A feed-forward pathway drives LRRK2 kinase membrane recruitment and activation
Figures
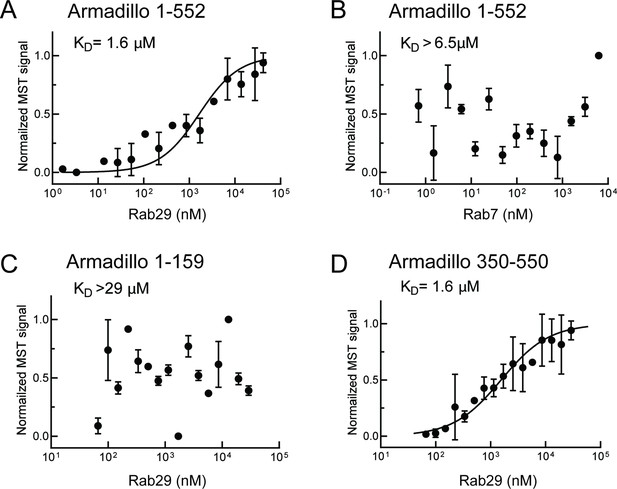
Rab29 binds to the C-terminal portion of the LRRK2 Armadillo domain.
Microscale thermophoresis of full-length (residues 1–552), labeled LRRK2 Armadillo domain with His-Rab29 (A) or with His-Rab7 (B). (C, D) Microscale thermophoresis of labeled LRRK2 Armadillo domain residues 1–159 (C) or 350–550 (D) with Rab29. Purified Rab29 was serially diluted and then NHS-RED-labeled-LRRK2 Armadillo (final concentration 100 nM) was added. Graphs show mean and SEM from three independent measurements, each from a different set of protein preparations. Data are summarized in Table 1.
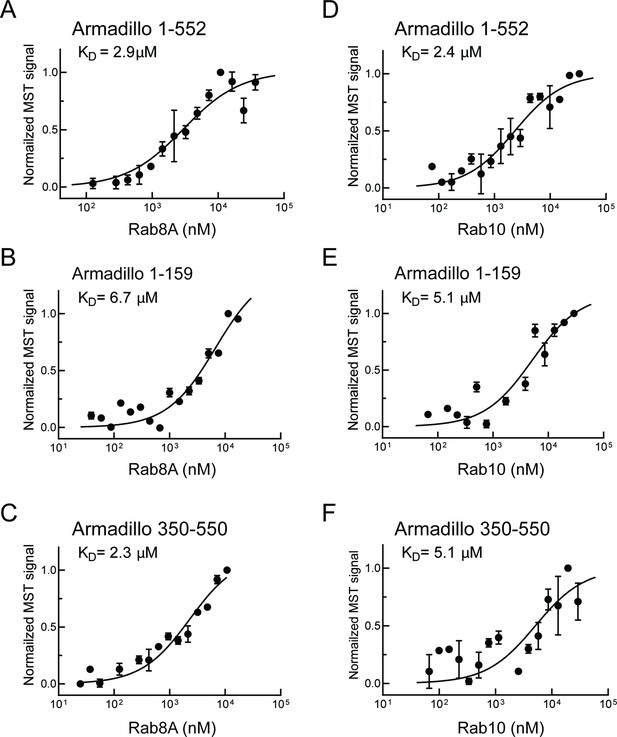
Rab8A and Rab10 bind to the LRRK2 Armadillo domain.
(A–C) Microscale thermophoresis of labeled, LRRK2 Armadillo domain fragments comprised of residues 1–552, 1–159, or 350–550 with Rab8A Q67L as indicated. (C–E) Microscale thermophoresis for Rab10 Q68L (1–181) with indicated LRRK2 Armadillo fragments, as in (A). Purified Rab proteins were serially diluted and then NHS-RED-labeled LRRK2 Armadillo domain (final concentration 100 nM) was added. Graphs show mean and SEM from three independent measurements, each from a different set of protein preparations. Data are summarized in Table 1.
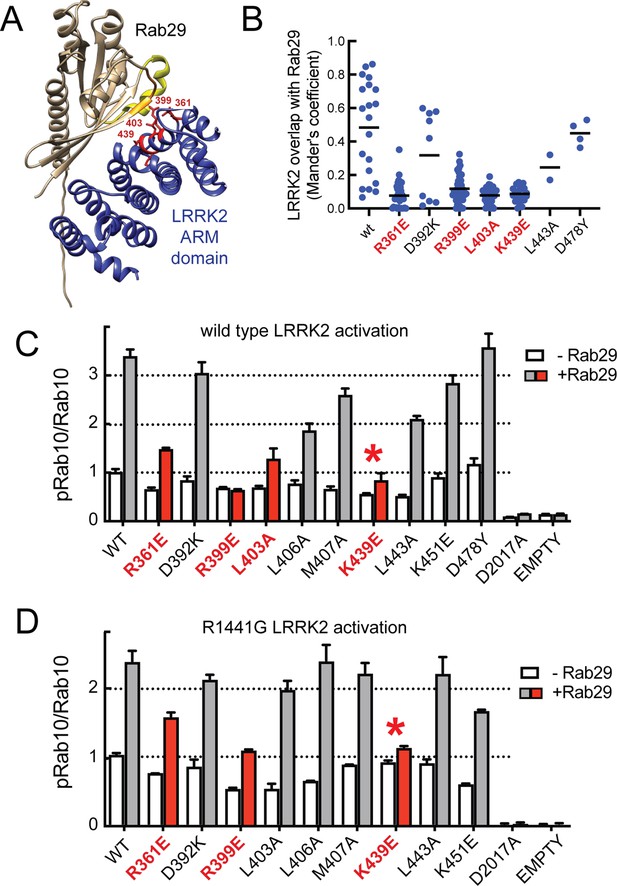
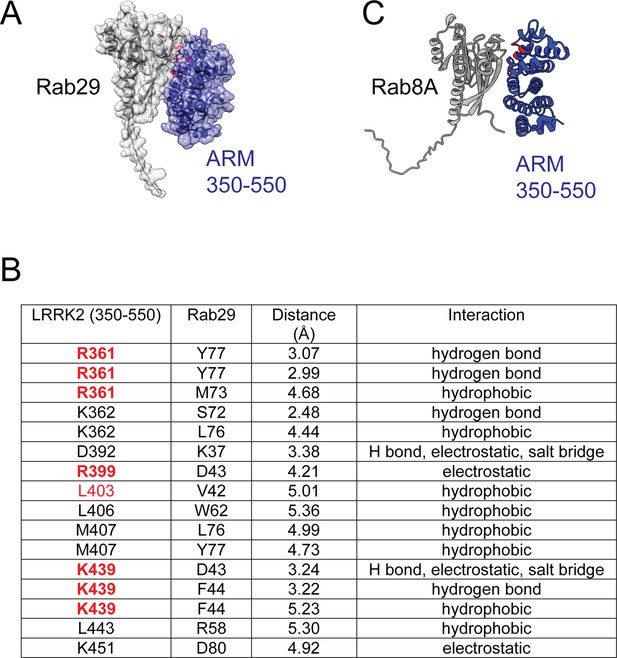
Characterization of critical LRRK2 residues mediating binding to Rab29.
