VPS9D1-AS1 overexpression amplifies intratumoral TGF-β signaling and promotes tumor cell escape from CD8+ T cell killing in colorectal cancer
Figures
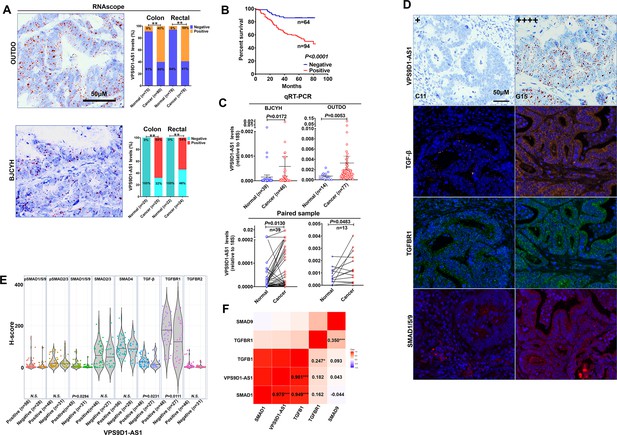
VPS9D1-AS1 is significantly upregulated in colorectal cancer (CRC) and activates the TGF-β signaling pathway.
(A) RNAscope stained VPS9D1-AS1 in CRC tissues that were enrolled in OUTDO (upper) and BJCYH cohorts (lower). Semiquantitative analyses of the levels of VPS9D1-AS1 in cancer and normal tissues of CRC patients (right). (B) Kaplan-Meier overall survival curves of VPS9D1-AS1-positive and VPS9D1-AS1-negative CRC patients. (C) qRT-PCR evaluation of the mRNA levels of VPS9D1-AS1 (upper). Expression of VPS9D1-AS1 was compared in paired normal and cancer tissues (lower). (D) Representative pictures of VPS9D1-AS1-negative (+, C11) and VPS9D1-AS1-positive (G15, ++++) and multispectral fluorescence immunohistochemistry (mfIHC)-stained TGF-β, TGFBR1, and SMAD1/5/9 in the same CRC patients. (E) Integrative analysis of RNAscope and mfIHC data indicates that cancer tissues with high levels of VPS9D1-AS1 had higher levels of TGF-β, TGFBR1, and SMAD1/5/9 than these with low levels of VPS9D1-AS1. (F) Pearson correlation analyses investigated the mRNA levels of VPS9D1-AS1, TGF-β, TGFBR1, SMAD1, and SMAD9. p-Values were obtained by chi-square (A), log-rank test (B), unpaired t nonparametric test (C, E), paired t test (C), and Pearson correlation test (F). Data are shown as data points with mean ± standard deviation of mean (SEM) (C), data are depicted by violin and scatter plots with mean value (E). * p<0.05, ** p<0.01, *** p<0.001.
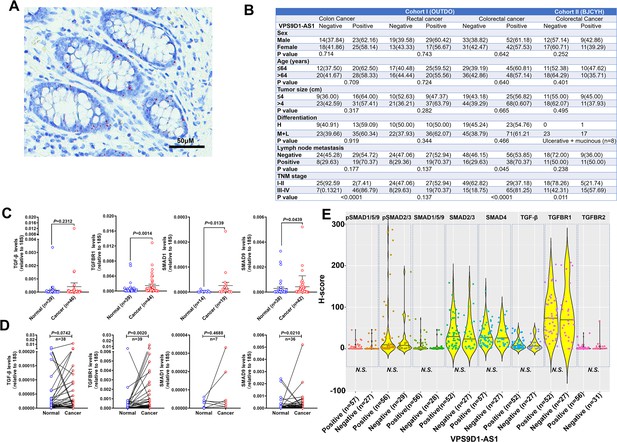
Levels of VPS9D1-AS1 were not related to TGF-β signaling in cancer stromal cells.
(A) Representative image of VPS9D1-AS1 expressed in normal colonic epithelial cells. (B) Clinical pathologic analyses demonstrated that VPS9D1-AS1 was correlated with lymph node metastasis and TNM stage in the OUTDO and BJCYH cohorts. (C, D) qRT-PCR was used to determine the mRNA levels of TGF-β, TGFBR1, SMAD1, and SMAD9 in the BJCYH cohort. (E) There were no significant relationships between VPS9D1-AS1 levels and TGF-β signaling in cancer stromal cells in OUTDO cohort. p-Values were obtained by Wilcoxon rank-sum test (B), unpaired t nonparametric test (C), and paired t nonparametric test (D). Data points are presented as the mean ± SEM (C) and the minimum, first quartile, median, third quartile, and maximum (E). N.S. not significant.
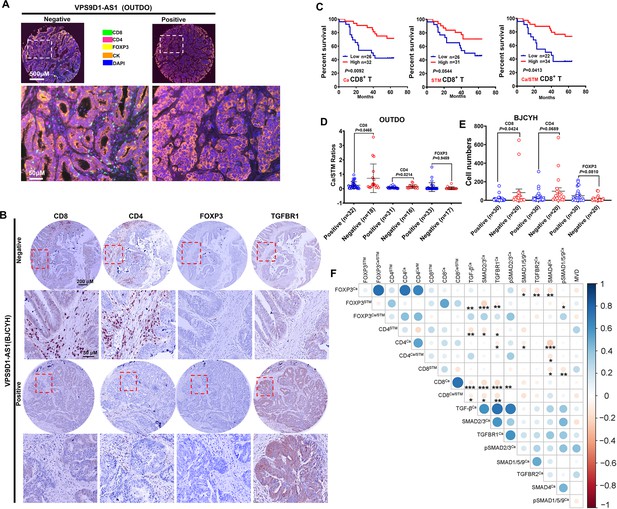
VPS9D1-AS1 is associated with reduced T lymphocyte infiltration.
(A) Representative pictures of T cell infiltration (CD4, CD8, FOXP3) in colorectal cancer (CRC) tissues for VPS9D1-AS1 quantification. Tumor cells are marked by cytokeratin. (B) Representative pictures of CD8+, CD4+, FOXP3+ T cells, and TGFBR1 stained by immunohistochemistry (IHC) in the BJCYH cohort. (C) The overall survival curves depicting the percentage of surviving CRC patients stratified by the levels of CD8+ T cell infiltration in cancerous (Ca) tissues, cancer stroma (STM), and the Ca/STM ratio. (D) Ca/STM ratios of CD4+, CD8+, and FOXP3+ T cells were calculated to identify the difference between VPS9D1-AS1 negative and positive populations in the OUTDO cohort. (E) The numbers of CD4+, CD8+, and FOXP3+ T cells in cancer tissues of BJCYH cohort were compared between VPS9D1-AS1 negative and positive tissues. (F) Pearson correlation analyses investigated the relationships between T-infiltrating lymphocytes and TGF-β signaling in cancer tissues. Eight protein levels were investigated by multispectral fluorescence IHC assays in same samples, and fluorescence intensity of each protein level was transferred into quantitative data for Pearson correlation analyses. p-Values were obtained by log-rank test (C), unpaired t nonparametric test (D, E), and Pearson correlation test (F). Data are shown by mean ± SEM (D, E). * p<0.05, ** p<0.01, *** p<0.001.
