Cellular reprogramming with ATOH1, GFI1, and POU4F3 implicate epigenetic changes and cell-cell signaling as obstacles to hair cell regeneration in mature mammals
Figures
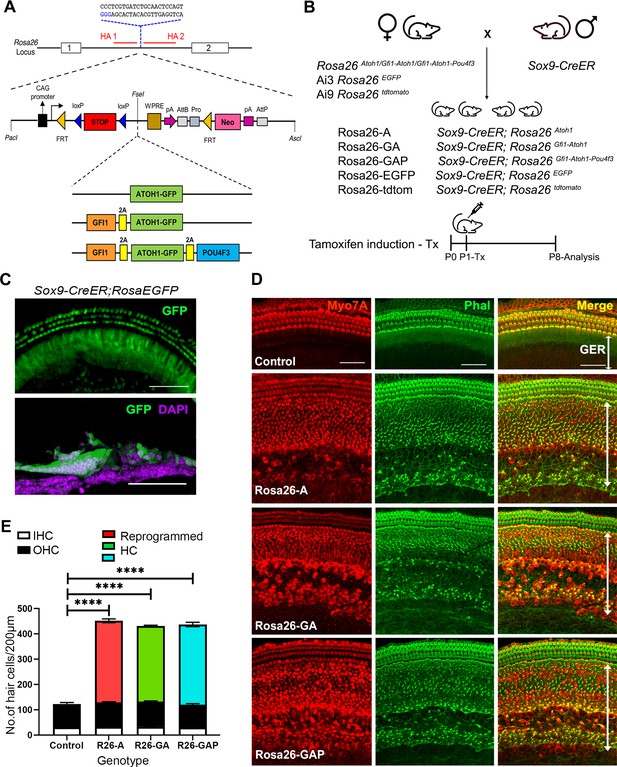
Non-sensory cells of the neonatal mouse cochlea can be efficiently reprogrammed to a hair cell fate with combinations of Atoh1, Gfi1 and Pou4f3 transcription factors.
(A) Schematic representation of the strategy to target the Rosa26 locus to generate three conditional mouse lines for transcription factor overexpression. A modified Ai3 vector (Madisen et al., 2010) was used to target different transcription factor combinations to the ROSA26 locus. ES cell targeting was enhanced using CRISPR-mediated cleavage with a sgRNA sequence targeting the ROSA26 locus between the targeting homology arms (HA1 and 2). The transcription factor coding sequences were separated by GSG-T2A self-cleaving peptide sequences to generate multiple proteins from a single primary transcript. (B) Mating schemes to express different transcription factor combinations in the cochlea. The Sox9-CreER mouse was bred to the three Rosa26 overexpression lines and reporters to generate experimental animals of the following genotypes: Rosa26-A (Sox9-CreER; RosaAtoh1GFP), Rosa26-GA (Sox9-CreER; RosaGfi1-Atoh1GFP), Rosa26-GAP (Sox9-CreER; RosaGfi1-Atoh1GFP-Pou4f3), Rosa26EGFP (Sox9-CreER; Rosa26EGFP), and Rosa26-tdtomato (Sox9-CreER; Rosa26tdtomato). Animals received tamoxifen (25 mg/kg body weight) at P1 and were sacrificed at P8. (C) GFP reporter expression obtained from mating Sox9-CreER mice with Rosa26EGFPmice. Fluorescence is seen in all GER cells in whole mounts and 16 µm sections. Images show GFP (green) and a DAPI nuclear stain (magenta). Scale bar: 50 µm. (D) Large numbers of reprogrammed hair cells (white arrows) are seen in P8 cochleae extending from the organ of Corti to the interdental cell region in Rosa-A, Rosa-GA, and Rosa-GAP mice, revealed by immunostaining for Myosin VIIA (red) and Phalloidin (green). (E) Quantification of hair cells in the P8 reprogrammed cochleae. The number of Myosin VIIA + cells per 200 µm length of the cochlea was measured (IHC – Inner hair cells, OHC – Outer hair cells). Compared to controls, significant numbers of reprogrammed cells (300–320 per 200 µm) were seen in Rosa26-A, Rosa26-GA and Rosa26-GAP genotypes (n=3 per genotype). An unpaired t-test was performed to compare hair cell numbers between genotypes. The significant differences are represented. ****P<0.00001. Data are presented as mean ± SEM.
-
Figure 1—source data 1
Overexpression of the ROSA-A, ROSA-GA, and ROSA-GAP transcription factor combinations from the Rosa26 locus was verified by culturing ES cells used to generate the three lines of mice with membrane soluble TAT-Cre.
Western blotting was performed after 48h with antibodies specific to ATOH1, GFP, POU4F3 , and GAPDH as a loading control. The raw blots are shown.
- https://cdn.elifesciences.org/articles/79712/elife-79712-fig1-data1-v1.zip
-
Figure 1—source data 2
Overexpression of the ROSA-A, ROSA-GA, and ROSA-GAP transcription factor combinations from the Rosa26 locus was verified by culturing ES cells used to generate the three lines of mice with membrane soluble TAT-Cre.
Western blotting was performed after 48h with antibodies specific to ATOH1, GFP, POU4F3 , and GAPDH as a loading control. The raw blots are shown with labels attached to indicate the relevant bands and the conditions used.
- https://cdn.elifesciences.org/articles/79712/elife-79712-fig1-data2-v1.zip
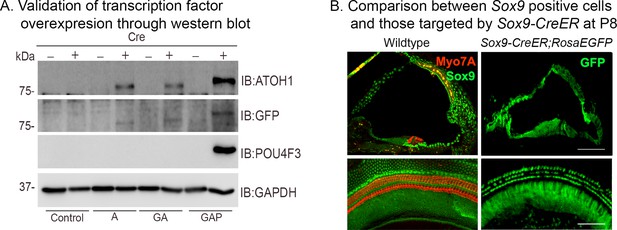
Validation of transcription factor expression in ES cell lines used to generate ROSA26-targeted mice, and cochlear expression of the Sox9-CreER line.
