RNA localization mechanisms transcend cell morphology
Figures
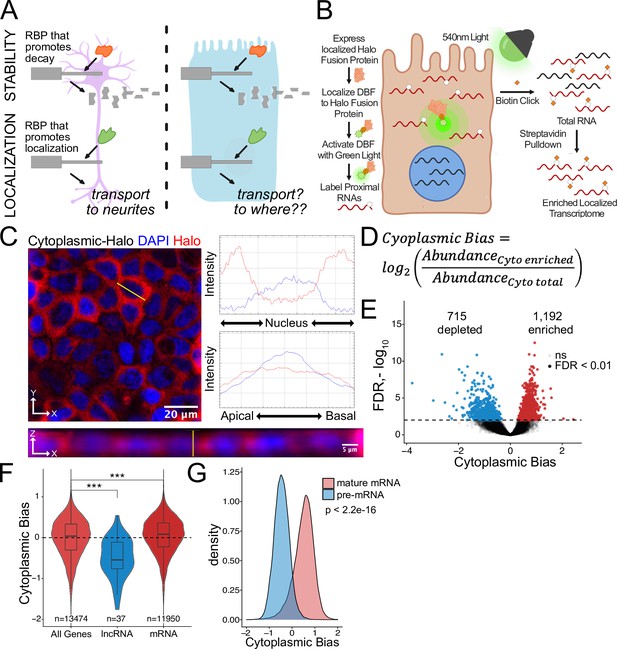
Halo-seq enriches cytoplasmic localized RNA molecules in C2bbe1 monolayers.
(A) RNA regulatory processes are generally driven through the interaction of an RBP and an RNA sequence element. Because RBPs are often widely expressed, many of these processes (exemplified here by RNA decay) operate across cellular contexts. The orange RBP binds a particular sequence within a transcript to promote its degradation in both cell types. However, because RNA localization is intimately linked with cell morphology, if an RBP/RNA interaction promotes RNA localization in one cell type, how the same interaction would affect RNA localization in another cell type is unclear. (B) Overview of the Halo-seq method. A HaloTag is genetically fused to a protein with specific localization. DBF-conjugated Halo ligands are specifically recruited to the Halo fusion proteins due to their high affinity and covalent binding to HaloTag protein domains. When exposed to green light, DBF produces oxygen radicals that, in combination with propargylamine, alkynylate DBF-proximal RNAs. ‘Click’ chemistry covalently attaches biotin moieties to alkynylated RNAs in vitro, facilitating their enrichment via streptavidin pulldown, thereby enriching localized RNAs for high-throughput sequencing. (C) HaloTag protein fusions to P65 are localized to the cytoplasm. HaloTag fusions are visualized in fixed cells through the addition of a Halo ligand fluorophore shown in red. DAPI stain marks the nuclei in blue. Profile lines across the XY image demonstrate exclusion of Halo signal from the nucleus and in the XZ-projected image demonstrate no apical or basal bias. Images are an average projection through the XZ axis of approximately 50 cells. (D) Equation for Cytoplasmic Bias. Cytoplasmic RNA localization is calculated with a metric termed Cytoplasmic Bias as the log2 of a transcript’s abundance in the streptavidin enriched fraction divided by its abundance in the input total RNA. (E) Genes with differing Cytoplasmic Bias following Halo-seq RNA labeling using the cytoplasmic-Halo fusion. FDR calculated by DESeq2 (F) Cytoplasmic Bias for defined classes of RNAs. p-Values were calculated using a Wilcoxon rank-sum test. (G) Cytoplasmic Bias for unspliced, intron-containing pre-mRNAs and spliced mature mRNAs. Cytoplasmic Bias was calculated for the spliced and unspliced isoform of each transcript. p-Values were calculated using a Wilcoxon rank-sum test. ns (not significant) represents p>0.05, * p<0.05, ** p<0.01, *** p<0.001 and **** represents p<0.0001.
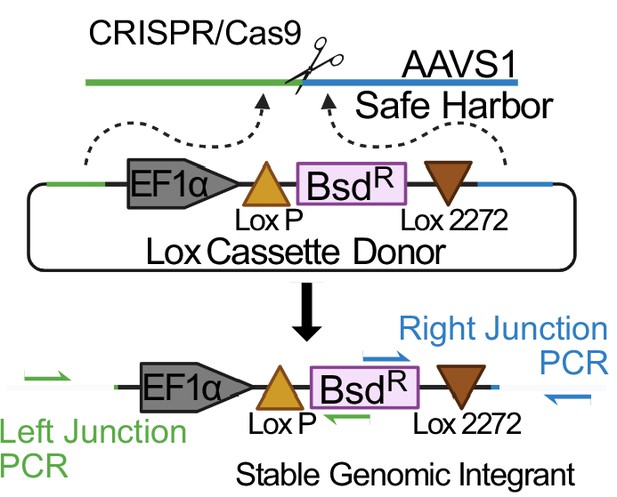
Schematic of Lox cassette knock-in to the AAVS1 safe harbor locus.
