Computationally defined and in vitro validated putative genomic safe harbour loci for transgene expression in human cells
Figures
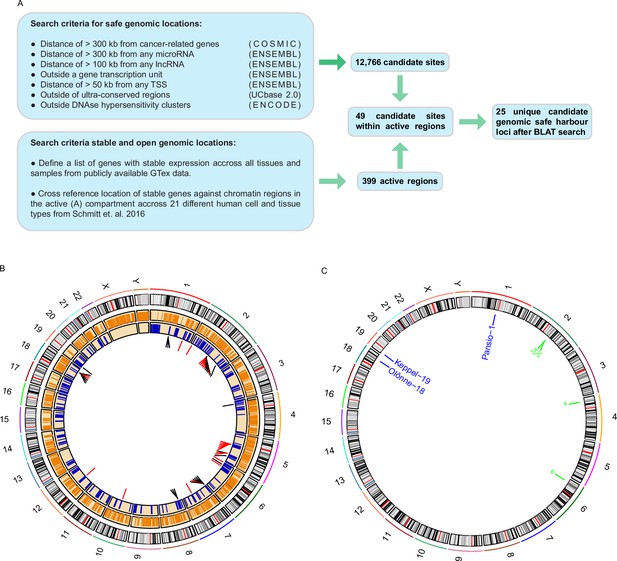
Computational search for candidate GSH.
(A) Schematic representation of the computational workflow for defining candidate GSH. (B) CIRCOS plot summarising computational search results. Ring 1: chromosome ideograms; ring 2: orange bars indicating safe sites; ring 3: blue bars indicating active regions; ring 4: candidate sites within active regions, red bars site failed BLAT screening, black bars site passed BLAT screening. (C) Locations of candidate GSH targeted in vitro. Blue labels: targeted clone established; green labels: no clone established.
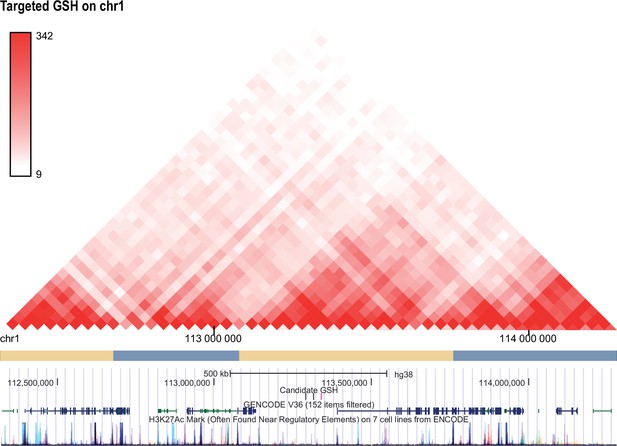
Hi-C profile for GSH candidate 1.
Screenshots of Hi-C interaction matrices from H1 hESC for targeted GSH candidate locus on chromosome 1 (113339961-113340514). TADs are indicated by the ‘pyramids’ of high interaction observed in the Hi-C matrices. UCSC genome browser track annotating the candidate GSH (targeted GSH in pink), GENCODE v36 and H3K27Ac mark from ENCODE shown below.
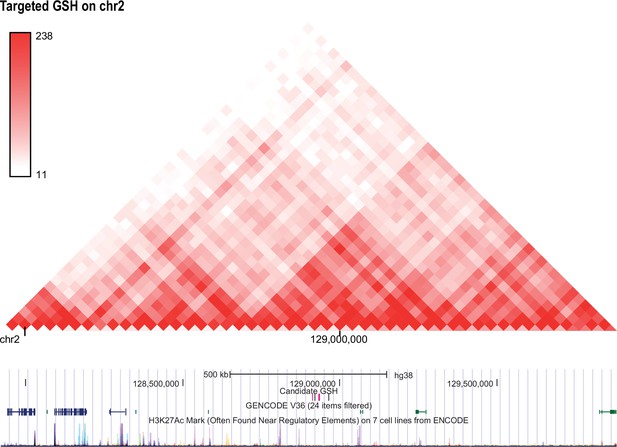
Hi-C profile for GSH candidate 2a.
As in Figure 1—figure supplement 1 but for GSH targeted GSH candidate locus on chromosome 2 (128912721-128914814).
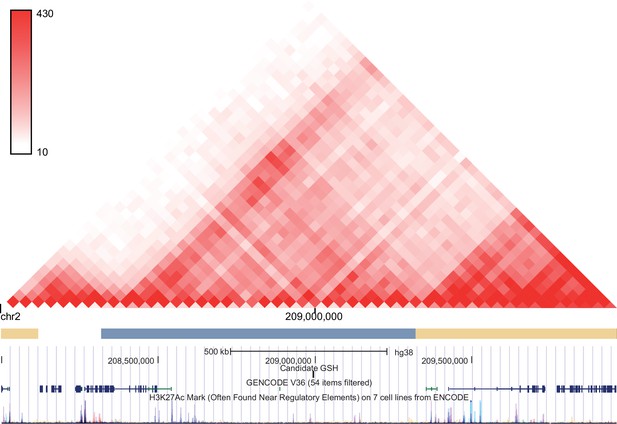
Hi-C profile for GSH candidate 2c.