(A) Overexpression of the Rosa-A, Rosa-GA, and Rosa-GAP transcription factor combinations from the Rosa26 locus was verified by culturing ES cells used to generate the three lines of mice with membrane-soluble TAT-Cre. Western blotting was performed after 48 hr with antibodies specific to ATOH1 (lane 1), GFP (lane 2), POU4F3 (lane 3), and GAPDH (loading control; lane 4). (B) Validation of the Sox9-CreER transgenic mice by crossing them to the Ai3 ROSAEGFP reporter line. The left panels show immunostaining for SOX9 protein (green) and Myosin VIIA (red) in the P8 control. The right panels show EGFP reporter expression (green) in the Rosa26EGFP cochlea at P8 following tamoxifen administration at P1. Images show 16 µm sections and 200 µm lengths of cochlear whole mounts. Scale bar: 50 µm.
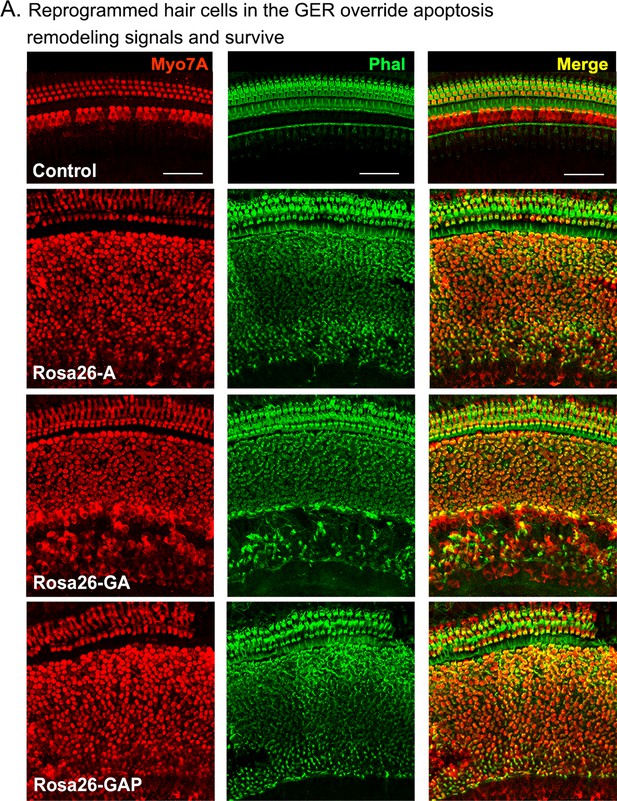
Reprogrammed hair cells at neonatal ages can survive until at least P15.
Reprogrammed hair cells obtained by administering tamoxifen at P1 survive until at least P15. They are not subject to the apoptosis-mediated GER remodeling event that normally occurs between day 7 and day 15 (Peeters et al., 2015). Images show 200 µm lengths of P15 control and experimental cochleae immunostained for Myosin VIIA (red) and fluorescently labeled phalloidin (green).
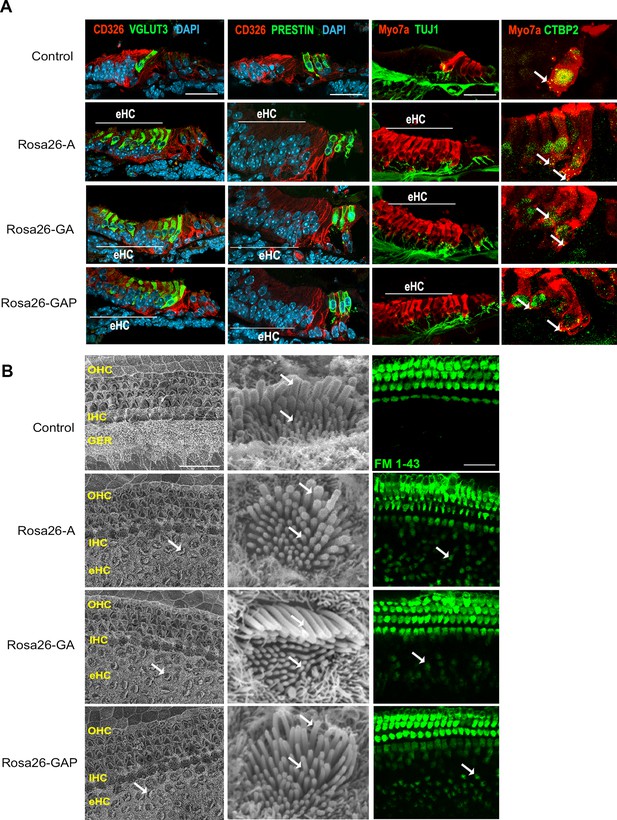
Day 8 reprogrammed hair cells are inner hair cell-like, innervated, form ribbon synapses, possess stereociliary bundles, and show evidence of mechanotransduction activity.
(A) Control and reprogrammed cochleae were immunostained for an inner hair cell-specific marker, VGLUT3, an outer hair-cell-specific marker, PRESTIN, a GER specific marker, CD326/EpCAM, a neuronal marker TuJ1, a ribbon synapse-specific marker, and a hair cell marker, Myosin VIIA. The reprogrammed hair cell region is indicated (white line; eHC – ectopic hair cells). Arrows indicate individual ribbon synapse structures observed in the cell bodies of hair cells. Scale bar: 50 µm. (B) Scanning electron micrographs (SEM) of the control and reprogrammed cochleae (500 X; scale bar- 50 µm). OHC: Outer hair cell region, IHC: Inner hair cell region, GER: greater epithelia ridge region. Arrows indicate individual reprogrammed hair cells in the GER. SEM mages at 10,000 X show the arrangement of stereocilia in control and reprogrammed hair cells. Arrows indicate variations in the length of individual stereocilia which are similar between control and reprogrammed hair cells. The presence of mechanotransduction activity in the induced hair cells was tested by uptake of FM1-43 dye after 10 s of exposure. Reprogrammed hair cells in the GER take up the dye to a lesser extent than endogenous hair cells (arrows), but more than controls, indicating some mechanotransduction channel activity.