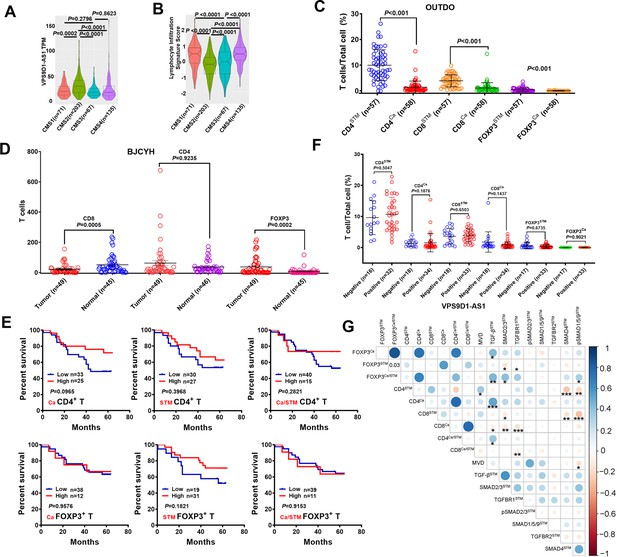
Integrative analysis of the relationship between VPS9D1-AS1, TGF-β signaling, and T-infiltrating lymphocytes (TILs).
(A) The Cancer Genome Atlas analysis confirmed that the highest VPS9D1-AS1 expression was predominantly in consensus molecular subtype 2 (CMS2)-type colorectal cancer patients. (B) CMS2 patients showed the lowest lymphocyte infiltration signal score. (C) Comparison of the percentages of CD4+, CD8+, and FOXP3+ T cells in cancer and cancer stromal tissues of OUTDO cohort. (D) Comparison of the number of CD4+, CD8+, and FOXP3+ T cells in cancer and normal tissues of the BJCYH cohort by immunohistochemistry assays. (E) Kaplan-Meier overall survival curves showed that CD4+ T and FOXP3+ T cells had no prognostic significance. (F) Proportions of CD4+, CD8+, and FOXP3+ T cell out of the total cells of Ca and STM tissues were compared according to the levels of VPS9D1-AS1 in BJCYH cohort. (G) Pearson correlation analyses investigated the relationships between TILs and TGF-β signaling in cancer stromal tissues of BJCYH cohort. p-Values were obtained by unpaired t nonparametric test (A, B, C, D, F), log-rank test (E), and Pearson correlation test (G). Data are shown as data points with mean ± SEM (C, D, F). *p<0.05, **p<0.01, ***p<0.001.
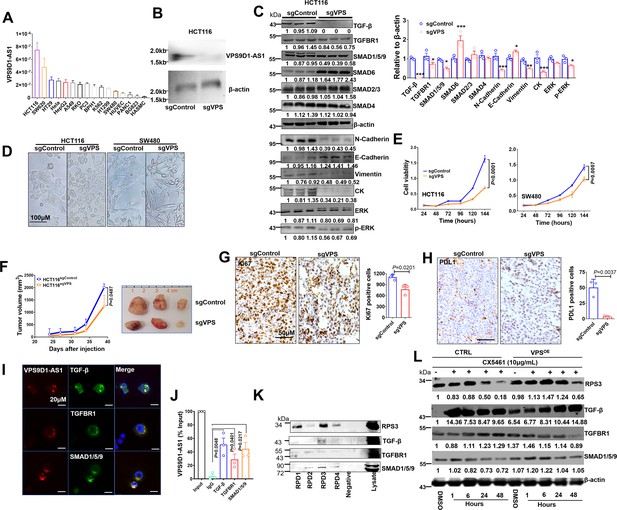
VPS9D1-AS1 controls TGF-β signaling and drives cell proliferation and metastasis.
(A) The levels of VPS9D1-AS1 were determined by qRT-PCR in 16 cell lines. (colorectal cancer: HCT116, SW620, HT29, RKO, SW480; cervical cancer: Hela; lung cancer: A549, H1299; gastric cancer: BGC823; prostatic cancer: PC3, BPH1; leukemia: K562; pancreatic cancer: PANC1; live cancer: HepG2; HUVEC: human umbilical vein endothelial cell; HASMC: human atrial smooth muscle cell). (B) Northern blotting validated the knockout (KO) of VPS9D1-AS1. (C) Western blotting measured the levels of proteins involved in TGF-β, EMT, and ERK signaling pathways. (D) Representative pictures show the cell morphologies of HCT116 and SW480 cell lines. (E) The proliferation of HCT116/SW480 sgControl and sgVPS cells was determined by Cell counting kit-8 (CCK8) assays. (F) Proliferation of xenograft tumors derived from HCT116 sgControl and sgVPS cells. (G) Immunohistochemistry determined the levels of Ki67 and (H) PDL1 in xenograft tissues. (I) RNA fluorescence in situ hybridization (FISH)-immunofluorescence (IF) and (J) RNA immunoprecipitation (RIP) assays showed the interaction between VPS9D1-AS1 and proteins that included TGF-β, TGFBR1, and SMAD1/5/9. (K) RNA pulldown-Western blotting assays detected the interaction between VPS9D1-AS1 and the intended proteins. (L) Western blotting determined the changes in RPS3, TGF-β, TGFBR1, and SMAD1/5/9 in HCT116 control (CTRL) and VPS9D1-AS1 (VPS)-overexpressing (OE) cells treated with CX5461. RPD, RNA pull down probe. p-Values were obtained by two-way ANOVA (E, F) and paired or unpaired t tests (C, G, H, J). Data are shown as the mean ± SEM (C, E, F, G, H, J). *p<0.05, ** p<0.01, *** p<0.001.
-
Figure 3—source data 1
TGF-β, TGFBR1, SMAD4, SMAD1/5/9, SMAD6, SMAD2/3, β-actin, N-cadherin, E-cadherin, vimentin, CK, ERK, and p-ERK western blot for Figure 3C.