(A). Predicted interactions between Rab29 and the LRRK2 Armadillo domain using AlphaFold docking (Jumper et al., 2021), ColabFold (Mirdita et al., 2022), and the AlphaFold2_advanced.ipynb notebook default settings. Residues identified in red show key contacts between LRRK2 and Rab29; orange and yellow coloring indicates the Switch I and Switch II domains of Rab29. (B) The wild-type and indicated mutants of full length of GFP-LRRK2 were co-expressed with HA-Rab29 in HeLa cells. 24 hr post transfection, cells were fixed and localization assessed by confocal microscopy. LRRK2 overlap with Rab29 is presented as a Mander’s coefficient determined using CellProfiler software (McQuin et al., 2018). (C, D) Wild-type and indicated mutants of full length of GFP-LRRK2 (C) or GFP-LRRK2 R1441G (D) were co-expressed with HA-Rab29 in HEK293T cells. 24 hr post transfection, cells were lysed and extracts immunoblotted with the indicated antibodies. Shown are the averages and standard deviations of duplicate determinations; red asterisks indicate preferred mutant.
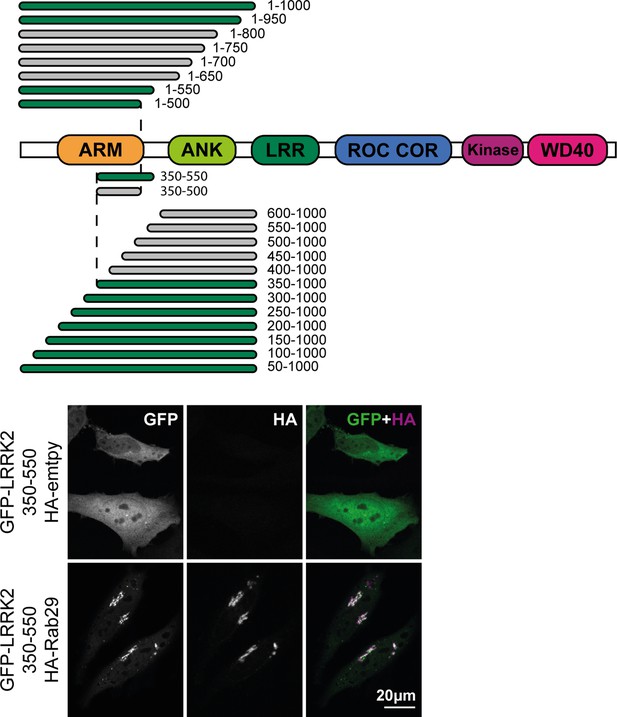
Top: Fragments of GFP-LRRK2 that were co-expressed with HA-Rab29 in HeLa cells.
24 hr post transfection, cells were fixed and localization assessed by confocal microscopy. Fragments that co-localized with Rab29 at the Golgi are shown in green and those that failed to co-localize in gray. The smallest fragment of LRRK2 that co-localized with Rab29 encompassed residues 350–550 (shown below). Magnification bar, 20 µm.
Residues predicted to be critical for Rab29 interaction with part of the LRRK2 Armadillo domain and comparison with an AlphaFold model for the complex with Rab8A.
(A) AlphaFold model of a complex of Rab29 (gray) bound to the 350–550 fragment of LRRK2 (purple) as in Figure 3A. (B) Table of site #1 residues predicted to lie within the interface of LRRK2 and Rab29. Highlighted in red are residues that when mutated suppress interaction of LRRK2 with Rab29 and inhibit Rab29-mediated LRRK2 activation in cells. (C) AlphaFold model of a complex of Rab8A (gray) bound to the 350–550 fragment of LRRK2 (navy); red residues are R361, R399, L403, and K439.