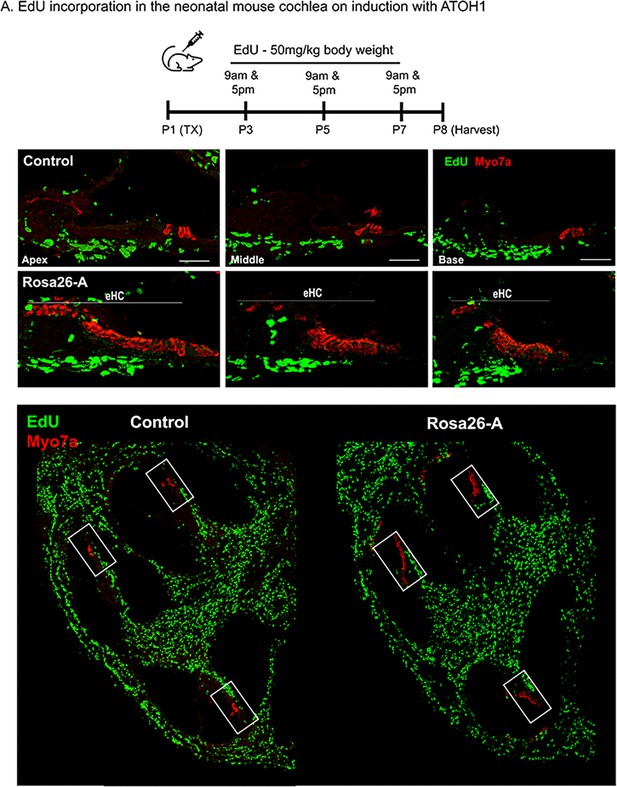
Transcription factor induction at P1 does not influence cell proliferation in the reprogrammed cochlea.
Control and Rosa26-A animals received tamoxifen (25 mg/kg body weight) at P1, EdU (50 mg/kg body weight) at regular intervals (P3, P5, P7 – twice per day). Animals were sacrificed at P8 for histological analysis. (B) EdU incorporation (green) was observed only in cells of the spiral ganglion neuron and basal lamina regions, but not the organ of Corti (apex, middle, and base) in control and Rosa26-A animals. Images are from stained 16 µm sections with Myosin VIIA (red) indicating hair cells. No Edu+/ Myosin VIIA + hair cells can be seen in either control or experimental animals. Scale bar: 50 µm. (C) Lower power images of the temporal bones analyzed at P8 show that in addition to the SGN and basal lamina regions, extensive proliferation is observed in most parts of the temporal bone except for the cochlea epithelium.
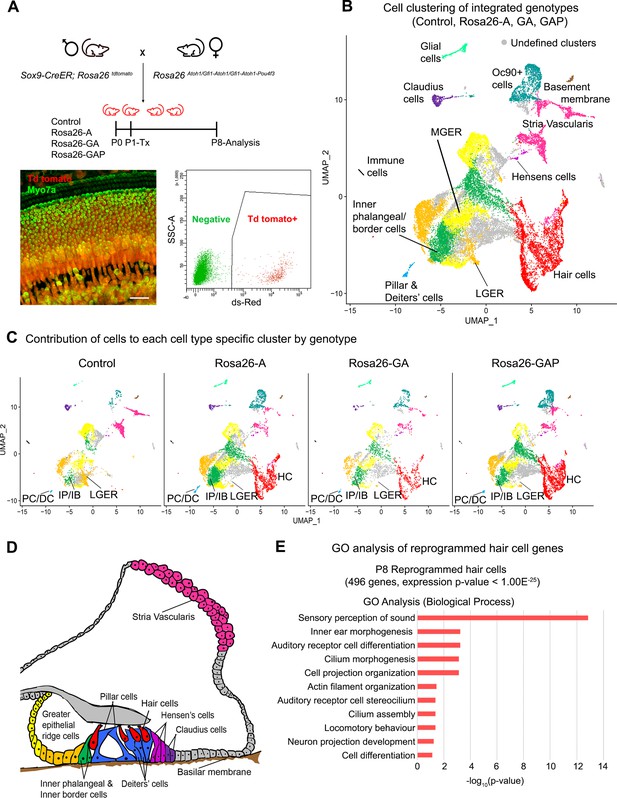
Single-cell transcriptomic analysis of control and reprogrammed P8 GER cells confirm the presence of a large number of reprogrammed cells that possess hair cell-like gene signatures.
(A) Mice carrying a Sox9-CreER allele, a ROSA26tdTomato reporter allele, and a modified ROSA26 allele containing reprogramming factors received tamoxifen at day 1 and tdTomato + cells were purified by FACS sorting one week later. A representative whole-mount image of a day 8 cochlea shows reprogrammed hair cells and the tdTomato reporter (scale bar: 50 µm). A representative FACS plot of dissociated cochlear cells is shown. tdTomato + cells were collected and analyzed by scRNA-seq. (B) UMAP plot for cells integrated and analyzed from all four genotypes (Control, Rosa26-A, Rosa26-GA, and Rosa26-GAP) purified in (A). Each identified cluster has been labeled and the anatomical location of each cluster is shown color-coded in panel (D). (C) Genotype-wise UMAP plots highlighting the contribution of cells from each genotype in every identified cluster. The GER cluster (particularly LGER) in the control is prominent and the hair cell cluster is present only in the reprogrammed cochlear genotypes as the Sox9-CreER line does not label endogenous hair cells. IP/IB – Inner phalangeal/border, PC/DC – Pillar/Deiters’ cells, HC – Hair cells, LGER – Lateral Greater epithelial ridge (D) Schematic of an organ of Corti cross-section at P8. Unique cell types identified in the scRNA-seq clustering have been color-coded and correspond to the cluster colors in (B) and (C). (E) Gene ontology analysis of the top differentially expressed genes in the reprogrammed hair cell-like cells from all three conditions (with respect to their expression in other cell clusters). A list of ~500 significantly expressed genes (P<1.00E–25) was analyzed and GO terms (Biological process; -log10 (p-value)>1) are shown.
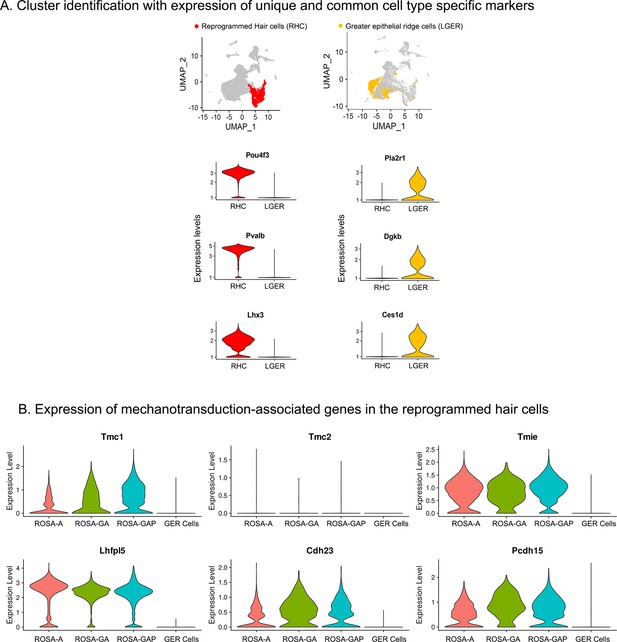
Examples of hair cell and LGER marker genes confirm cluster assignments in P8 cochlear cell clusters.