- https://cdn.elifesciences.org/articles/79811/elife-79811-fig3-data1-v2.zip
-
Figure 3—source data 2
RPS3 RNA-pull-down western blot for Figure 3K.
- https://cdn.elifesciences.org/articles/79811/elife-79811-fig3-data2-v2.zip
-
Figure 3—source data 3
SMAD1/5/9, RPS3, TGF-β, and TGFBR1 western blot for Figure 3L.
- https://cdn.elifesciences.org/articles/79811/elife-79811-fig3-data3-v2.zip
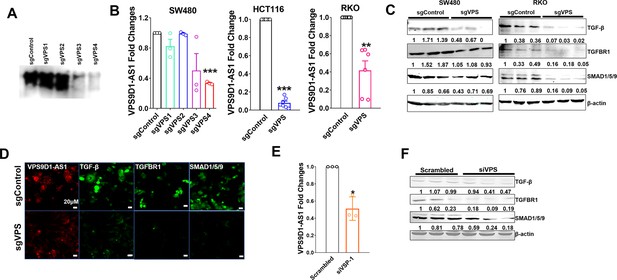
VPS9D1-AS1 activated TGF-β signaling.
(A) Northern blotting detected the effectiveness of sgRNA targeting VPS9D1-AS1 in SW480 cells. (B) qRT-PCR was used to determine the levels of VPS9D1-AS1 in stable knockout cells (sgControl vs. sgVPS). (C) Western blotting identified the levels of TGF-β, TGFBR1, and SMAD1/5/9. (D) RNA FISH-IF showed the levels of VPS9D1-AS1 and proteins that included TGF-β, TGFBR1, and SMAD1/5/9 in HCT116 cells. (E) qRT-PCR showed the downregulation of VPS9D1-AS1 after siRNA (siVPS) transfection in HCT116 cells. (F) Changes in TGF-β, TGFBR1, and SMAD1/5/9 after siRNA transfection in HCT116 cells. p-Values were obtained by paired t tests (B, E). Data are shown as the mean ± SEM (B, E). * p<0.05, ** p<0.01, *** p<0.001.
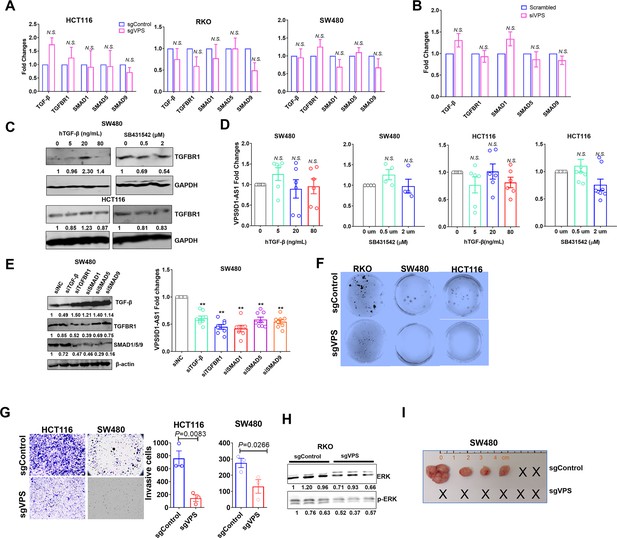
VPS9D1-AS1 regulated TGF-β signaling and promoted tumor proliferation and migration.
(A, B) mRNA levels of TGF-β, TGFBR1, SMAD1, ~5, and ~9 after VPS9D1-AS1 knockout or knockdown. (C) Human recombinant (h) TGF-β- and SB431542-treated SW480 and HCT116 cells. (D) hTGF-β and SB431542 had no effect on VPS9D1-AS1 levels. (E) VPS9D1-AS1 levels were decreased by siTGF-β, siTGFBR1, and siSMAD1, ~5, ~9. (F) Clone forming assay results of RKO/SW480/HCT116 sgControl and sgVPS cells after culturing for 14 days. (G) Transwell assays determined the migration of HCT116 and SW480 cells. (H) The levels of ERK and pERK in RKO cells (the interference gene was same with used as Figure 3—figure supplement 1C). (I) SW480 sgControl and sgVPS cells were separately transplanted subcutaneously into BALB/c nude mice. p-Values were obtained by paired or unpaired t tests (A, B, D, E, G). Data are shown as the mean ± SEM (A, B, D, E, G). N.S. not significant, ** p<0.01.
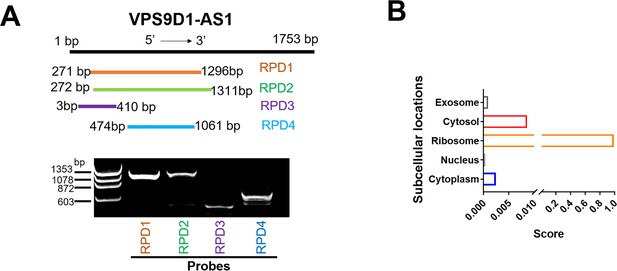
VPS9D1-AS1 functions as the scaffolding lncRNA.
(A) Synthesized probes used in RNA pulldown (RPD) assays for targeting the sequence of VPS9D1-AS1. (B) Subcellular localizations of VPS9D1-AS1 were analyzed by online tools (http://www.csbio.sjtu.edu.cn/bioinf/lncLocator/).
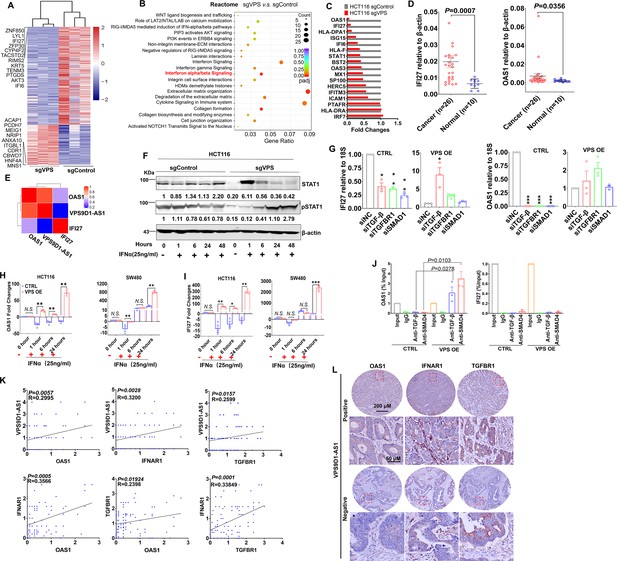
VPS9D1-AS1 regulates interferon signaling.