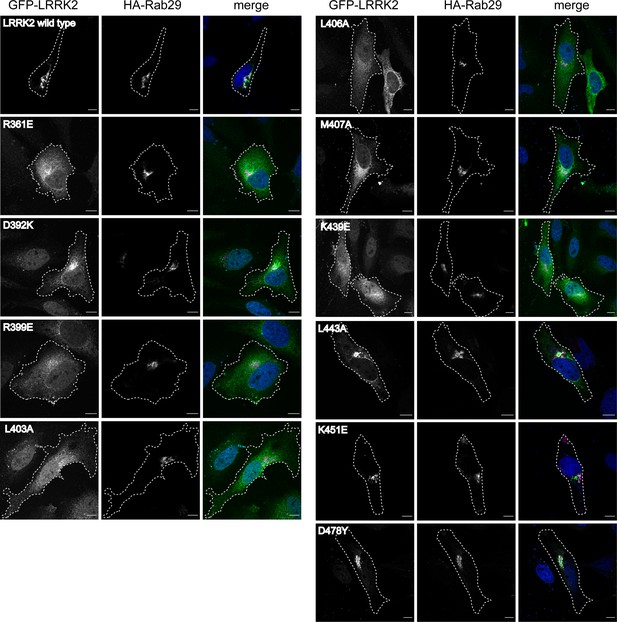
Examples of micrographs used to create Figure 3B.
Mutants (indicated) of full-length GFP-LRRK2 were co-expressed with HA-Rab29 in HeLa cells. 24 hr post transfection, cells were fixed and localization assessed by confocal microscopy. Magnification bars, 20 µm.
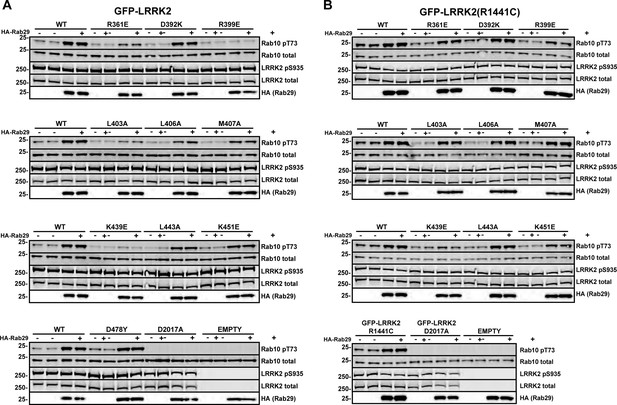
Immunoblots used to obtain Figure 3C and D.
Wild-type and indicated mutants of full length of GFP-LRRK2 (A) or R1441G LRRK2 (B) were co-expressed with HA-Rab29 in HEK293T cells. 24 hr post transfection, cells were fixed and samples analyzed for immunoblotting. Each membrane was probed with anti-pRab10 (rabbit), anti-Rab10 (mouse), and anti-HA (rat) antibodies. pRab10 and Rab10 signals were detected using 800 (anti-rabbit) and 680 (anti-mouse) channels in LI-COR, whereas the HA (showing Rab29 expression) was developed using ECL (anti-rat). Numbers at the left of each gel represent the mobility of molecular weight markers in kilodaltons.
-
Figure 3—figure supplement 4—source data 1
Raw data for gels.
- https://cdn.elifesciences.org/articles/79771/elife-79771-fig3-figsupp4-data1-v2.zip
-
Figure 3—figure supplement 4—source data 2
Annotated gels.
- https://cdn.elifesciences.org/articles/79771/elife-79771-fig3-figsupp4-data2-v2.pdf
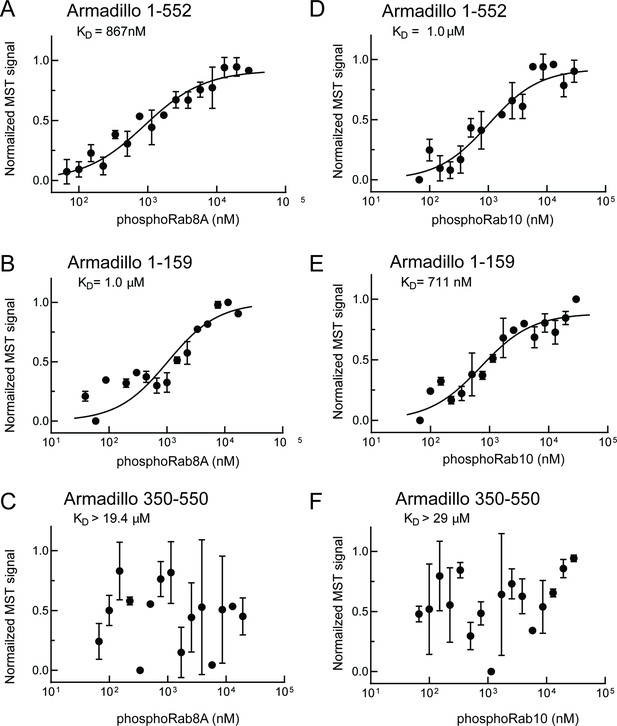
PhosphoRab8A and phosphoRab10 bind with high affinity to the N-terminal portion of the LRRK2 Armadillo domain.
(A–F) Microscale thermophoresis of labeled, indicated, LRRK2 Armadillo fragments with His-phosphoRab8A Q67L (A–C) or with His phosphoRab10 Q68L 1–181 (pRab10; D–F). Purified Rab proteins were phosphorylated with Mst3 kinase at 27°C for 2 hr and then serially diluted; NHS-RED-labeled Armadillo (final concentration 100 nM) was then added. Graphs show mean and SEM from three independent measurements, each from a different set of protein preparations.
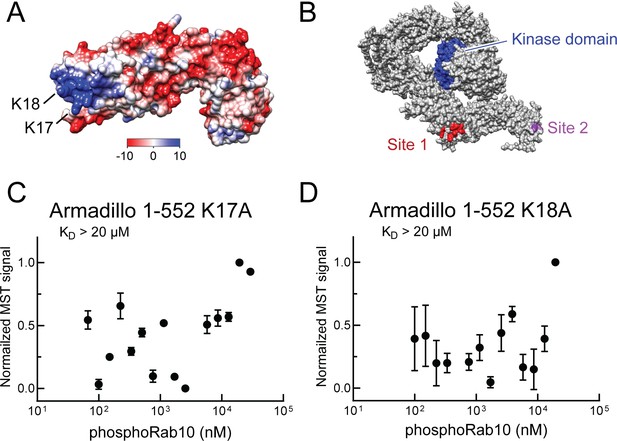
Identification of a basic patch at the N-terminus of LRRK2 that is needed for phosphoRab interaction.