(A) UMAP plots (all four genotypes integrated) highlighting reprogrammed hair cell (RHC; red) and lateral GER cell (LGER; yellow) clusters. Violin plots show normalized log-transformed expression values for hair cell marker genes Pou4f3, Pvalb, Lhx3, and LGER marker genes Pla2r1, Dgkb, Ces1d in the two clusters of interest. (B) Violin plots showing normalized expression of six components of the hair cell mechanotransduction channel in hair cells derived in the three reprogramming conditions at P8, compared to wild type GER cells. The plots show normalized log-transformed expression values.
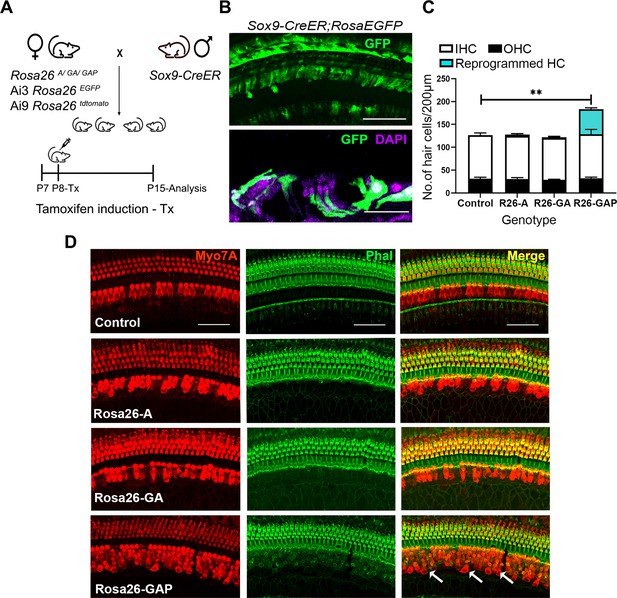
Gfi1, Atoh1, Pou4f3, but not Atoh1 or Gfi1+Atoh1, can reprogram 8-day-old GER cells into hair cell-like cells.
(A) Mating scheme for the targeting of transcription factors to the greater epithelial ridge and all supporting cells. The Sox9-CreER mouse is bred to the three Rosa26 overexpression lines in a similar manner to Figure 1A. Animals received tamoxifen (25 mg/kg body weight) at P8 and were sacrificed at P15. (B) GFP reporter expression in some lateral GER cells and all supporting cells detected by immunostaining in the organ of Corti of the Sox9-CreER; Rosa26EGFP cochlea. Images show detection of GFP (green) and nuclear stain, DAPI (magenta). Scale bar: 50 µm. (C) Quantification of hair cells in the P15 reprogrammed cochleae. The number of Myosin VIIA + cells per 200 µm length of the organs of Corti from control, Rosa26-A, Rosa26-GA and Rosa26-GAP genotypes (n=3 per genotype) are represented. Rosa-GAP mice show approximately 50–60 ectopic hair cells, whereas Rosa-A and Rosa-GA show less than 5 ectopic cells per 200 µm. An unpaired t-test was performed to compare hair cell numbers between genotypes. Significant differences are represented. **p<0.001. Data are presented as mean ± SEM. (D) Rosa26-GAP mice can reprogram GER cells to hair cell-like cells. Immunostaining for Myosin VIIA (red) and Phalloidin (green) in the P15 cochleae (whole-mount organ of Corti - 200 µm length) of control, Rosa26-A, Rosa26-GA, and Rosa26-GAP mice. Arrows point to the GER region in the Rosa26-GAP cochlea, where many reprogrammed hair cells are observed.
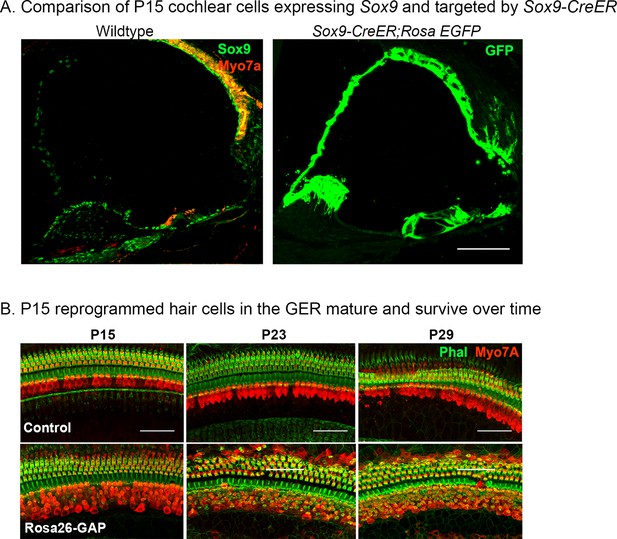
Validation of Sox9-CreER activity in the cochlea from P8 to P15 and survival of reprogrammed hair cells from P15 to P29.
(A) Immunostaining for SOX9 (green) and Myosin VIIA (red) in the P8 control cochlea shows widespread SOX9 protein expression throughout the cochlear duct (left panel). The right panel shows Sox9-CreER; ROSAEGFP mice that received tamoxifen at P8 and analyzed at P15. Most of the cochlear duct is labeled, apart from parts of the GER that undergo remodeling between P7 and P15. Images show 16 µm sections at 20 x. Scale bar: 50 µm. (B) Reprogrammed Rosa26-GAP hair cells obtained by administering tamoxifen at P8 show ectopic hair cells in the LGER region at P15, P23, and P29. By P29, most of the ectopic hair cells have organized actin bundles, revealed by enhanced phalloidin staining in the ectopic Myosin VIIA cells. Images show 200 µm lengths of P15, P23, P29 control, and Rosa26-GAP cochleae immunostained for Myosin VIIA (red) and fluorescently labeled phalloidin (green).
