864 | |
| Genetic reagent (D. melanogaster) | UAS-Synj-RNAi | Bloomington Drosophila Stock Center | BDSC:27489; FBst0027489; RRID:BDSC_27489 | |
| Genetic reagent (D. melanogaster) | UAS-Pi3K68D-RNAi | Bloomington Drosophila Stock Center | BDSC:31252; FBst0031252; RRID:BDSC_31252 | |
| Genetic reagent (D. melanogaster) | UAS-Pten-RNAi | Bloomington Drosophila Stock Center | BDSC:25967; FBst0025967; RRID:BDSC_25967 | |
| Genetic reagent (D. melanogaster) | UAS-Pi3K92E-RNAi | Bloomington Drosophila Stock Center | BDSC:27690; FBst0027690; RRID:BDSC_27690 | |
| Genetic reagent (D. melanogaster) | UAS-norpA-RNAi | Bloomington Drosophila Stock Center | BDSC:31197; FBst0031197; RRID:BDSC_31197 | |
| Genetic reagent (D. melanogaster) | UAS-fwd-RNAi | Bloomington Drosophila Stock Center | BDSC:31187; FBst0031187; RRID:BDSC_31187 | |
| Genetic reagent (D. melanogaster) | UAS-Plc21C-RNAi | Bloomington Drosophila Stock Center | BDSC:31269; FBst0031269; RRID:BDSC_31269 | |
| Genetic reagent (D. melanogaster) | UAS-sl-RNAi | Bloomington Drosophila Stock Center | BDSC:32906; FBst0032906; RRID:BDSC_32906 | |
| Genetic reagent (D. melanogaster) | UAS-CG3530-RNAi | Bloomington Drosophila Stock Center | BDSC:38340; FBst0038340; RRID:BDSC_38340 | |
| Genetic reagent (D. melanogaster) | UAS-CG5026-RNAi | Bloomington Drosophila Stock Center | BDSC:57020; FBst0057020; RRID:BDSC_57020 | |
| Genetic reagent (D. melanogaster) | UAS-PIP4K-RNAi | Bloomington Drosophila Stock Center | BDSC:65891; FBst0065891; RRID:BDSC_65891 | |
| Genetic reagent (D. melanogaster) | UAS-Sac1-RNAi | Bloomington Drosophila Stock Center | BDSC:56013; FBst0056013; RRID:BDSC_56013 | |
| Genetic reagent (D. melanogaster) | UAS-Pi3K59F-RNAi | Bloomington Drosophila Stock Center | BDSC:64011; FBst0064011; RRID:BDSC_64011 | |
| Genetic reagent (D. melanogaster) | UAS-Pi3K68D-RNAi | Bloomington Drosophila Stock Center | BDSC:35265; FBst0035265; RRID:BDSC_35265 | |
| Genetic reagent (D. melanogaster) | UAS-PIP5K59B-RNAi | Bloomington Drosophila Stock Center | BDSC:62855; FBst0062855; RRID:BDSC_62855 | |
| Genetic reagent (D. melanogaster) | UAS-Pi3K93E-RNAi | Bloomington Drosophila Stock Center | BDSC:61182; FBst0061182; RRID:BDSC_61182 | |
| Genetic reagent (D. melanogaster) | UAS-mtm-RNAi | Bloomington Drosophila Stock Center | BDSC:57298; FBst0057298; RRID:BDSC_57298 | |
| Genetic reagent (D. melanogaster) | UAS-Pi3K21B-RNAi | Bloomington Drosophila Stock Center | BDSC:38991; FBst0038991; RRID:BDSC_38991 | |
| Genetic reagent (D. melanogaster) | UAS-Pi3K59F-RNAi | Bloomington Drosophila Stock Center | BDSC:36056; FBst0036056; RRID:BDSC_36056 | |
| Genetic reagent (D. melanogaster) | UAS-FIG4-RNAi | Bloomington Drosophila Stock Center | BDSC:58063; FBst0058063; RRID:BDSC_5806335265 | |
| Cell line (Homo-sapiens) | HEK293 | ATCC | CRL-1573 | |
| Recombinant DNA reagent | P4M::GFP (plasmid) | Hammond et al., 2014 | ||
| Recombinant DNA reagent | P4M × 2::GFP (plasmid) | Hammond et al., 2014 | ||
| Recombinant DNA reagent | PLC-PH::GFP (plasmid) | Hammond et al., 2014 | ||
| Recombinant DNA reagent | pGU (plasmid) | Lu et al., 2021 | ||
| Recombinant DNA reagent | MaLionR (plasmid) | Addgene | Addgene #113908 | |
| Recombinant DNA reagent | pNP (plasmid) | Qiao et al., 2018 | ||
| Recombinant DNA reagent | pNP-fwd-KD (plasmid) | This paper | ||
| Recombinant DNA reagent | pNP-PI4KII-KD (plasmid) | This paper | ||
| Recombinant DNA reagent | pNP-PI4KIIIα-KD (plasmid) | This paper | ||
| Recombinant DNA reagent | pNP-PI4K-2KD (plasmid) | This paper | ||
| Recombinant DNA reagent | pNP-PI4K-3KD (plasmid) | This paper | ||
| Commercial assay or kit | Plasmid midi kid | Qiagen | Cat#12,143 | |
| Commercial assay or kit | Plasmid mini kit | Thermo Scientific | Cat#K0503 | |
| Commercial assay or kit | Gel extraction | Thermo Scientific | Cat#K0692 | |
| Chemical compound, drug | Halocarbon oil | Halocarbon 95 oil | Cat#9002-83-9 | |
| Chemical compound, drug | X-treme Gene 9 DNA transfection reagent | Sigma | Cat#6365787001 | |
| Chemical compound, drug | Fluorescent PM dye | ThermoFisher | ThermoFisher, Cat#C10046 | |
| Chemical compound, drug | 2-Deoxy-D-glucose | Sigma | Cat#D8375 | |
| Chemical compound, drug | Antimycin A | Sigma | Cat#A8674 | |
| Chemical compound, drug | Membrane dye | Invitrogen CellMask | Cat# C10046 | |
| Chemical compound, drug | Wortmannin | Sigma | Cat# 19545-26-7 | |
| Software, algorithm | Fiji (ImageJ) | https://imagej.net/Fiji | https://imagej.net/Fiji | |
| Software, algorithm | GraphPad Prism 8.0 | GraphPad Software | http://www.graphpad.com/ | |
| Other | air-permeable membrane | YSI Inc | YSI Membrane Model #5,793 | YSI Inc, Yellow Springs, OH |
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/79582/elife-79582-mdarchecklist1-v2.pdf
-
Supplementary file 1
Candidate RNAi screen on regulators controlling hypoxia-induced acute and reversible loss of PM Lgl.
- https://cdn.elifesciences.org/articles/79582/elife-79582-supp1-v2.xlsx
-
Supplementary file 2
Primers used in this study.
- https://cdn.elifesciences.org/articles/79582/elife-79582-supp2-v2.docx