(A) Electrostatic surface potential of LRRK2 Armadillo domain residues 1–552 modeled using Chimera 2 software (Pettersen et al., 2004); blue indicates a positively charged surface. LRRK2 K17 and K18 are indicated. (B) AlphaFold (Jumper et al., 2021) structure of putative, active LRRK2 with residues that mediate Rab29 binding shown in red (site #1) and the K17/K18 residues that are required for phosphoRab10 binding (site #2) shown in magenta; the kinase domain is shown in blue. (C, D) Microscale thermophoresis of labeled, full-length LRRK2 K17A or K18A Armadillo 1–552 with His phosphoRab10 Q68L 1–181. Purified Rab10 protein was phosphorylated with Mst3 kinase at 27°C for 2 hr and then serially diluted; NHS-RED-labeled Armadillo (final concentration 100 nM) was then added. Graphs show mean and SEM from three independent measurements, each from a different set of protein preparations.
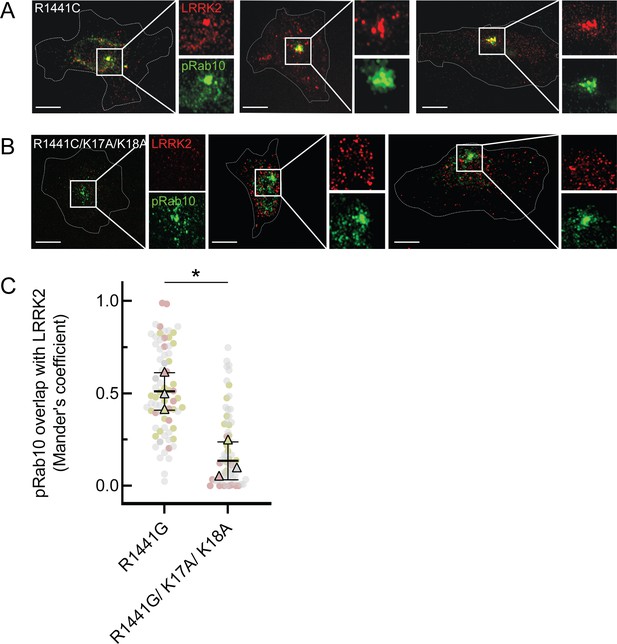
LRRK2 K17 and K18 are critical for pRab10 interaction in cells.
(A) FLAG-LRRK2 R1441G (red) was transfected into HeLa cells plated on collagen-coated coverslips and co-localized with endogenous wild-type pRab10 (green). Cells on coverslips were dipped in liquid nitrogen to deplete cytosol and enhance membrane-bound signal. Insets show enlargements of boxed areas representing peri-centriolar LRRK2 and pRab10. (B) FLAG-LRRK2 R1441G/K17A/K18A (red) was transfected into HeLa cells plated on collagen-coated coverslips and stained and localized with pRab10 (green) as in (A). Scale bars, 10µm. (C) Quantification of pRab10 overlap with LRRK2 by Mander’s coefficient. Error bars represent SEM of means from three different experiments (represented by colored dots), each with >40 cells per condition. Significance was determined by t-test, *p=0.0108.
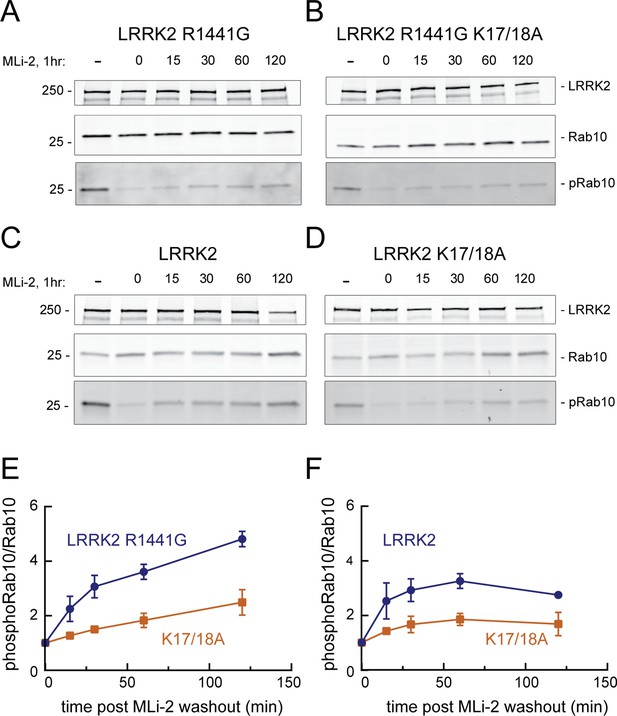
LRRK2 K17 and K18 increase endogenous pRab10 recovery after LRRK2 inhibitor washout.
(A–D) FLAG-LRRK2 R1441G, FLAG-LRRK2 R1441G/K17A/K18A, LRRK2, or LRRK2 K17A/K18A was transfected into HeLa cells. 48 hr post transfection, cells were treated with 200 nM of MLi-2 for 1 hr. The MLi-2 was then removed by multiple washes and incubated for the indicated times prior to cell lysis. Whole-cell extracts (20 µg) were subjected to quantitative immunoblot analysis using anti-LRRK2, anti-Rab10, and anti-pRab10 antibodies. Numbers at the left of the gels represent the mobilities of molecular weight markers in kilodaltons. (E, F) Quantification of pRab10/total Rab10 fold change and normalized to no MLi2 control. Error bars represent mean ± SD from two different experiments per condition.