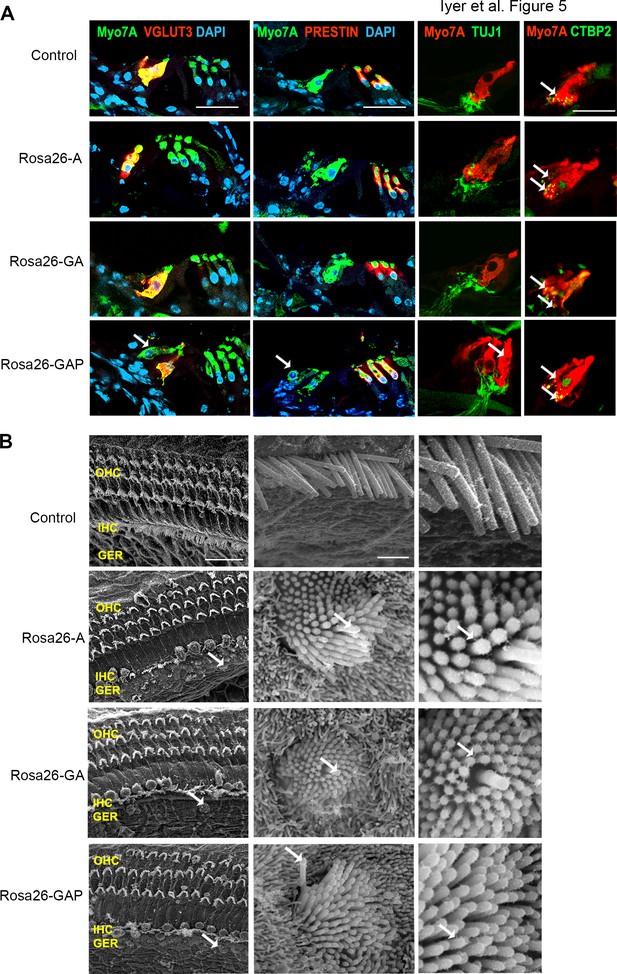
Postnatal (P15) Rosa26-GAP reprogrammed hair cells are innervated and form ribbon synapses, but possess immature stereociliary bundles.
(A) Control and reprogrammed cochleae immunostained for the inner hair cell-specific marker, VGLUT3, outer hair-cell-specific marker, PRESTIN, neuronal marker TuJ1, ribbon synapse-specific marker CTBP2 and hair cell marker, Myosin VIIA. Arrows point to reprogrammed hair cells that are positive for Myosin VIIA in the Rosa26-GAP condition, innervation of the reprogrammed hair cells, and individual ribbon synapse structures observed in the cell bodies of endogenous and reprogrammed hair cells (Rosa-GAP). Images show detection of described markers on a 16 µm section of the organ of Corti (control and reprogrammed). Scale bar: 50 µm. (B) Scanning electron micrographs of reprogrammed hair cells from all three genotypes show similar hair cell-like structural features. Scanning electron micrographs (SEM) of the control and reprogrammed cochleae at 1000 X (scale bar- 50 µm). Arrows indicate individual reprogrammed hair cells. OHC: Outer hair cell region, IHC: Inner hair cell region, GER: greater epithelia ridge region. SEM images at 50,000 X show the kinocilium on individual hair cells and side link structures connecting hair cell stereocilia as indicated by arrows.
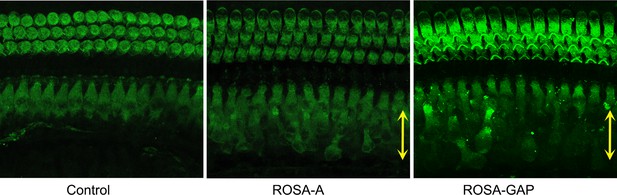
FM1-43X labeling of P15 reprogrammed hair cells.
The presence of mechanotransduction activity in the induced hair cells was tested by uptake of FM1-43X dye. Temporal bones were dissected and fixable FM1-43X dye was perfused into the cochlea of samples, followed rapidly by flushing of the cochlea with 4% paraformaldehyde. Reprogrammed hair cells in the GER (yellow arrows) from Rosa-A and Rosa-GAP mice take up the dye to a lesser extent than endogenous hair cells, but more than controls, indicating some mechanotransduction channel activity. Similar results were seen with Rosa-GA mice (not shown).
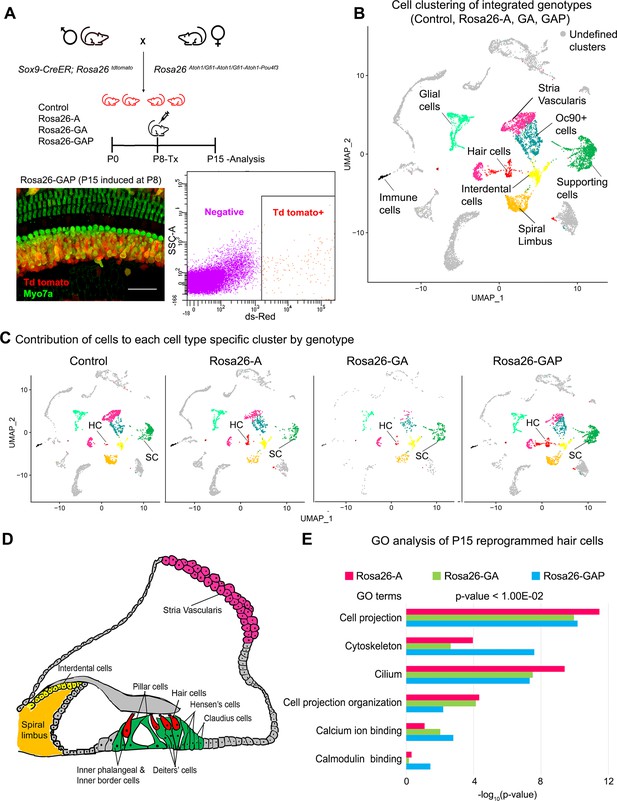
Single-cell transcriptomic analysis of control and reprogrammed cochlear cells at P15.