As in Figure 1—figure supplement 1 but for GSH targeted GSH candidate locus on chromosome 2 (128932307-128935799).
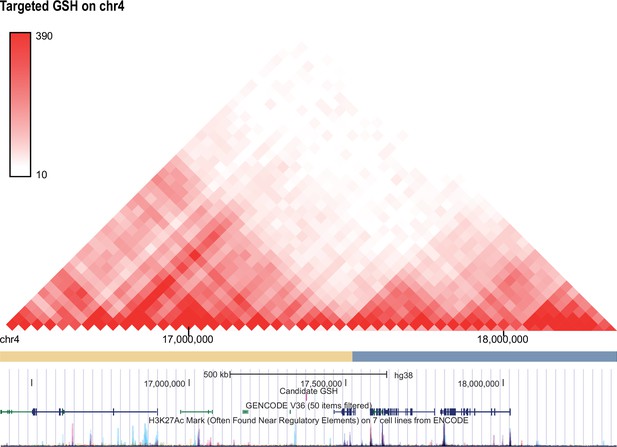
Hi-C profile for GSH candidate 4.
As in Figure 1—figure supplement 1 but for GSH targeted GSH candidate locus on chromosome 4 (17373361-17374159).
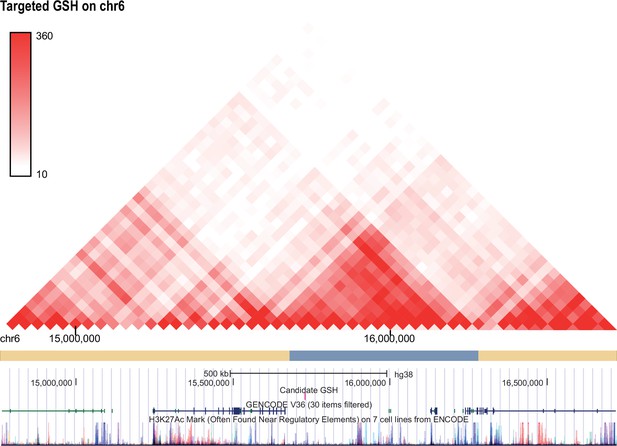
Hi-C profile for GSH candidate 6.
As in Figure 1—figure supplement 1 but for GSH targeted GSH candidate locus on chromosome 6 (15727241-15727490).
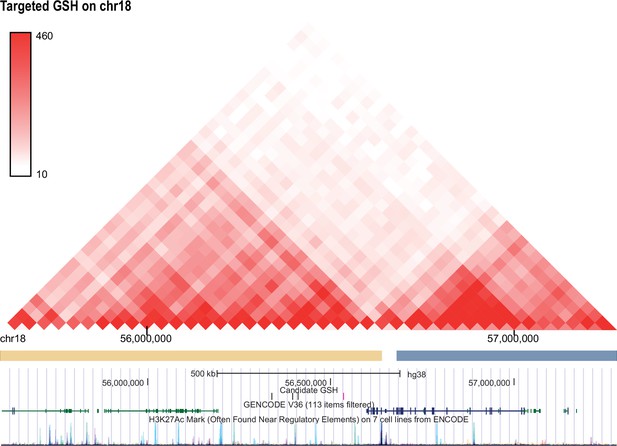
Hi-C profile for GSH candidate 18.
As in Figure 1—figure supplement 1 but for GSH targeted GSH candidate locus on chromosome 18 (56534775-56536439).
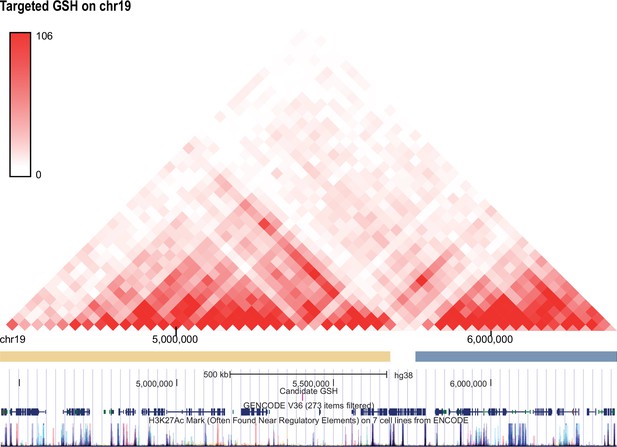
Hi-C profile for GSH candidate 19.