-
Figure 7—source data 1
Raw data for gels.
- https://cdn.elifesciences.org/articles/79771/elife-79771-fig7-data1-v2.zip
-
Figure 7—source data 2
Annotated gels.
- https://cdn.elifesciences.org/articles/79771/elife-79771-fig7-data2-v2.pdf
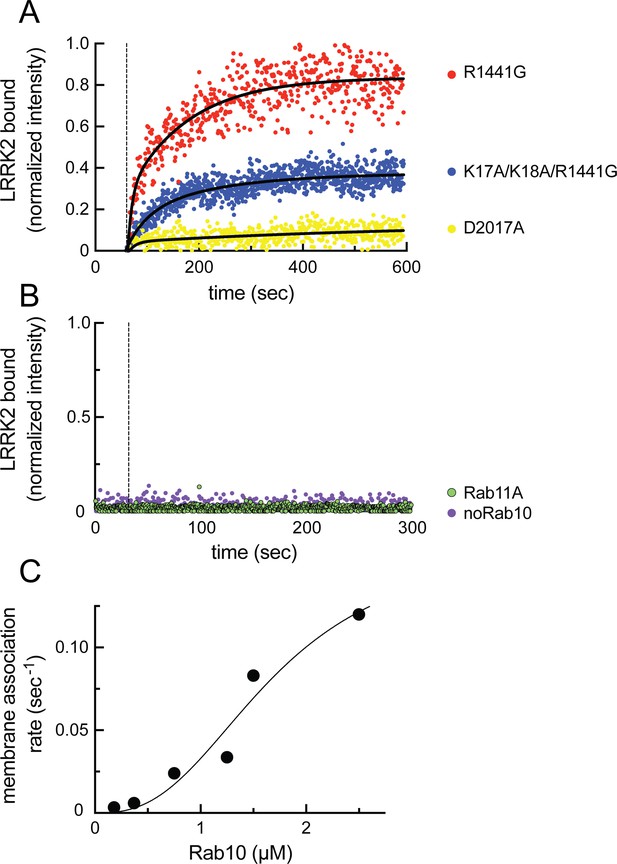
Feed-forward pathway for Rab10 phosphorylation is dependent on LRRK2 kinase activity.
(A) Fluorescence intensity traces of individual, single molecules of 7 nM CF633-labeled FLAG-LRRK2 R1441G on a substrate-supported lipid bilayer decorated with lipid-anchored GFP-Rab10 Q68L-His across 600 s of live total internal reflection (TIRF) microscopy. Red, R1441G; blue, K17A/K18A/R1441G; yellow, D2017A. (B) Reactions were carried out as in (A) except Rab10 was omitted (purple) or Rab10 was replaced with Rab11 (green). Dashed lines in (A) and (B) represent time of addition of fluorescently labeled LRRK2 at 60 s; shown are representative experiments carried out at least three times for each condition. Fluorescence intensity was fitted by a nonlinear regression curve for two-phase association. Fold change was calculated by dividing the average fluorescence intensity at steady state and subtracting background fluorescence intensity average determined from 60 s prior to LRRK2 addition. (C) Rate of membrane association of LRRK2 as a function of Rab10 concentration. This curve was fitted by a nonlinear regression fit using PRISM software (MathWorks) to determine a Hill coefficient. Data are from two independent experiments plotted together.
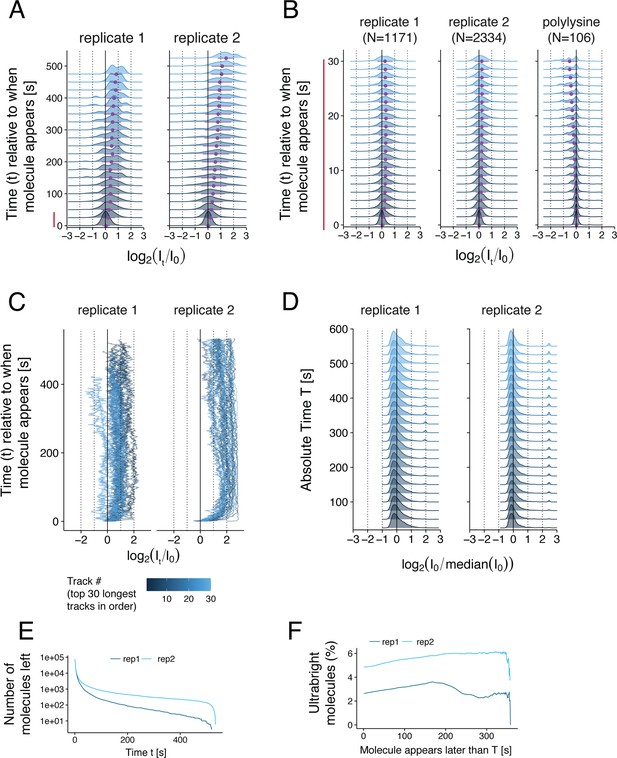
Quantitative analysis of total internal reflection (TIRF) images of LRRK2 recruitment on planar lipid bilayers.