(A) Heatmap illustrating the results of RNA sequencing of the genes regulated by VPS9D1-AS1. (B) VPS9D1-AS1 regulated the pathways associated with interferon signaling. (C) Differential expression of 17 genes in the IFNα/β signaling pathway was validated in HCT116 cells. (D) Colorectal cancer (CRC) tissue mRNA levels of IFI27 and OAS1 were determined by qRT-PCR, and (E) their relationships with VPS9D1-AS1 were calculated with Pearson correlation analysis. (F) Effect of VPS9D1-AS1 on STAT1 pathway activation induced by human recombinant IFNα. (G) VPS9D1-AS1 overexpression (OE) increased the expression of IFI27 and OAS1 through activated TGF-β signaling. (H) OAS1 and (I) IFI27 levels exhibited disparate changes upon IFNα stimulation in VPS9D1-AS1 OE cells and control (CTRL) cells. (J) Chromatin immunoprecipitation (ChIP) assays demonstrated the interactions between SMAD4 and the promoter regions of OAS1. (K) Pearson correlation analyses investigated the relationships among VPS9D1-AS1, OAS1, IFNAR1, and TGFBR1 in CRC tissues. (L) Immunohistochemistry assays showed the levels of OAS1, IFNAR1, and TGFBR1 in patients with negative or positive expression of VPS9D1-AS1. p-Values were obtained by unpaired t test (D, J), two-way ANOVA (G, H, I), and Pearson correlation (K). Data are shown as the mean ± SEM (D, G, H, I, J). * p<0.05, ** p<0.01, *** p<0.001. N.S., not significant.
-
Figure 4—source data 1
STAT1 and pSTAT1 western blot for Figure 4F.
- https://cdn.elifesciences.org/articles/79811/elife-79811-fig4-data1-v2.zip
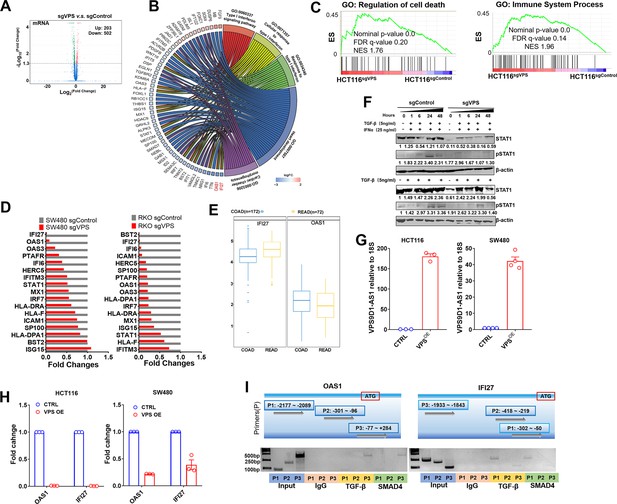
VPS9D1-AS1 plays a role on interferon signaling.
(A) Volcano plot displaying the differential mRNA profiles of HCT116sgControl and HCT116sgVPS cells. (B) Gene ontology (GO) analyses explored the roles of VPS9D1-AS1 in regulating IFNα/β signaling. (C) Gene Set Enrichment Analysis (GSEA) revealed that VPS9D1-AS1 was involved in pathways related to cell death and immune system processes. (D) Differential expression of 17 genes in the IFNα/β signaling pathway was validated in RKO and SW480 cells. (E) TCGA COAD and READ datasets confirmed the overexpression (OE) of OAS1 and IFI27. The fold changes for OAS1 and IFI27 expression in cancer tissues relative to normal tissues were shown. (F) hTGF-β prevents hIFNα from activating STAT1 phosphorylation in HCT116 cells. (G) Fold changes of VPS9D1-AS1 in HCT116 and SW480 VPS9D1-AS1 OE stale cell lines relative to control (CTRL) stale cell lines. ROI, region of interest. (H) Levels of IFI27 and OAS1 in cells with VPS9D1-AS1 OE. (I) ChIP assays determined the interaction of antibodies against TGF-β and SMAD4 with the promoter regions of OAS1 and IFI27 in HCT116 cells. Data are shown as the mean ± SEM (G, H).
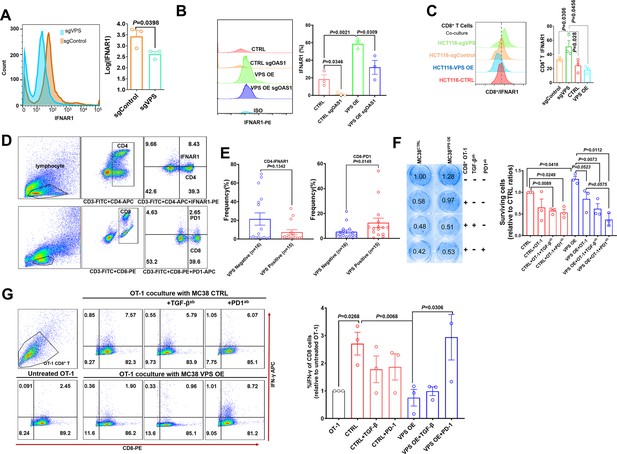
VPS9D1-AS1 mediates crosstalk between T cells and cancer cells by regulating IFNAR1.
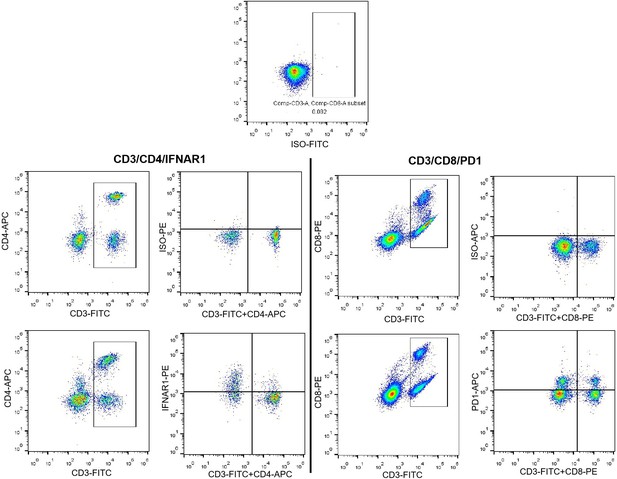
(A) Flow cytometry (FCM) revealed the decrease in IFNAR1 after VPS9D1-AS1 knockout in HCT116 cells and (B) in HCT116 sgControl and sgVPS cells after CRISPR/Cas9-mediated inhibition of OAS1. Experiments were repeated three times. (C) Levels of IFNAR1 on the surface of CD8+ T cells cocultured with HCT116 control (CTRL), VPS overexpression (OE), sgControl, and sgVPS cells were detected by FCM in three independent assays. (D), (E) FCM determination demonstrated that colorectal cancer patients with positive VPS9D1-AS1 expression had lower levels of IFNAR1 in CD4+ and higher levels of PD1 in CD8+ T cells in peripheral blood than these patients with negative VPS9D1-AS1 expression. (F) T cell cytotoxicity assays against CTRL and VPS9D1-AS1-OE MC38-OVA cell lines by OT-1 CD8+ T cells. VPS9D1-AS1 OE reduced the cytotoxicity of activated OT-1 CD8+ T cells. (G) FCM determined the IFN-γ levels in OT-1 CD8+ T cells after exposure to CTRL and VPS-OE MC38 cells. Antibodies against TGF-β and PD1 were added to the medium. The experiments were repeated three times. p-Values were obtained by unpaired t test (A, B, C, E, F, G). Data are shown as the mean ± SEM (A, B, C, E, F, G).