As in Figure 1—figure supplement 1 but for GSH targeted GSH candidate locus on chromosome 19 (5400761-5402139).
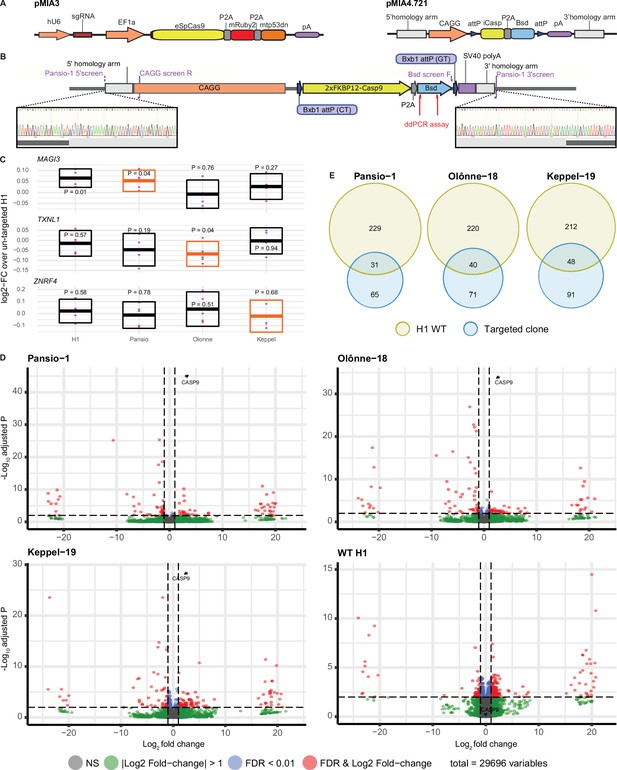
GSH targeting and transcriptomic analysis of H1 hESC.
(A) Schematic representation of CRISPR/Cas9 plasmid (pMIA3) and homology directed repair donor (pMIA4.721) used for targeting with functional components annotated. (B) Schematic of integrated landing pad expression construct. Positions of primers for junction-PCR as well as of ddPCR assay are indicated. Representative junction-PCR Sanger sequencing reads from Pansio-1 targeted clones shown in expanded view. (C) Log2-FC of mRNA expression levels against un-targeted H1 hESC samples for the nearest genes of Pansio-1, Olônne-18, and Keppel-19 candidate GSH. Evaluated samples: H1=un-targeted hESC, Pansio-1=landing pad construct integrated to Pansio-1 GSH in H1 hESC, Olônne-18=landing pad construct integrated to Olônne-18 GSH in H1 hESC and Keppel-19=landing pad construct integrated to Keppel-19 GSH in H1 hESC. Box plots representing 95% confidence intervals of mean log2-FC. Nearest gene for each GSH indicated in orange. Individual data points shown in pink with p-value for each comparison shown above. (D) Volcano plots of RNA-seq analysis against un-targeted H1 hESC. Samples analysed as in (C). Differentially expressed (DE) genes with FDR ≤0.01 and |logFC|≥1 in pink, genes with |logFC|≥1 in green, genes with FDR ≤0.01 in blue, others in grey. (E) Venn-diagrams illustrating the overlap of DE genes between un-targeted H1 hESC and the three GSH targeted H1 hESC lines.
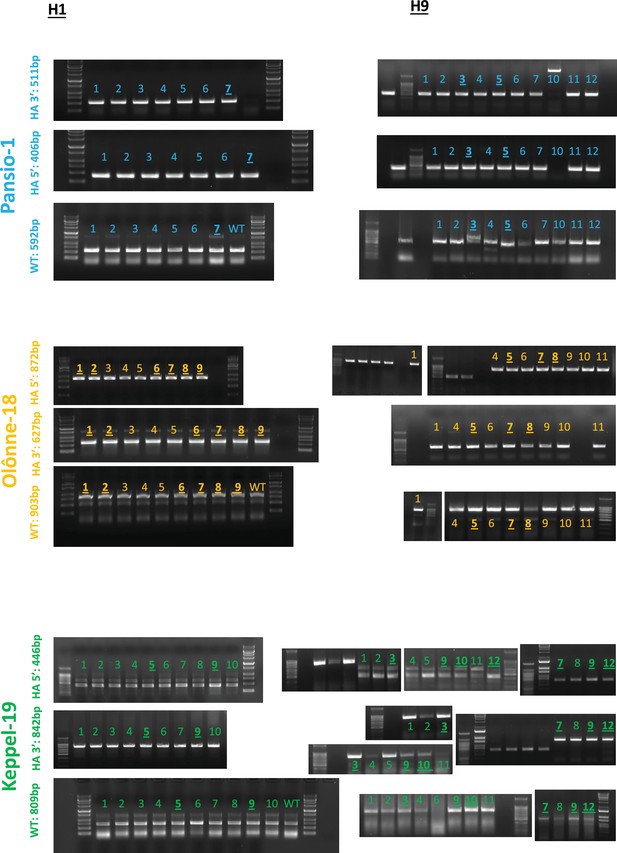
PCR gel images of junction PCR and wild type allele PCR reactions for screened clones.