(A) FACS-based enrichment of cochlear cells targeted for transcription factor overexpression. The breeding scheme with an experimental timeline is described followed by a representative whole mount image (bar: 50 µm) from the Rosa26-GAP cochlea. The scheme is similar to that shown in Figure 3, except that tamoxifen is injected to induce reprogramming at 8 days after birth, followed by analysis at day 15. All cells targeted for TF overexpression are tdTomato positive (red), including reprogrammed hair cells (green). A representative FACS scatter plot of dissociated induced cochlear cells is shown. (B) UMAP plot for cells integrated and analyzed by scRNA-seq from all four genotypes (control, Rosa26-A, Rosa26-GA, and Rosa26-GAP) purified in (A). Each identified cluster has been labeled. (C) Genotype-wise UMAP plots highlighting the contribution of cells from each genotype in every identified cluster. (D) Schematic of the organ of Corti cross-section at P15. Unique cell types have been color-coded and correspond to cluster colors in (B) and (C). (E) Gene ontology analysis of the top differentially expressed genes in reprogrammed hair cells from each condition (with respect to their expression in other cell clusters for that genotype). A list of ~200 significantly expressed genes (p<1.00E–02) was analyzed and GO terms (Biological process, Cellular component, Molecular function; -log10 (p-value)>1) are represented.
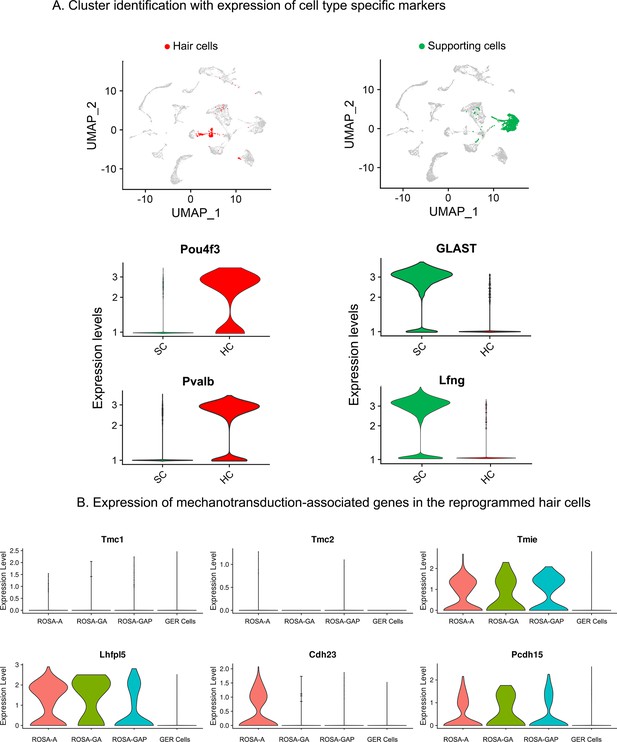
Examples of hair cell and supporting cell marker genes confirm cluster assignments in P15 cochlear cell clusters.
(A) UMAP plots (all four genotypes integrated) highlighting the hair cell (HC; red) and supporting cell (SC; green) clusters. Violin plots show normalized log-transformed expression values for hair cell marker genes Pou4f3, Pvalb, and SC marker genes GLAST, Lfng in the two clusters. (B) Violin plots showing normalized expression of six components of the hair cell mechanotransduction channel in hair cells derived in the three reprogramming conditions at P15, compared to wild-type GER cells. The plots show normalized log-transformed expression values.
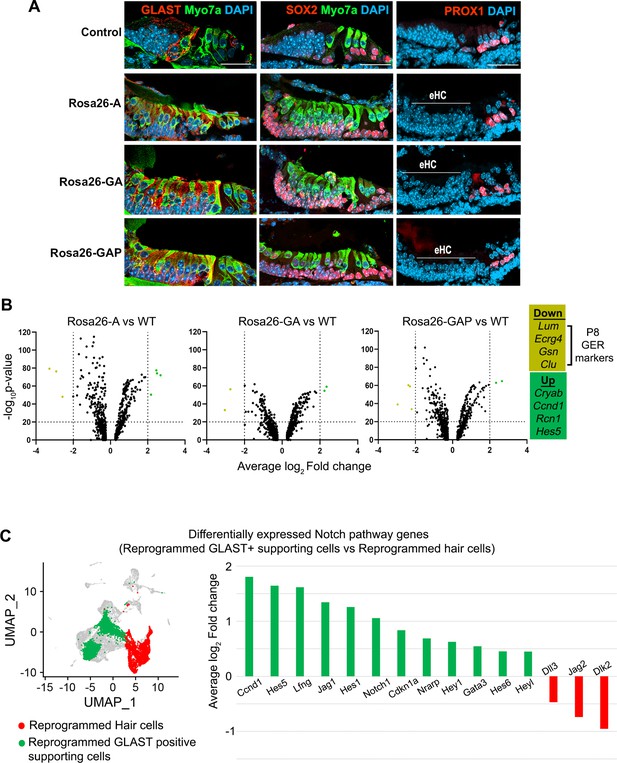
GLAST+, SOX2+supporting cells are induced adjacent to reprogrammed hair cells in the GER.
(A) Control and reprogrammed cochleae immunostained for markers specific to inner phalangeal and border cells (GLAST), a general supporting cell marker (SOX2), pillar and Deiters’ cells (PROX1), and the hair cell marker, Myosin VIIA. The reprogrammed hair cell region is indicated (line; eHC – ectopic hair cells). Images show 16 µm sections of the organ of Corti (control and reprogrammed). Scale bar: 50 µm. (B) Differentially expressed genes from our P1-P8 scRNA-seq experiments in reprogrammed GLAST + supporting cells are compared to control inner phalangeal/border cells. Volcano plots show common upregulated genes Cryab, Ccnd1, Rcn1, Hes5, and downregulated genes Lum, Ecrg4, Gsn, Clu (GER specific genes). (C) Notch pathway genes are upregulated in the reprogrammed GLAST + cells and hair cells in response to transcription factor induction at P8. UMAP plot of cells integrated from all genotypes is shown with the reprogrammed hair cells (red) and GLAST positive supporting cells (green). Average log10 fold change in the expression of supporting cell-specific Notch genes – Ccnd1, Hes5, Lfng, Jag1, Hes1, Notch1, Cdkn1a, Nrarp, Hey1, Gata3, Hes6, Heyl and hair-cell-specific Notch genes – Dll3, Jag2, Dlk2 is represented.