(A) Ridge plot showing the distribution of the fluorescence intensity of CF633-labeled LRRK2 molecules (or complexes) as a function of the time elapsed since the molecule first appeared on the surface. Two replicates of LRRK2 are shown. For each molecule, the fluorescence intensity It at each time point t was normalized with respect to its initial intensity I0 during the first frame when the molecule first appeared on the surface. The intensity distributions are shown in log scale (x axis). Intensity distributions were computed using the average intensity of each molecule over 25 s increments. Purple data points and error bars below each distribution show its mean and standard deviation. For each time point t, all the molecules with a fluorescence lifetime greater than t were used to compute the distribution. The number of such molecules at each time point is shown in panel (E). (B) Same ridge plot as in (A) showing the evolution of the fluorescence intensity with greater temporal resolution during the first 30 s after a molecule appeared on the surface. Here, the intensity distributions were computed using the average intensity of each molecule over 1.5 s increments. Only molecules with a fluorescence lifetime larger than 30 s were included in this ridge plot (N = 1171, 2334, and 106) molecules respectively for the three conditions shown (left to right). (C) Individual fluorescence intensity over time for the 30 molecules with the longest fluorescence lifetimes as in (A). (D) Ridge plot showing the distribution of the initial fluorescence intensity I0 of individual CF633-labeled LRRK2 molecules or complexes when they first appeared on the surface as a function of the time elapsed since the first molecule was detected (referred to as the absolute time T). These initial intensities were normalized to the median initial intensity of all molecules. (E) Inverse cumulative distribution of the fluorescence lifetime of individual molecules. (F) Percentage of molecules that were ‘ultrabright’ when they first appeared on the surface as a function of the absolute time T at which they appeared. Ultrabright refers to molecules with an initial intensity greater than 21.5 fold the median initial intensity across all molecules (log2[I0/median(I0)] > 1.5).
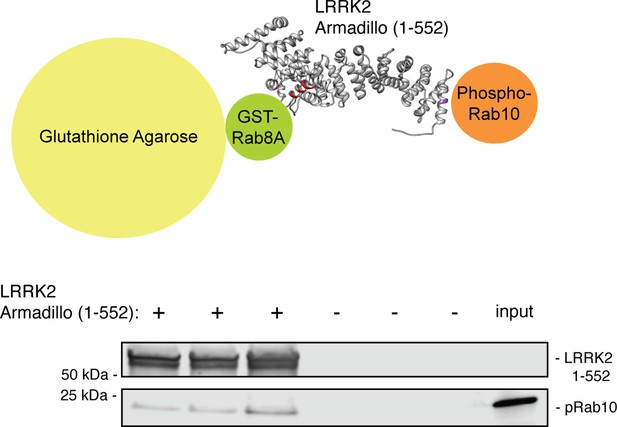
The LRRK2 Armadillo domain can bind phosphorylated Rab10 and unphosphorylated Rab8A simultaneously.
GST-Rab8A Q67L was immobilized on glutathione agarose, then LRRK2 Armadillo (or buffer) was added; beads were washed and His-phosphoRab10 Q68L was then added. Bead-bound material (triplicates shown) was eluted with reduced glutathione and analyzed by immunoblotting. Input, 50% of that used in each binding reaction. PhosphoRab10 (5% of input) was detected only in ARM domain-containing samples, consistent with the KD values.
-
Figure 8—figure supplement 2—source data 1
Raw data for gels.
- https://cdn.elifesciences.org/articles/79771/elife-79771-fig8-figsupp2-data1-v2.zip
-
Figure 8—figure supplement 2—source data 2
Annotated gels.
- https://cdn.elifesciences.org/articles/79771/elife-79771-fig8-figsupp2-data2-v2.pdf
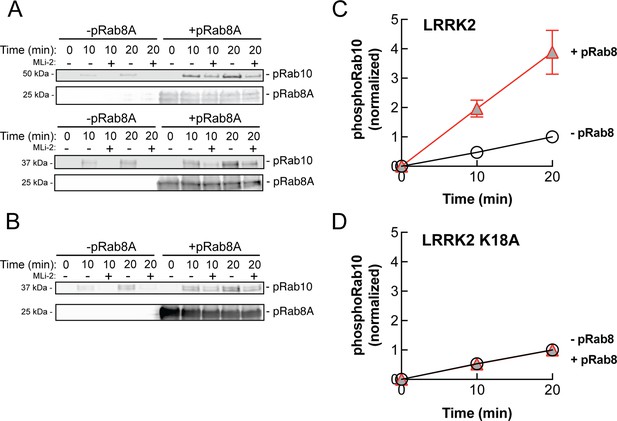
PhosphoRab8A activates LRRK2 phosphorylation of Rab10 in solution.
(A) Immunoblot analysis of the kinetics of LRRK2 G2019S phosphorylation of Rab10 with and without additional pRab8. Upper gel: GFP-Rab10 Q68L His substrate. Lower gel: His-Sumo-Rab10 wild-type full-length substrate. Indicated reactions contained 200 nM MLi-2. pRab8A was detected with anti-phosphoRab8A antibody. (B) Same as panel (A) with K18A-LRRK2-R1441G and His-Sumo-Rab10 wild-type full-length as substrate. PhosphoRab8A was detected with total Rab8 antibody. (C) Kinetics of phosphoRab10 production as in (A). Shown are the combined means of independent, quadruplicate determinations ± SEM, as indicated. (D) PhosphoRab10 production as in (B). Shown are the combined means of independent duplicate determinations,± SEM, as indicated. Background signal in the presence of pRab8A is likely due to trace MST3 contamination that is not sensitive to MLi-2 inhibition and was subtracted. pRab8 preparation was by method #1 for (A), upper gel, and (B), and method #2 was used in panel (A), lower gel.
-
Figure 9—source data 1
Raw data for gels.
- https://cdn.elifesciences.org/articles/79771/elife-79771-fig9-data1-v2.zip
-
Figure 9—source data 2
Annotated gels.