Off-target screening for Pansio-1.
Sanger sequencing traces of the top five predicted gRNA off-target sites for un-targeted wild type H1 & H9 hESC and Pansio-1 GSH targeted clones. All sequencing traces are ordered as follows: H1 WT, H1 clone, H9 WT & H9 clone.

Off-target screening for Olônne-18.
As in Figure 2—figure supplement 2, but for Olônne-18.

Off-target screening for Keppel-19.
As in Figure 2—figure supplement 2, but for Keppel-19.
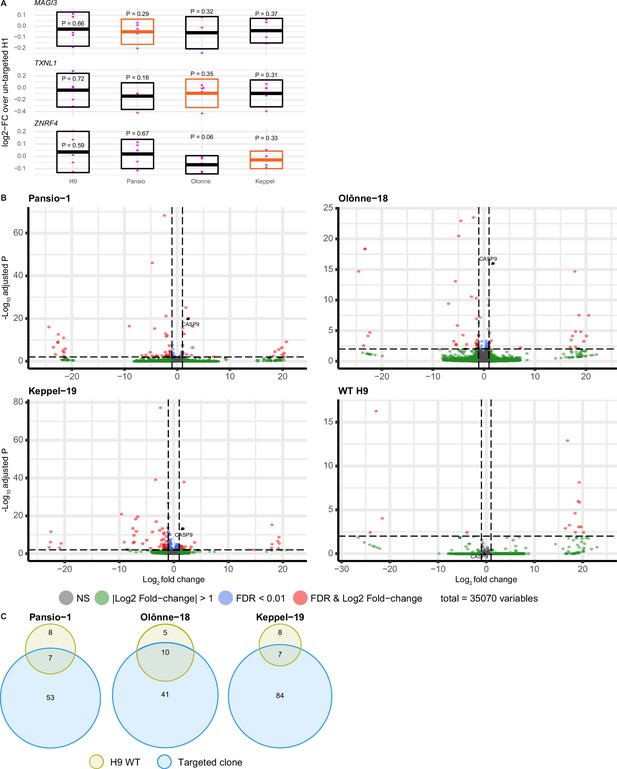
Transcriptomic analysis of GSH targeted H9 hESC.
(A) Log2-FC of mRNA expression levels against un-targeted H9 hESC samples for the nearest genes of Pansio-1, Olônne-18, and Keppel-19 candidate GSH. Evaluated samples: H9=un-targeted hESC, Pansio-1=landing pad construct integrated to Pansio-1 GSH in H9 hESC, Olônne-18=landing pad construct integrated to Olônne-18 GSH in H9 hESC and Keppel-19=landing pad construct integrated to Keppel-19 GSH in H9 hESC. Box plots representing 95% confidence intervals of mean log2-FC. Nearest gene for each GSH indicated in orange. Individual data points shown in pink with P-value for each comparison shown above. (B) Volcano plots of RNA-seq analysis against un-targeted H9 hESC. Samples analysed as in (A). Differentially expressed (DE) genes with FDR ≤0.01 and |logFC|≥1 in pink, genes with |logFC|≥1 in green, genes with FDR ≤0.01 in blue, others in grey. (C) Venn-diagrams illustrating the overlap of DE genes between un-targeted H9 hESC and the three GSH targeted H9 hESC lines.
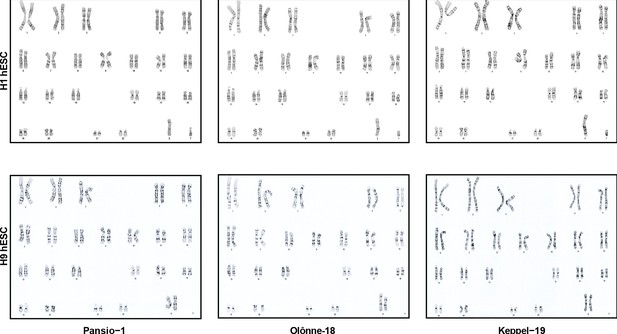
Representative images of metaphase spreads used for karyotyping the GSH targeted H1 and H9 cell lines.