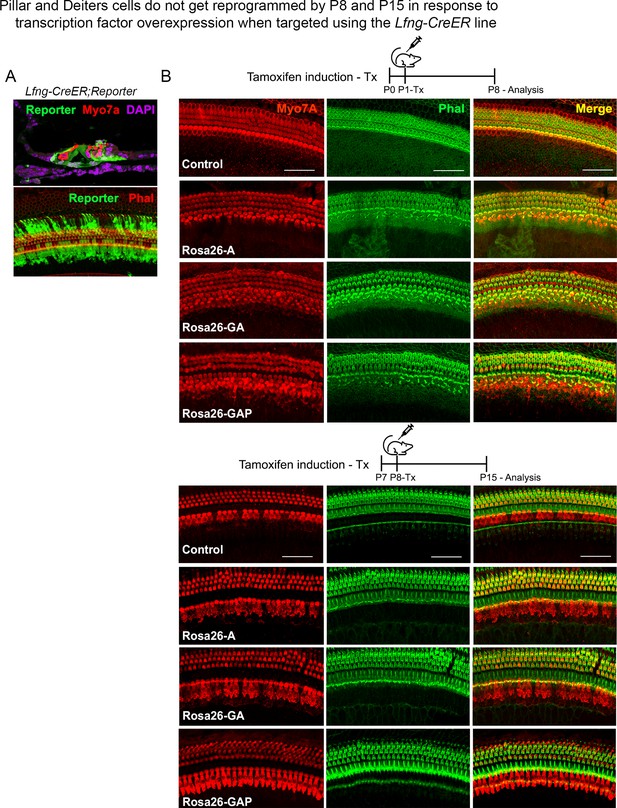
Use of the a second Cre line, Lfng-CreER, confirms that pillar and Deiters’ cells do not get reprogrammed into hair cell-like cells in response to transcription factor overexpression.
(A) Lfng-CreER mice crossed to ROSAEGFP reporter mice show most supporting cells are labeled when induced with tamoxifen at P1 and analyzed at P8. A representative image of a 200 µm cochlear length is shown with the reporter (green), hair cells (red – Myosin VIIA, upper panel; and phalloidin, lower panel), and DAPI (magenta). (B) Control mice and Rosa26-A, Rosa26-GA, and Rosa26-GAP received tamoxifen at P1 or P8 and were analyzed 7 days later. Samples were labeled for Myosin VIIA (red) and Phalloidin (green). 200 µm lengths are shown in all conditions. No ectopic hair cells are seen in the pillar or Deiters’ cell regions in any condition. Scale bar: 50 µm.
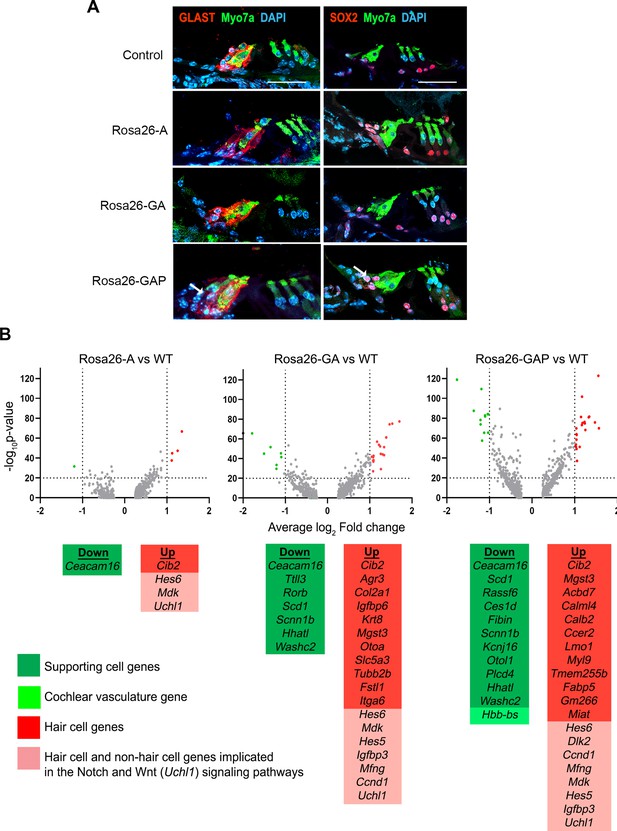
Rosa26-GAP reprogramming from day 8 to day 15 induces ectopic GLAST + supporting cells and upregulates some hair cell genes in endogenous supporting cells.
(A) Control and reprogrammed cochleae immunostained for markers specific to inner phalangeal and border cells (GLAST), a general supporting cell marker (SOX2), and a hair cell marker, Myosin VIIA. A reprogrammed GLAST positive supporting cell in Rosa26-GAP condition is indicated with arrows. Images show a 16 µm section of the organ of Corti (control and reprogrammed). Scale bar: 50 µm. (B) Single-cell RNA seq analysis of supporting cells under reprogramming conditions (induction at day 8, analysis at day 15). Volcano plots show that several hair cell-specific genes and Notch pathway genes are upregulated by reprogramming factors, while several supporting cell genes are downregulated.
-
Figure 8—source data 1
List of genes downregulated in supporting cells in response to transcription factor reprogramming at P15.
- https://cdn.elifesciences.org/articles/79712/elife-79712-fig8-data1-v1.docx
-
Figure 8—source data 2
List of genes upregulated in supporting cells in response to transcription factor reprogramming at P15.
- https://cdn.elifesciences.org/articles/79712/elife-79712-fig8-data2-v1.docx
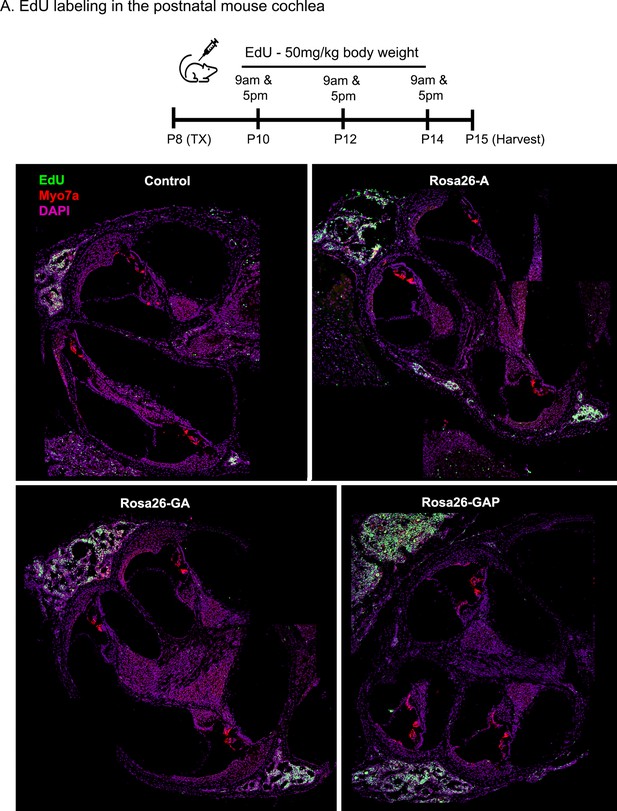
Transcription factor induction at P8 does not influence cell proliferation status in the control and reprogrammed cochlea at P15.