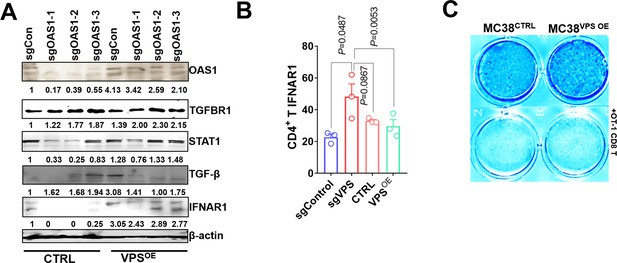
VPS9D1-AS1 mediates the crosstalk between tumors and T cells through TGF-β and IFN signaling.
(A) Western blotting assay to determine the levels of OAS1, TGFBR1, STAT1, TGF-β, and IFNAR1 in HCT116 cells. (B) IFNAR1 levels in CD4+ cells after coculturing with sgControl, sgVPS, CTRL, and VPS overexpression (OE) cells. (C) Representative results of T cell cytotoxicity assays of the indicated MC38CTRL and MC38VPS OE cell lines after exposure to OT-1 CD8+ T cells. p-Values were obtained by unpaired t test (B). Data are shown as the mean ± SEM (B).
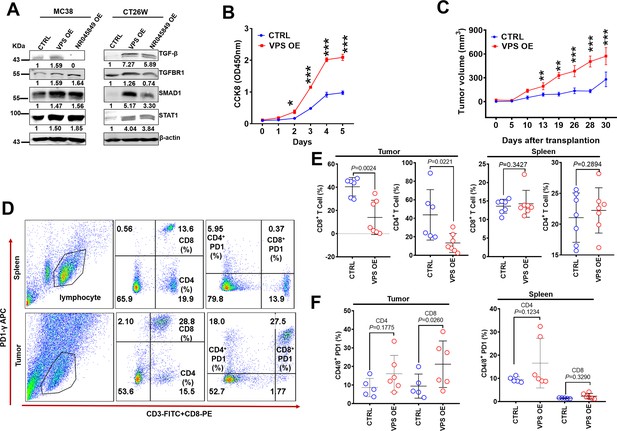
VPS9D1-AS1 overexpression (OE) cells inhibited T cell function in vivo.
(A) VPS9D1-AS1 OE promoted the expression of TGF-β, TGFBR1, SMAD1, and STAT1 in murine cells. (B) MC38 VPS9D1-AS1 OE and control (CTRL) cells were determined proliferation using CCK8 assays. (C) Growth curves of MC38 allograft tumors (n=7 per group). (D) Plots represent the percentages of CD8+ and CD4+ T cells and PD1 frequencies in allograft tumors and spleens. (E) Levels of CD8+ and CD4+ T cells were compared in the tumor and spleen. (F) Levels of PD1 in CD4+ T and CD8+ T cells were compared between CTRL and VPS OE allograft tumors (left) and spleens (right). p-Values were obtained by two-way ANOVA (B) and unpaired t nonparametric tests (E, F). Data are shown as the mean ± SEM (E, F). **p<0.01, ***p<0.001.
-
Figure 6—source data 1
TGF-β, SMAD1, STAT1, TGFBR1, and β-actin western blot for Figure 6A.
- https://cdn.elifesciences.org/articles/79811/elife-79811-fig6-data1-v2.zip
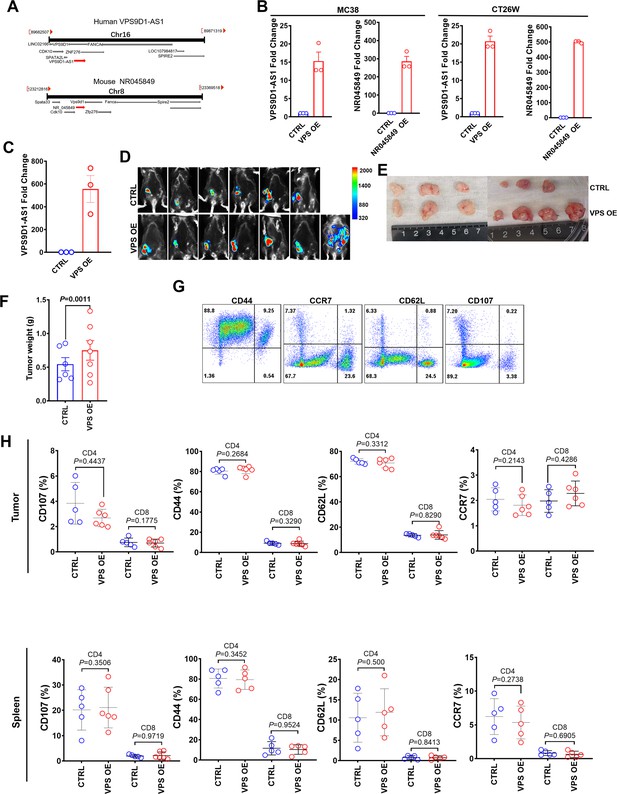
VPS9D1-AS1 inhibited T-infiltrating lymphocyte in allograft tumors.