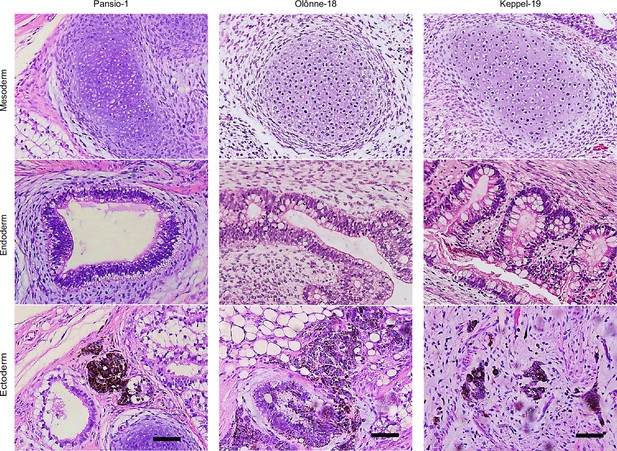
Representative images of haematoxylin and eosin stained teratoma from Pansio-1, Olônne-18, and Keppel-19 H1 cells.
Examples of mesodermal, endodermal, and ectodermal tissues are presented. Scale bar 100 µm.
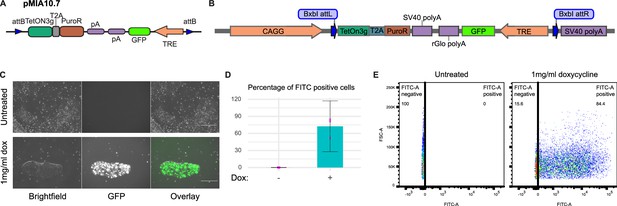
Inducible expression from GSH integrated cassette.
(A) Schematic representation of ‘all-in-one’ inducible transposon donor construct (pMIA10.7). (B) Schematic of landing pad construct with integrated ‘all-in-one’ inducible cassette. (C) Representative images of H1 Pansio-1 cells targeted with pMIA10.7 untreated or treated with 1 mg/ml doxycycline for 24 hr. Brightfield and GFP channels are shown. Scale bars for all images equal to 150 µm. (D) Bar plots representing the mean percentage of FITC-A positive cells from flow cytometry analysis of three replicate treatments as described in (A). Error bars indicate 95% confidence intervals. Individual data points shown in pink. (E) Representative scatter plots flow analysis described in (D).
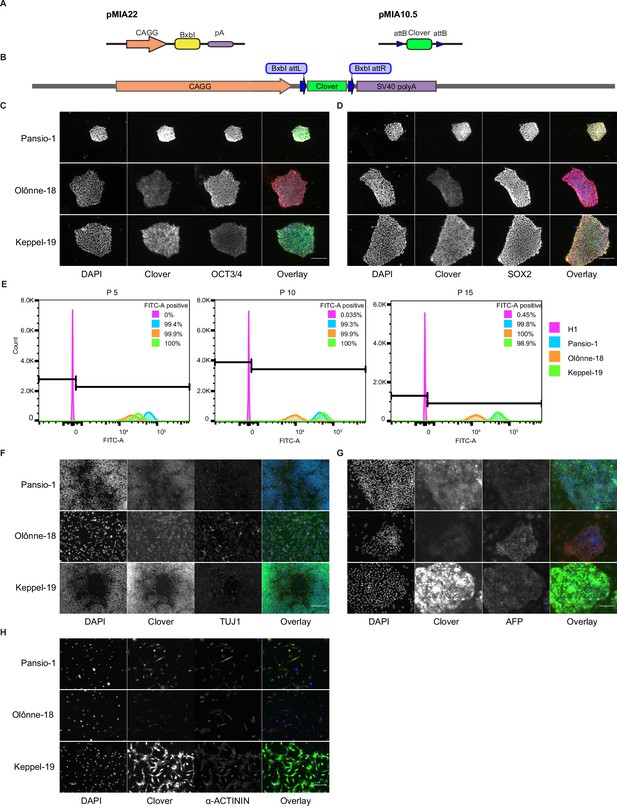
Integration and validation of transgene expression in GSH targeted H1 hESC and their differentiated progeny.
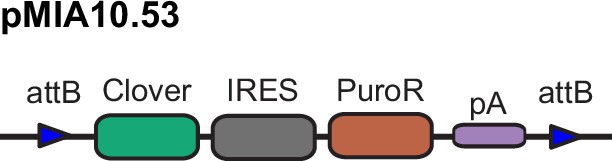
(A) Schematic representation of integrase expression construct (pMIA22) and transposon donor construct (pMIA10.5). (B) Schematic of landing pad construct with integrated Clover transgene. (C) Representative immunofluorescence images of Clover-integrated GSH H1 cells. DAPI = nuclear staining with 4′,6-diamidino-2-phenylindole, Clover = fluorescence from Clover transgene, OCT3/4=antibody staining against OCT3/4, Overlay = overlay of the three imaged channels. (D) As in (C) apart from antibody staining against SOX2. (E) Histograms of flow cytometry analysis for FITC-A channel of un-targeted H1 hESC, and the three GSH targeted hESC lines over 15 passages. Percentages of FITC-A-positive cells according to the indicated gating. (F) Representative immunofluorescence images of Clover-integrated GSH H1 cells differentiated to neuronal-like cells. Channels imaged as in (C) apart from antibody staining against TUJ1. (G) As in (F) for cells differentiated to hepatocyte-like cells, antibody staining against AFP. (H) As in (F) for cells differentiated to cardiomyocyte-like cells, antibody staining against sarcomeric α-ACTININ. Scale bars for all immunofluorescence images equal to 150 µm.