A loxP-flanked blasticidin resistance gene driven off a constitutively active EF1α promoter is inserted into the AAVS1 safe harbor. Primer positions to PCR amplify the junctions of this insertion are notated.
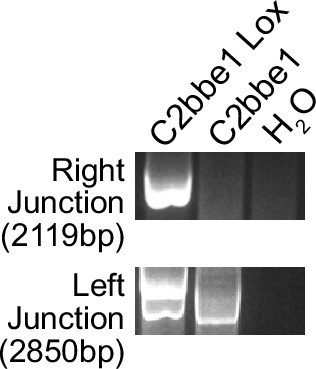
PCR amplification of knock-in junctions.
A cell line without the knocked in cassette and water included as template controls. Right junction PCR product is 2119 bp, and Left Junction PCR product is 2850 bp. The presence of a PCR product demonstrates correct integration.
-
Figure 1—figure supplement 2—source data 1
1% Agarose gel of PCR products using primers flanking the AAVS1 insertion site for the LoxP cassette.
Where targeted insertion is performed (AAVS1 lox) a unique band is observed. When the LoxP cassette is not targeted (Random Lox) we do not observe specific insertion into the AAVS1 site. A no template water control is also included. The ladder used is a 1 kb Plus ladder (NEB N3200).
- https://cdn.elifesciences.org/articles/80040/elife-80040-fig1-figsupp2-data1-v1.zip
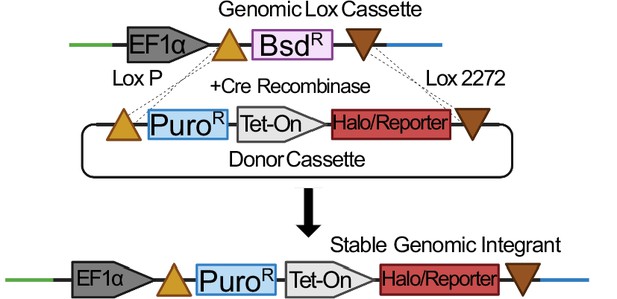
Schematic of cre-mediated cassette switching.
Through cre recombinase activity, the floxed blasticidin gene is replaced with a puromycin resistance gene as well as any additional reporter or HaloTag fusion proteins. Stable cassette switching events are then selected with puromycin to produce stable cell lines.
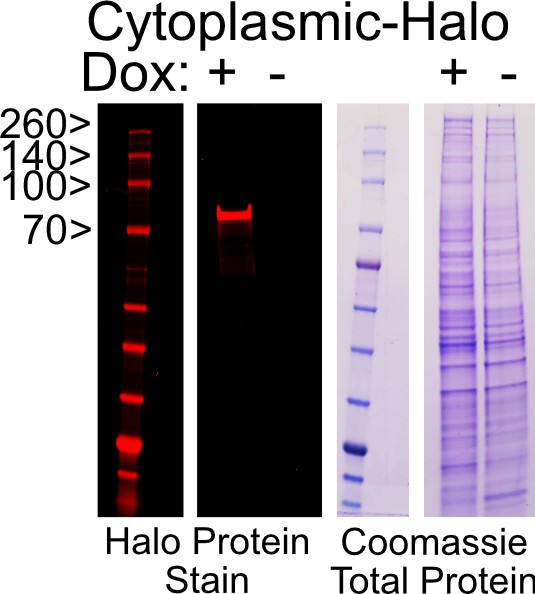
SDS-PAGE gel of doxycycline-induced cytoplasmic-Halo construct (95 kDa) visualized with Halo ligand fluorophore in red (JF646).
Total protein is visualized with a Coomassie stain.
-
Figure 1—figure supplement 4—source data 1
GoPAGE Bis-Tris Precast Gel, 4–12% with Halo Ligand JF646 visualizing Halo tagged proteins in doxycycline induced cell lysates.
The ladder is a broad-spectrum protein ladder (Fisher Scientific PI26623).
- https://cdn.elifesciences.org/articles/80040/elife-80040-fig1-figsupp4-data1-v1.zip
-
Figure 1—figure supplement 4—source data 2
GoPAGE Bis-Tris Precast Gel, 4–12% with coomassie visualizing total protein extracted from the halo tag expressing cell lysates.
The ladder is a broad-spectrum protein ladder (Fisher Scientific PI26623).