(A) Schematic of human VPS9D1-AS1 and murine NR045849 locations on the chromosome. (B) Fold changes in the expression levels of VPS9D1-AS1 and NR045849 in MC38 and CT26W cells. (C) qRT-PCR confirmed the overexpression of VPS9D1-AS1 after three transfections with lentivirus vectors in MC38 cells. (D) In vivo imaging showed transplanted MC38CTRL and MC38VPS-OE allograft tumors. (E) Allograft tumors were harvested 35 days after injection (one mouse in the control [CTRL] group did not form tumors). (F) Allograft tumor weights for MC38CTRL and MC38VPS OE. (G) Representative flow cytometry results of CD44, CD62L, CD107, and CCR7 in CD4+ and CD8+ T cells. (H) Comparisons of the levels of CD44, CD62L, CD107, and CCR7. p-Values were obtained by unpaired t test (F, H). Data are shown as the mean ± SEM (F, H).
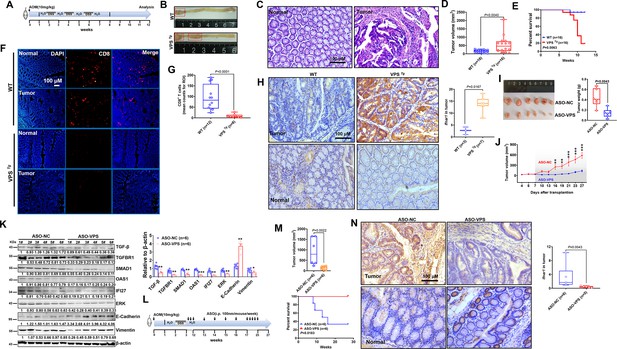
Transgenic mice validated the inhibitory role of VPS9D1-AS1 on CD8+ T cell infiltration and demonstrated the antitumor effects of targeting VPS9D1-AS1 with an antisense oligonucleotide (ASO) drug.
(A) Treatment scheme of the azoxymethane/dextran sulfate sodium salt (AOM/DSS) colorectal cancer (CRC) model. Endpoint at 12 weeks. (B) Representative images of colorectal tumors in mice of the indicated genotypes. (C) Hematoxylin (H) and eosin (E) staining showing the AOM/DSS-induced murine tumors. (D) Tumor volumes are plotted for wild-type (WT) and VPS Tg mice. (E) The overall survival (OS) curve depicts the difference in survival rate for WT and VPS Tg mice. (F), (G) VPS9D1-AS1 suppressed CD8+ T cell infiltration in AOM/DSS-induced CRC tissues. (ROI, region of interest). (H) The levels of Ifnar1 were upregulated in VPS Tg tumors compared with WT tumors. (I) Tumors, their weights, and (J) growth curves of HCT116 xenograft tumors after injecting ASO-VPS (n=6) and ASO-NC (n=6). (K) Western blotting assays of the proteins involved in TGF-β, IFN, ERK, and EMT signaling in xenograft tumors. (L) Treatment scheme of ASO-treated mice with AOM/DSS-induced CRC. (M) ASO-VPS treatment decreased tumor volumes and increased OS of VPS Tg mice with AOM/DSS-induced CRC. (N) Ifnar1 expression upon ASO-VPS and ASO-NC treatment. p-Values were obtained by unpaired t nonparametric test (D, G, H, I, K, M, N), log-rank test (E, M), and two-way ANOVA tests (J). Data are shown as the mean ± SEM (D, G, H, I, K, M, N). * p<0.05, ** p<0.01, *** p<0.001.
-
Figure 7—source data 1
TGF-β, TGFBR1, SMAD1, OAS1, IFI27, ERK, E-cadherin, vimentin, and β-actin western blot for Figure 7K.
- https://cdn.elifesciences.org/articles/79811/elife-79811-fig7-data1-v2.zip
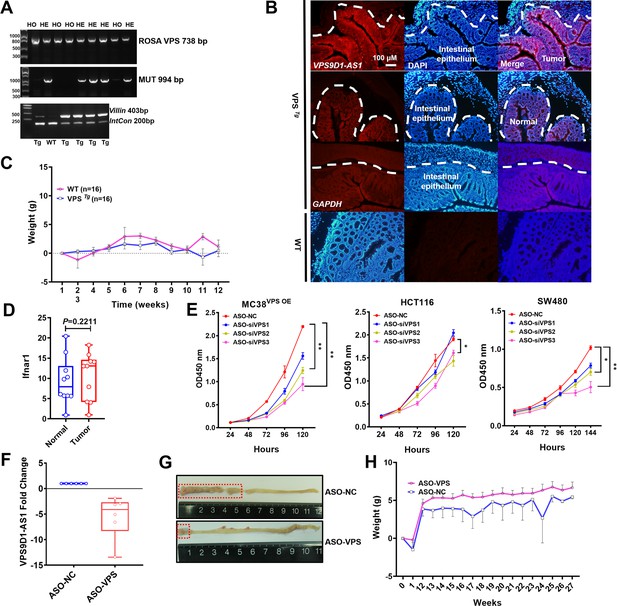
VPS9D1-AS1 drives azoxymethane/dextran sulfate sodium salt (AOM/DSS)-induced mouse model of colorectal cancer.
(A) Genotyping of VPS Tg mice. Internal control (IntCon). (B) RNA FISH identified VPS9D1-AS1 expression in intestinal epithelium of VPS Tg and wild-type (WT) mice. A probe targeting GAPDH mRNA was used as a positive control. (C) Body weight changes upon AOM/DSS treatment. (D) Comparison of Ifnar1 expression in normal tissue and tumor tissue. (E) CCK-8 assays for evaluating the proliferation of MC38VPS OE, HCT116, and SW480 cells after antisense oligonucleotide (ASO) treatment. (F) Fold changes in the levels of VPS9D1-AS1 in xenograft tumors. (G) Representative pictures of the mouse intestine treated with ASO-NC and ASO-VPS. (H) VPS Tg mice were treated with ASO-VPS and ASO-NC, and body weight changes were measured weekly. p-Values were obtained by one-way ANOVA (E) or unpaired t nonparametric test (D). Data are shown as the mean ± SEM (C, E, H) or box plots with minimum, first quartile, mean, third quartile, and maximum values (D, F).
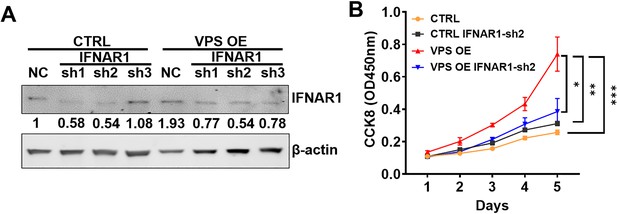
IFNAR1 knockdown (KD) inhibits tumor proliferation in VPS9D1-AS1 overexpression (OE) cells.