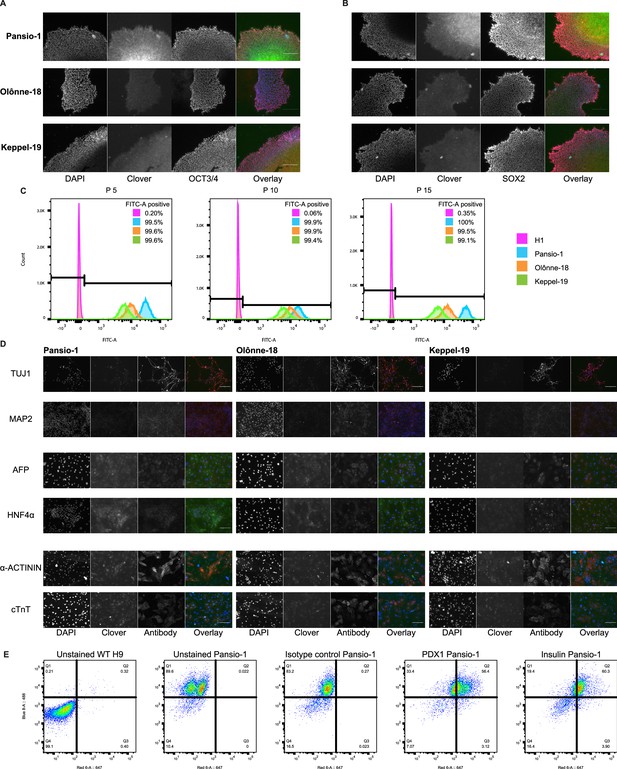
Integration and validation of transgene expression in GSH targeted H9 hESC and their differentiated progeny.
(A) Representative immunofluorescence images of Clover-integrated GSH H9 cells. DAPI = nuclear staining with 4′,6-diamidino-2-phenylindole, Clover = fluorescence from Clover transgene, OCT3/4=antibody staining against OCT3/4, Overlay = overlay of the three imaged channels. (B) As in (A) apart from antibody staining against SOX2. (C) Histograms of flow cytometry analysis for FITC-A channel of un-targeted H9 hESC, and the three GSH targeted hESC lines over 15 passages. Percentages of FITC-A positive cells according to the indicated gating. (D) Representative immunofluorescence images of Clover-integrated GSH H9 cells differentiated to neuronal cells. stained for TUJ1 and MAP2, hepatic cells, stained for AFP and HNF4α and cardiac cells stainde for sarcomeric α-ACTININ and cardiac TROPONIN-T. Channels imaged as in (C) apart from respective antibody stain. Scale bars for all immunofluorescence images equal to 150 µm. (E) Flow cytometry analysis scatter plots of hESC-derived β-like cells at D35 of β cell differentiation of H9 wild-type and Pansio-1 Clover-targeted cells. Channels used in analysis were 488 for Clover and 647 for antibody staining. PDX1, Insulin and isotype control antibodies were used.
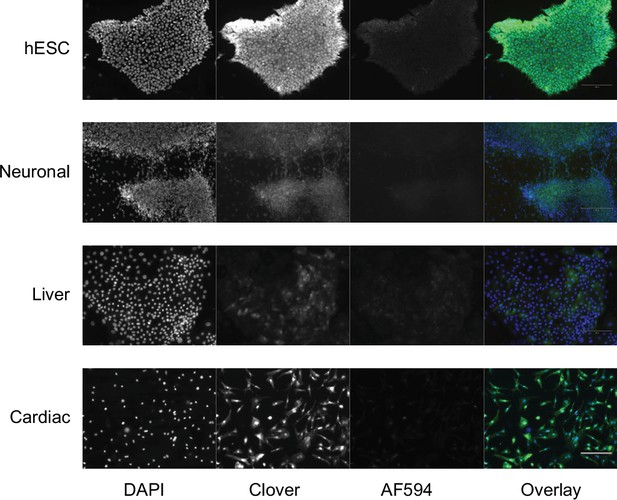
Representative images of H1 Pansio-1 line immunofluorescence staining with AF594 secondary antibody alone.
DAPI = nuclear staining with 4′,6-diamidino-2-phenylindole, Clover = fluorescence from Clover transgene, AF594=staining with secondary antibody alone, Overlay = overlay of the three imaged channels.