- https://cdn.elifesciences.org/articles/79771/elife-79771-fig9-data2-v2.pdf
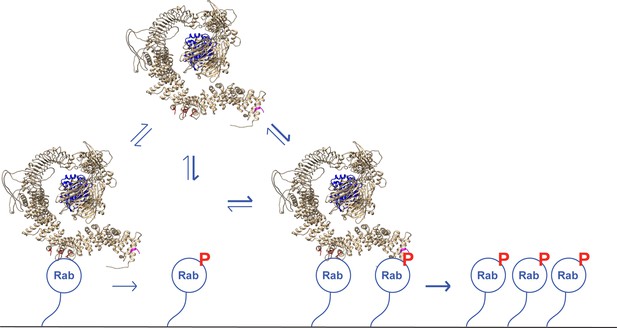
A model for LRRK2 membrane recruitment.
LRRK2 can interact with non-phosphorylated Rab GTPases via site #1. Once membrane bound, it can generate phosphoRabs that can now engage site #2. Rab binding to both sites increases the avidity of LRRK2 for membranes and retains LRRK2 on the membrane surface to phosphorylate more Rab substrates. We have shown that LRRK2 binding to phosphoRabs also activates the kinase, likely by altering its oligomeric state.
Videos
Total internal reflection (TIRF) microscopy of R1441G LRRK2 binding to Rab10-lipid bilayers.
Captured at 1 frame/s and compressed 20×.
Total internal reflection (TIRF) microscopy of R1441G LRRK2 binding to lipid bilayers without Rab10.
Captured at 1 frame/s and compressed 20×.
Total internal reflection (TIRF) microscopy of R1441G LRRK2 binding to Rab11-lipid bilayers.
Captured at 0.5 frame/s and compressed 40×.
Total internal reflection (TIRF) microscopy of D2017A LRRK2 binding to Rab10-lipid bilayers.
Captured at 1 frame/s and compressed 20×.
Total internal reflection (TIRF) microscopy of K17A/K18A/R1441G LRRK2 binding to Rab10-lipid bilayers.
Captured at 0.5 frame/s and compressed 40×.
Tables
Summary of binding affinities.
Note that these values are likely underestimates of affinities as typical preparations of the indicated, purified Rab proteins contained ~50% bound GDP and ~50% bound GTP by mass spectrometry. Non-phosphorylated Rab interaction with Armadillo 1–159 is shown in parentheses as it likely reflects binding to an AlphaFold-predicted site near the C-terminus of this fragment that will not be accessible in full-length LRRK2 protein.
| Armadillo1–159(site #2-containing) | Armadillo1–552 | Armadillo350–550(site #1-containing) | Armadillo1–552 K17A | Armadillo1–552 K18A | |
|---|---|---|---|---|---|
| Rab29 | >29 | 1.6 ± 0.9 | 1.6 ± 0.5 | - | - |
| Rab10-Q68L | (5.1 ±3.1) | 2.4 ± 0.6 | 5.1 ± 2.5 | - | - |
| pRab10-Q68L | 0.71 ± 0.3 | 1.0 ± 0.4 | >29 | >20 | >20 |
| Rab8A-Q67L | (6.7 ± 3.6) | 2.9 ± 1.2 | 2.3 ± 1.0 | - | - |
| pRab8A-Q67L | 1.0 ± 0.6 | 0.87 ± 0.4 | >19.4 | - | - |
| Rab7 | - | >6.5 | - | - | - |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-LRRK2 (mouse monoclonal) | NeuroMab RRID:AB_2877351 | N241A/34 | (1:1000) |
| Antibody | Anti-LRRK2 phospho S935 (rabbit monoclonal) | Abcam RRID:AB_2904231 | UDD2 | (1:1000) |
| Antibody | Anti-Rab10 (mouse monoclonal) | Nanotools RRID:AB_2921226 | 0680-100/Rab10-605B11 | (1:1000) |
| Antibody | Anti-Rab10 (phospho T73) (rabbit monoclonal) | Abcam RRID:AB_2811274 | ab230261 | (1:1000) |
| Antibody | Anti-FLAG M2 (mouse monoclonal) | MilliporeSigma RRID:AB_262044 | F-1804 | (1:2000) |
| Strain, strain background (Escherichia coli) | E. coli DH5α | Thermo Fisher | 18258012 | |
| Strain, strain background (E. coli) | E. coli STBL3 | Thermo Fisher | C737303 | |
| Strain, strain background (E. coli) | E. coli Rosetta DE3 pLys | Millipore | 70956 | |
| Cell line (Homo sapiens) | HeLa | ATCC | CCL-2 | |
| Cell line (H. sapiens) | HEK293T | ATCC | CRL-3216 | |
| Chemical compound, drug | MLi-2 | MRC PPU | ||
| Chemical compound, drug | Creatine phosphate | Fluka Analytical | #27920 | 20 mM |
| Commercial assay or kit | RED-NHS 2nd Generation (Amine Reactive) Protein Labeling Kit | NanoTemper Technologies | MO-L011 | |
| Commercial assay or kit | CF 633 Succinimidyl Ester Protein Labeling Kit | Biotium | #92217 | |
| Other | Creatine Phosphokinase | Sigma | C3755 | 30U |
| Chemical compound, drug | 18:1 (Δ9-Cis) PC (DOPC) | Avanti Polar Lipids | #850375 | 11 µmol |
| Chemical compound, drug | 18:1 PS (DOPS) | Avanti Polar Lipids | #840035 | 5 µmol |
| Chemical compound, drug | 18:1 DGS-NTA(Ni) | Avanti Polar Lipids | #790404 | 0.85 µmol |
| Chemical compound, drug | 18:1 PI(4)P | Avanti Polar Lipids | #850151 | 0.15 µmol |
| Chemical compound, drug | DiD | Thermo Fisher | D7757 | 0.01 µmol |
| Recombinant DNA reagent | pNIC Bsa-4 His-Sumo Rab10 Q68L 1–181 | Gift of Amir Khan | Human | |
| Recombinant DNA reagent | pET15b His-Mst3 | Gift of Amir Khan | Human | |
| Recombinant DNA reagent | pET21b GFP-Rab10 Q68L-His | Addgene RRID:Addgene_186015 | 186015 | Human |
| Recombinant DNA reagent | pET21b His Rab8A Q67L | Addgene RRID:Addgene_186014 | 186014 | Human |
| Recombinant DNA reagent | pQE-80L 2xHis-Rab29 | Addgene RRID:Addgene_186021 | 186021 | Human |
| Recombinant DNA reagent | pGEB GST-Rab8A-Q67L | Addgene RRID:Addgene_86079 | 86079 | Human |
| Recombinant DNA reagent | His-Rab11 | Gift of Marino Zerial | Canine | |
| Recombinant DNA reagent | pQE-80L 2xHis-LRRK2 Armadillo 1–552 | Addgene RRID:Addgene_186017 | 186017 | Human |
| Recombinant DNA reagent | pQE-80L 2xHis-LRRK2-Armadillo 1–159 | Addgene RRID:Addgene_186016 | 186016 | Human |
| Recombinant DNA reagent | pQE-80L 2xHis-LRRK2-Armadillo 350–550 | Addgene RRID:Addgene_186018 | 186018 | Human |
| Recombinant DNA reagent | pQE-80L 2xHis-LRRK2-Armadillo K17A | Addgene RRID:Addgene_186019 | 186019 | Human |
| Recombinant DNA reagent | pQE-80L 2xHis-LRRK2-Armadillo K18A | Addgene RRID:Addgene_186020 | 186020 | Human |