(A) Western blotting identified the levels of IFNAR1 after transfection of three shRNAs targeting IFNAR1 in HCT116 control (CTRL) and VPS9D1-AS1 OE cells. (B) CCK8 assays determined the cell proliferated rates for HCT116 CTRL cells, VPS OE cells, and these cells with stably IFNAR1KD.
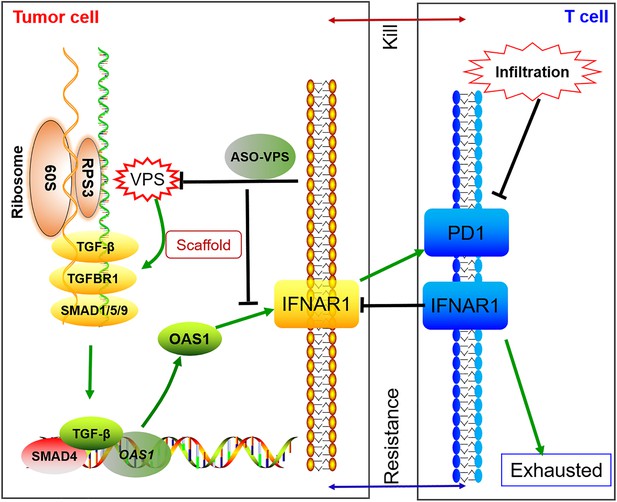
Illustration of the mechanism of VPS9D1-AS1 on promoting immune evasion via TGF-β signaling and interferon (IFN)-stimulated genes (OAS1 and IFNAR1).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | TGF-β (Rabbit polyclonal) | Cell Signaling Technology | Cat# 3711 s | mfIHC(1:800) WB(1:200) IF (1:100) |
| Antibody | TGFBR1 (Rabbit polyclonal) | Abcam | Cat# ab31013 | IF (1:1000) mfIHC(1:2000) WB(1:500) IHC(1:400) |
| Antibody | TGFBR2 (Mouse monoclonal) | Abcam | Cat# ab78419 | WB(1:500) |
| Antibody | SMAD1/5/9 (Rabbit polyclonal) | Abcam | Cat# ab66737 | mfIHC(1:1000) WB(1:400) |
| Antibody | pSMAD1/5/9 (Ser 463/Ser465) (Rabbit polyclonal) | Santa Cruz Biotechnology | Cat# sc-12353 | mfIHC(1:500) |
| Antibody | SMAD4 (Mouse monoclonal) | Cell Signaling Technology | Cat# 46535 | mfIHC(1:1000) |
| Antibody | SMAD2/3 (Rabbit polyclonal) | Santa Cruz Biotechnology | Cat# sc-8332 | mfIHC(1:400) |
| Antibody | pSMAD2/3 (Ser 423/425) (Rabbit polyclonal) | Santa Cruz Biotechnology | Cat# sc-11769 | mfIHC(1:1000) |
| Antibody | SMAD6 (Rabbit polyclonal) | Santa Cruz Biotechnology | Cat# sc-25321 | WB(1:200) |
| Antibody | Cytokeratin (CK) (Mouse monoclonal) | Santa Cruz Biotechnology | Cat# sc-57004 | mfIHC(1:200) WB(1:500) |
| Antibody | STAT1 (Rabbit polyclonal) | Cell Signaling Technology | Cat# 14994 | WB(1:500) |
| Antibody | pSTAT1(Tyr701) (Rabbit polyclonal) | Cell Signaling Technology | Cat# 9167 | WB(1:500) |
| Antibody | ERK (Rabbit polyclonal) | Cell Signaling Technology | Cat# 4695 S | WB(1:200) |
| Antibody | pERK (Rabbit polyclonal) | Cell Signaling Technology | Cat# 4376 S | WB(1:200) |
| Antibody | Vimentin (Rabbit monoclonal) | Abcam | Cat# ab92547 | WB(1:100) |
| Antibody | E-cadherin (Rabbit polyclonal) | Abcam | Cat# ab15148 | WB(1:100) |
| Antibody | N-cadherin (Rabbit polyclonal) | Abcam | Cat# ab12221 | WB(1:100) |
| Antibody | GAPDH (Mouse monoclonal) | Beyotime | Cat# AG019-1 | WB(1:1000) |
| Antibody | β-actin (Mouse monoclonal) | Beyotime | Cat# AF0003 | WB(1:1000) |
| Antibody | CD4 (Rabbit monoclonal) | Abcam | Cat# ab133616 | IHC(1:200) mfIHC(1:1000) |
| Antibody | CD8 (Mouse monoclonal) | Santa Cruz Biotechnology | Cat# sc-53212 | IHC(1:500) mfIHC(1:200) |
| Antibody | FOXP3 (Rabbit polyclonal) | Cell Signaling Technology | Cat# 98377 | IHC(1:200) mfIHC(1:800) |
| Antibody | OAS1 (Rabbit polyclonal) | Cell Signaling Technology | Cat# 14498 | IHC(1:100) |
| Antibody | IFI27 (Rabbit polyclonal) | Abclonal | Cat# A14174 | WB(1:100) |
| Antibody | RPS3 (Rabbit monoclonal) | Abcam | Cat# ab128995 | WB(1:200) |
| Antibody | IFNAR1 (Rabbit polyclonal) | Abcam | Cat# ab45172 | WB(1:100) |
| Antibody | PDL1 (Rabbit monoclonal) | Abcam | Cat# ab205921 | WB (1:100) |
| Antibody | Ki67 (Rabbit polyclonal) | ZSGB-Bio | Cat# ZM-0166 | IHC (1:1000) |
| Antibody | Human CD3 (OKT3) FG (Mouse monoclonal) | eBioscience | Cat# 16-0037-85 | (1:100) |
| Antibody | Human CD28 FG 16-0289-85 (Mouse monoclonal) | eBioscience | Cat# 16-0289-85 | |
| Antibody | Human FITC CD3 (Mouse monoclonal) | BD Pharmingen | Cat# 555332 | FCM(1:100) |
| Antibody | Human APC CD4 (Mouse monoclonal) | BD Pharmingen | Cat# 555349 | FCM(1:100) |
| Antibody | Human PE CD8 (Mouse monoclonal) | BD Pharmingen | Cat# 555637 | FCM(1:100) |
| Antibody | Ultra-LEAF purified anti-mouse CD3 Antibody (Rabbit monoclonal) | BioLegend | Cat# 100239 | FCM (1:100) |
| Antibody | Ultra-LEAF purified anti-mouse CD28 Antibody (Mouse monoclonal) | BioLegend | Cat# 122021 | (1:100) |
| Antibody | Mouse FITC CD3 (Rabbit monoclonal) | BioLegend | Cat# 100204 | FCM(1:100) |
| Antibody | Mouse PE CD8a (Rabbit monoclonal) | BioLegend | Cat# 100708 | FCM(1:100) |
| Antibody | Mouse APC CD44 (Rabbit monoclonal) | BioLegend | Cat# 103012 | FCM(1:100) |
| Antibody | Mouse BV421 CD62L (Rabbit monoclonal) | BioLegend | Cat# 104435 | FCM(1:100) |
| Antibody | Mouse APC CD279 (PD-1) (Rabbit monoclonal) | BioLegend | Cat# 135209 | FCM(1:100) |