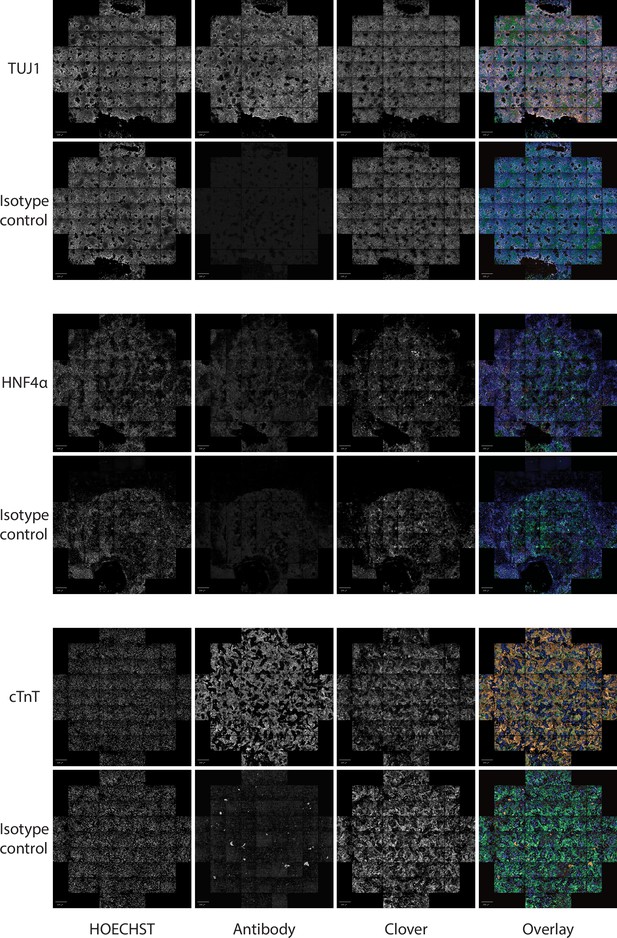
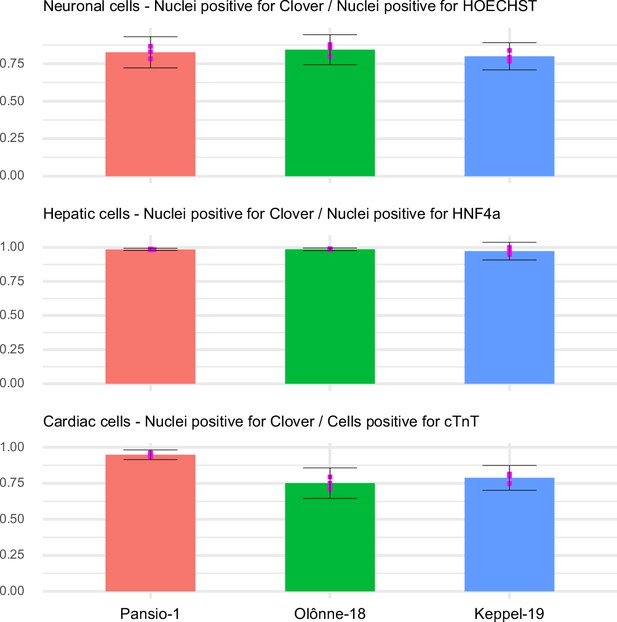
Representative images of Pansio-1 H9 cells differentiated to neuronal, hepatic, and cardiac cell types.
Staining for respective lineage markers TUJ1, HNF4α and cTnT and isotype controls is shown as well as nuclear staining with HOECHST and the channel for Clover-transgene. Images are composites of 61 individual images from HCI. Scale bars in all images equal to 500 µm.
Tables
Coordinates of candidate GSH, their associated active chromosome regions & housekeeping gene, and BLAT score against the most similar region.