Control, Rosa26-A, Rosa26-GA, and Rosa26-GAP animals received tamoxifen (25 mg/kg body weight) at P8 and EdU (50 mg/kg body weight) at regular intervals (P10, P12, P14 – twice per day). Animals were sacrificed at P15. EdU incorporation (green) was observed in a small number of mesenchymal cells of the temporal bone wall in all four genotypes/samples at this age, but no labeling is seen in the organ of Corti. Images are from 16 µm sections stained with Myosin VIIA (red) and DAPI (magenta).
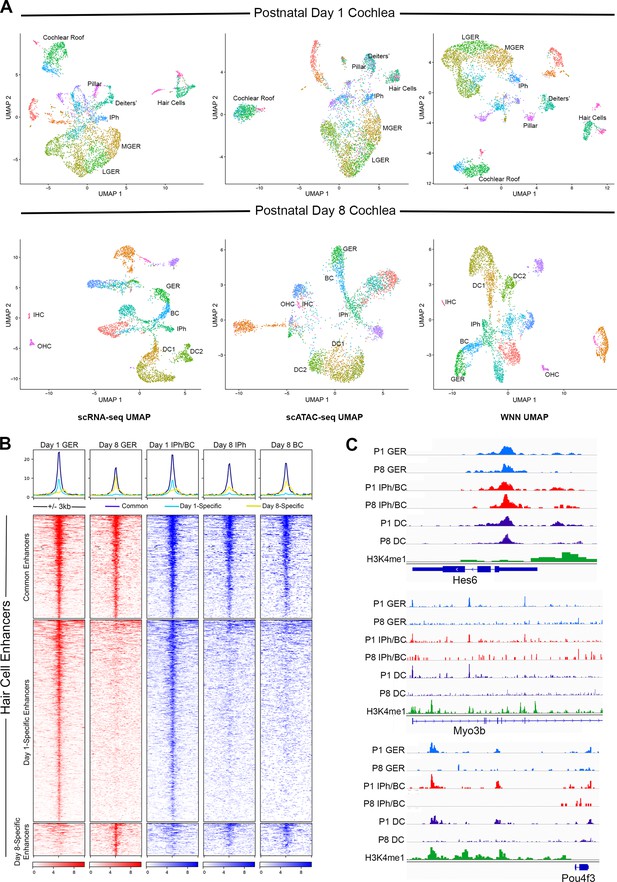
Multiomic analysis of 1 and 8-day-old mouse cochlea shows a loss of epigenetic accessibility of hair cell loci in GER and supporting cells.
(A) Clustering of P1 and P8 cochlear cells on the basis of scRNA-seq, scATAC-seq and weighted-nearest neighbor analyses. Different cochlear cell types can be resolved at both ages. IPh: Inner phalangeal cells; MGER: Medial greater epithelial ridge; LGER: Lateral greater epithelial ridge; IHC: Inner hair cells; OHC: Outer hair cells; GER: Greater Epithelial Ridge; BC: Border cells; DC1 and 2: Deiters’ cells. (B) Heat map showing ATAC-seq peaks of 1627 distal regulatory elements identified in hair cell gene loci. ATAC-seq data was extracted from day 1 and day 8 GER cells, and inner phalangeal and border cells. (C) Examples of changes in the accessibility of three hair cell loci (Hes6, Myo3b, Pou4f3) in GER cells and supporting cells in P1 and P8 mouse cochlea, measured by scATAC-seq. H3K4me1 data for each locus is taken from Tao et al., 2021. Reductions in accessibility can be seen in all three loci between P1 and P8.
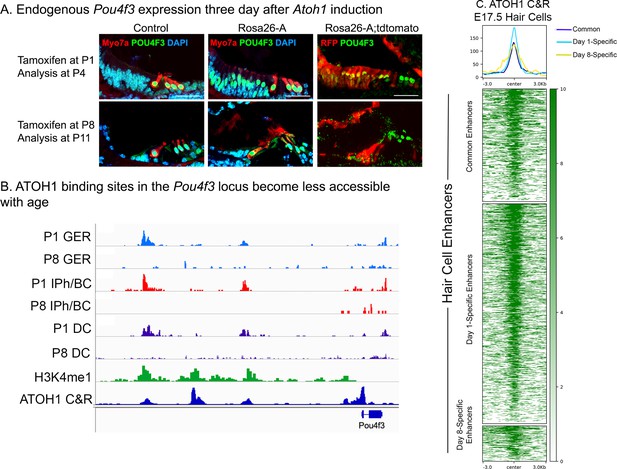
Endogenous Pou4f3 expression can be induced in 1-day-old, but not 8-day-old cochleae by reprogramming with Atoh1.
(A) Control and Rosa26-A animals were induced with tamoxifen at P1 or P8. Immunostaining for POU4F3 three days later at P4 (top panel) and P11 (bottom panel) revealed positive expression in the neonatal GER cells but not in the 11-day-old samples. These cells were confirmed to be targeted for Atoh1 overexpression by the Sox9-CreER, verified by staining for tdTomato expression in the control and Rosa26-A mice. Images show 16 µm sections at 20 x. Scale bar: 50 µm. (B) ATAC-seq data andH3K4me1 data from Figure 9C, combined with ATOH1 CUT&RUN data from E17.5 hair cells (Tao et al., 2021). ATOH1 binding peaks are accessible in GER cells, inner phalangeal cells and border cells and Deiters’ cells at P1, but significantly less accessible at P8. (C) Heat map of ATOH1 binding sites in E17.5 hair cells, viewed for all 1627 distal regulatory elements identified as being differentially accessible in the P1 vs P8 multii-omic analysis in Figure 9B. Many of these hair cell enhancers are bound by ATOH1.