| Antibody | Mouse PE/Cy7 CD107a(LAMP-1) (Rabbit monoclonal) | BioLegend | Cat# 121619 | FCM(1:100) |
| Antibody | Mouse PE/Cy7 CD197 (CCR7) (Rabbit monoclonal) | BioLegend | Cat# 353211 | FCM(1:100) |
| Antibody | InVivoPlus Anti-mouse PD-1 (Mouse monoclonal) | BiXcell | Cat# BP0273 | (1:100) |
| Antibody | InVivoPlus Anti-mouse/human TGF-β (Mouse monoclonal) | BiXcell | Cat# BP0057 | (1:100) |
| Antibody | Human IFN-alpha/beta R1 PE-conjugated antibody (Mouse monoclonal) | R&D | Cat# FAB245P | FCM(1:100) |
| Biological samples (Homo sapiens) | Human colon cancer tissue microarray | OUTDO | HCol-AS-180Su-14 | |
| Biological samples (H. sapiens) | Human rectal cancer tissue microarray | OUTDO | HRec-Ade180Sur-05 | |
| Biological samples (H. sapiens) | Human colorectal cancer tissue cDNA | OUTDO | HcolA30CS01 | |
| Cell lines (H. sapiens) | HCT116 | Dr. L. Yang (Beijing Chaoyang Hospital) | ||
| Cell lines (H. sapiens) | SW480 | Dr. L. Yang (Beijing Chaoyang Hospital) | ||
| Cell lines (H. sapiens) | RKO | Dr. L. Yang (Beijing Chaoyang Hospital) | ||
| Cell lines (H. sapiens) | HEK293T | Dr. L. Yang (Beijing Chaoyang Hospital) | ||
| cell lines (Mus musculus) | MC38 | PUMC | ||
| Cell lines (M. musculus) | CT26W | PUMC | ||
| Commercial assay or kit | QuantiNova SYBR Green PCR kit | Qiagen | Cat# 208054 | |
| Commercial assay or kit | QuantiTect Reverse Transcription kit | Qiagen | Cat# 205314 | |
| Commercial assay or kit | Poerce BCA Protein Assay Kit | Thermo | Cat# RB230514 | |
| Chemical compound, drug | Lipofectamine 3000 | Invitrogen | Cat# L3000015 | |
| Recombinant proteins | Human Interleukin-2 (hIL-2) | Cell Signaling Technology | Cat# 8907SC | |
| Commercial assay or kit | PerkinElmer Opal 7-color fIHC Kit | PerkinElmer | Cat# NEL797001KT | |
| Commercial assay or kit | Ficoll reagent | Solarbio | Cat# P8900 | |
| Commercial assay or kit | Tumor Dissociation Kit, mouse | Miltenyi Biotec | Cat# 130-091-376 | |
| Commercial assay or kit | CD8a+ T Cell Isolation Kit, mouse | Miltenyi Biotec | Cat# 130-104-075 | |
| Commercial assay or kit | Anti-Human CD8 Magnetic Particles-DM | BD Pharmingen | Cat# 557766 | |
| Commercial assay or kit | HiScribeTM T7 Quick High Yeild RNA Synthesis Kit | NEB | Cat# E2040S | |
| Commercial assay or kit | Monarch RNA Cleanup Kit | NEB | Cat# T2030S | |
| Commercial assay or kit | IP Lysis Buffer | Thermo Fisher | Cat# 87787 | |
| Commercial assay or kit | Pierce RNA 3’ Desthiobiotinylation kit | Thermo Fisher | Cat# 20163 | |
| Commercial assay or kit | PIERCE MAGNETIC RNA PULL-DOWN KIT | Thermo Fisher | Cat# 20164 | |
| Chemical compound, drug | CX-5461 | MCE | Cat# HY-13323 | |
| Recombinant proteins | Human Interferon-α1 | Cell Signaling Technology | Cat# 8927SC | |
| Commercial assay or kit | DIG Northern Starter | Roche | Cat# 12039672910 | |
| Commercial assay or kit | VPS9D1-AS1 FISH Probes | Biosearch | N/A | |
| Commercial assay or kit | Magna RIP RNA-Binding Protein Immunoprecipitation Kit | Millipore | Cat# 17–700 | |
| Commercial assay or kit | Positively Charged Biodyne Nylon membrane | Invitrogen | Cat# 10100 | |
| Commercial assay or kit | ChIP-IT Express Enzymatic Magnetic Chromatin Immunoprecipitation kit | Active Motif | Cat# 53009 | |
| Commercial assay or kit | Stellaris RNA FISH Wash Buffer A | BIOSEARCH | Cat# SMF-WA1-60 | |
| Commercial assay or kit | Stellaris RNA FISH Wash Buffer B | BIOSEARCH | Cat# SMF-WA1-20 | |
| Commercial assay or kit | Stellaris RNA FISH Hybridization buffers | BIOSEARCH | Cat# SMF-HB1-10 | |
| Commercial assay or kit | Stellaris FISH Probes with Quasar 570 Dye | BIOSEARCH | Cat# SMF-1063–5 | |
| Commercial assay or kit | FITC labeled Goat anti-rabbit secondly antibody | Byotime | Cat# 0652 | |
| Commercial assay or kit | FITC labeled Goat anti-mouse secondly antibody | Byotime | Cat# A0568 | |
| Commercial assay or kit | Antifade mounting medium with DAPI | Solarbio | Cat# S2110 |
Additional files
-
Supplementary file 1
Sequences for sgRNA, PCR primer, siRNA and shRNA.
(a) sgRNA sequence. (b) Primers for PCR. (c) siRNA and shRNA sequences. (d) The primer for RNA pull down (RPD) probe synthesized. (e) The Primers for ChIP-PCR.
- https://cdn.elifesciences.org/articles/79811/elife-79811-supp1-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/79811/elife-79811-mdarchecklist1-v2.pdf