| Chromosome | Start | End | Width | Active region start | Active region end | Housekeeping gene | BLAT-score |
|---|---|---|---|---|---|---|---|
| 1 | 113289036 | 113289342 | 307 | 113000001 | 114000000 | HIPK1 | 0.21 |
| 1 | 113314841 | 113318369 | 3529 | 113000001 | 114000000 | HIPK1 | 0.18 |
| 1 * | 113339961 | 113340514 | 554 | 113000001 | 114000000 | HIPK1 | 0.27 |
| 2 * | 128912721 | 128914814 | 2094 | 128000001 | 129000000 | UGGT1 | 0.08 |
| 2 | 128918961 | 128919839 | 879 | 128000001 | 129000000 | UGGT1 | 0.16 |
| 2* | 128932307 | 128935799 | 3493 | 128000001 | 129000000 | UGGT1 | 0.15 |
| 2 | 128963272 | 128965759 | 2488 | 128000001 | 129000000 | UGGT1 | 0.44 |
| 2 | 208992998 | 208997459 | 4462 | 208000001 | 209000000 | PIKFYVE | 0.28 |
| 4* | 17373361 | 17374159 | 799 | 17000001 | 18000000 | MED28 | 0.04 |
| 5 | 131058585 | 131058947 | 363 | 131000001 | 132000000 | FNIP1 | 0.41 |
| 5 | 148753741 | 148757219 | 3479 | 148000001 | 149000000 | FBXO38 | 0.10 |
| 6 * | 15727241 | 15727490 | 250 | 15000001 | 16000000 | DTNBP1 | 0.47 |
| 7 | 4314741 | 4315279 | 539 | 4000001 | 5000000 | FOXK1 | 0.15 |
| 7 | 4321017 | 4323839 | 2823 | 4000001 | 5000000 | FOXK1 | 0.09 |
| 7 | 4328040 | 4329659 | 1620 | 4000001 | 5000000 | FOXK1 | 0.21 |
| 7 | 4353504 | 4354219 | 716 | 4000001 | 5000000 | FOXK1 | 0.32 |
| 7 | 4454808 | 4456201 | 1394 | 4000001 | 5000000 | FOXK1 | 0.17 |
| 8 | 23945241 | 23945819 | 579 | 23000001 | 24000000 | R3HCC1 | 0.34 |
| 8 | 23986981 | 23988319 | 1339 | 23000001 | 24000000 | R3HCC1 | 0.06 |
| 8 | 23999628 | 24001194 | 1567 | 23000001 | 24000000 | R3HCC1 | 0.02 |
| 18 | 56339813 | 56340245 | 433 | 56000001 | 57000000 | TXNL1 | 0.06 |
| 18 | 56396821 | 56397319 | 499 | 56000001 | 57000000 | TXNL1 | 0.07 |
| 18 | 56410681 | 56411039 | 359 | 56000001 | 57000000 | TXNL1 | 0.14 |
| 18 * | 56534775 | 56536439 | 1665 | 56000001 | 57000000 | TXNL1 | 0.16 |
| 19 * | 5400761 | 5402139 | 1379 | 5000001 | 6000000 | SAFB | 0.18 |
-
*
= GSH shortlisted for in vitro validation.
| Olônne-18 GSH targeted H1 hESC |
| Keppel-19 GSH targeted H1 hESC |
| Control targeted H1 hESC (CRISPR and HDR of an expression cassette at a non-GSH locus) |
| Independent untargeted H1 hESC |
Additional files
-
Supplementary file 1
Output of safe and active searches and their overlap.
(1) Coordinates that pass safe site filters. (2) Regions that pass active filters. (3) Overlap of safe sites and active regions.
- https://cdn.elifesciences.org/articles/79592/elife-79592-supp1-v2.xlsx
-
Supplementary file 2
Oligos, primers and antibodies used, and details of beta cell differentiation.
(1) gRNAs used for targeting. (2) Primers used for plasmid construction. (3) Primers used for junction PCR and WT allele screening. (4) Primers used for predicted off-target screening. (5) Antibodies used for immunofluorescence staining and flow cytometry. (6) Details for pancreatic differentiation.
- https://cdn.elifesciences.org/articles/79592/elife-79592-supp2-v2.xlsx
-
Supplementary file 3
GSH targeted hESC screening.
(1) Summary of GSH targeted hESC clones screening. (2) ddPCR copy number analysis.
- https://cdn.elifesciences.org/articles/79592/elife-79592-supp3-v2.xlsx
-
Supplementary file 4
Source data for qPCR results.
(1) qPCR data source file for Figure 2C. (2) qPCR data source file for Figure 2–figure supplement 5A.
- https://cdn.elifesciences.org/articles/79592/elife-79592-supp4-v2.xlsx
-
Supplementary file 5
RNAseq analysis of GSH targeted H1 clones.
(1) Differentially expressed genes |logFC|≥1 and FDR ≤0.01. (2) Functional enrichment analysis results from g:GOSt.
- https://cdn.elifesciences.org/articles/79592/elife-79592-supp5-v2.xlsx
-
Supplementary file 6
RNAseq analysis of GSH targeted H9 clones.
(1) Differentially expressed genes |logFC|≥1 and FDR ≤0.01. (2) Functional enrichment analysis results from g:GOSt.
- https://cdn.elifesciences.org/articles/79592/elife-79592-supp6-v2.xlsx
-
Supplementary file 7
HCI data analysis source file for Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/79592/elife-79592-supp7-v2.xlsx
-
Supplementary file 8
Percentage of FITC-A positive cells from H1 Pansio inducible cells source file for Figure 2—figure supplement 8.
- https://cdn.elifesciences.org/articles/79592/elife-79592-supp8-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/79592/elife-79592-mdarchecklist1-v2.docx