| Recombinant DNA reagent | pCMV5 FLAG-LRRK2 K17A/K18A/R1441G | Addgene RRID:Addgene_186012 | 186012 | Human |
| Recombinant DNA reagent | pCMV5 FLAG-LRRK2 | MRC PPU Reagents and Services, University of Dundee (‘MRC PPU’) | DU6841 | Human |
| Recombinant DNA reagent | pCMV5 FLAG-LRRK2 R1441G | MRC PPU | DU13077 | Human |
| Recombinant DNA reagent | pCMV5 FLAG-LRRK2 D2017A | MRC PPU | DU52725 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 WT | MRC PPU | DU13363 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 R361E | MRC PPU | DU62605 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 D392K | MRC PPU | DU72261 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 R399E | MRC PPU | DU72262 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 L403A | MRC PPU | DU72263 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 L406A | MRC PPU | DU72266 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 M407A | MRC PPU | DU72267 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 K439E | MRC PPU | DU72268 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 L443A | MRC PPU | DU72270 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 K451E | MRC PPU | DU72271 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 D478Y | MRC PPU | DU68605 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 D2017A | MRC PPU | DU13364 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 R1441C | MRC PPU | DU13387 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 R1441C R361E | MRC PPU | DU72304 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 R1441C D392K | MRC PPU | DU72305 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 R1441C R399E | MRC PPU | DU72306 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 R1441C L403A | MRC PPU | DU72307 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 R1441C L406A | MRC PPU | DU72308 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 R1441C M407A | MRC PPU | DU72309 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 R1441C K439E | MRC PPU | DU72310 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 R1441C L443A | MRC PPU | DU72311 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 R1441C K451E | MRC PPU | DU72312 | Human |
| Recombinant DNA reagent | pCMV5D HA RAB29 | MRC PPU | DU50222 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 1–950 | MRC PPU | DU62702 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 1–900 | MRC PPU | DU62701 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 1–850 | MRC PPU | DU62700 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 1–800 | MRC PPU | DU62693 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 1–750 | MRC PPU | DU62726 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 1–700 | MRC PPU | DU62689 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 1–650 | MRC PPU | DU62678 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 1–600 | MRC PPU | DU62677 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 1–550 | MRC PPU | DU62676 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 1–500 | MRC PPU | DU62675 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 50–1000 | MRC PPU | DU62725 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 100–1000 | MRC PPU | DU62742 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 150–1000 | MRC PPU | DU62674 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 200–1000 | MRC PPU | DU62679 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 250–1000 | MRC PPU | DU62680 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 300–1000 | MRC PPU | DU62681 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 350–1000 | MRC PPU | DU62682 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 400–1000 | MRC PPU | DU62683 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 450–1000 | MRC PPU | DU62684 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 500–1000 | MRC PPU | DU62685 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 550–1000 | MRC PPU | DU62686 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 600–1000 | MRC PPU | DU62687 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 350–550 | MRC PPU | DU68397 | Human |
| Recombinant DNA reagent | pcDNA5D FRT TO GFP LRRK2 350–500 | MRC PPU | DU68398 | Human |
| Recombinant DNA reagent | His-SUMO Rab10 | MRC PPU | DU51062 | Human |
| Recombinant DNA reagent | His Rab7 | Gift of Marino Zerial | ||
| Software, algorithm | Fiji | PMID:29187165 | RRID:SCR_002285 | |
| Software, algorithm | CellProfiler | PMID:29969450 | RRID:SCR_007358 | |
| Software, algorithm | TrackIt | PMID:33947895 | ||
| Software, algorithm | Chimera 2 | PMID:15264254 | RRID:SCR_004097 | |
| Software, algorithm | ChimeraX | PMID:32881101 | RRID:SCR_015872 | |
| Software, algorithm | NanoTemper NTAAffinityAnalysis | MO.Affinity Analysis v2.2.5 | ||
| Software, algorithm | Prism | Prism 9 version 9.3.1 (350) | RRID:SCR_002798 | |
| Software, algorithm | R CRAN R package | Version 4.2.0 (2022-04-22) | RRID:SCR_003005 | |
| Software, algorithm | Dplyr_1.0.9 | RRID:SCR_016708 | ||
| Software, algorithm | ggridges_0.5.3 | |||
| Software, algorithm | ggplot_3.3.6 | RRID:SCR_014601 |