- https://cdn.elifesciences.org/articles/80040/elife-80040-fig1-figsupp4-data2-v1.zip
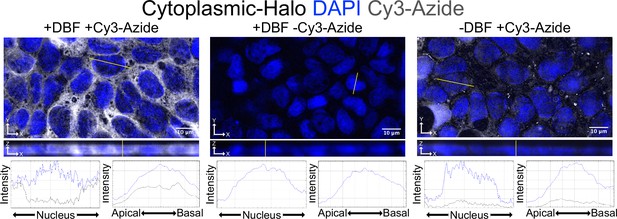
Fusing Cy3-Azide fluorophores to alkynylated biomolecules using Click chemistry allows for visualization of labeled molecules in fixed cells.
Alkynylated molecules are restricted to the cytoplasm in cells expressing cytoplasmic-Halo. This localization is dependent on both addition of DBF and Cy3-azide, demonstrating the ability of HaloTag-restricted DBF to induce alkynylation of biomolecules specifically in the cytoplasm.
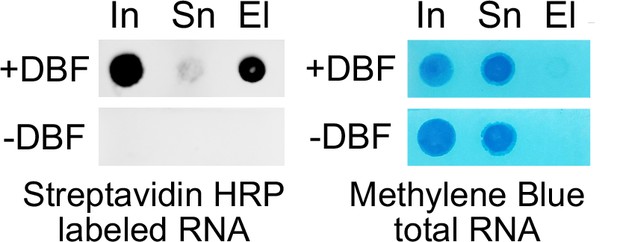
In vitro biotinylation of alkynyklated RNA labeled from cytoplasmic-Halo expressing cells is dependent on DBF addition as visualized by streptavidin-HRP on an RNA dot blot.
Biotinylated input (In) RNA are cleared from the supernatant (Sn) by streptavidin pull-downs but efficiently eluted (El) off the beads. Methylene Blue stains total RNA.
-
Figure 1—figure supplement 6—source data 1
RNA is spotted and crosslinked onto an Amersham Hybond-N +western blotting membrane.
Biotinylated RNA is visualized with Streptavidin HRP.
- https://cdn.elifesciences.org/articles/80040/elife-80040-fig1-figsupp6-data1-v1.zip
-
Figure 1—figure supplement 6—source data 2
RNA is spotted and crosslinked onto an Amersham Hybond-N +western blotting membrane.
Total RNA is visualized with 1% Methylene Blue.
- https://cdn.elifesciences.org/articles/80040/elife-80040-fig1-figsupp6-data2-v1.zip
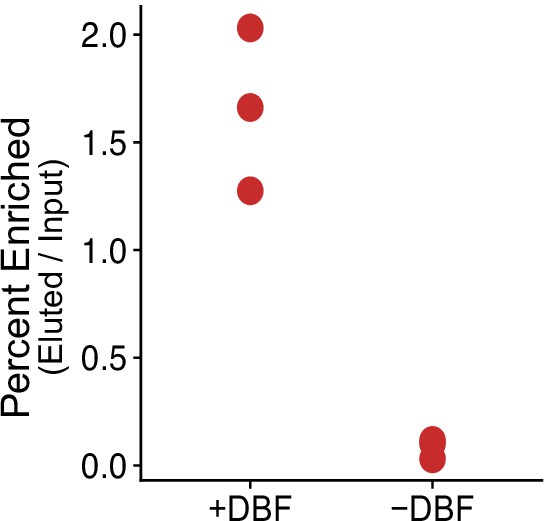
Percent enriched RNA after streptavidin pull-down of cytoplasmic-Halo-labeled RNAs.
Input and eluted RNA quantified by Qubit.
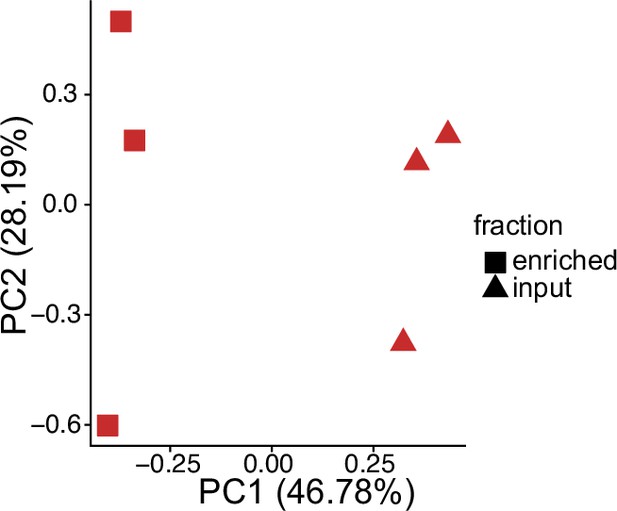
Principal component analysis of gene expression values from Halo-seq in cytoplasmic-Halo cells.
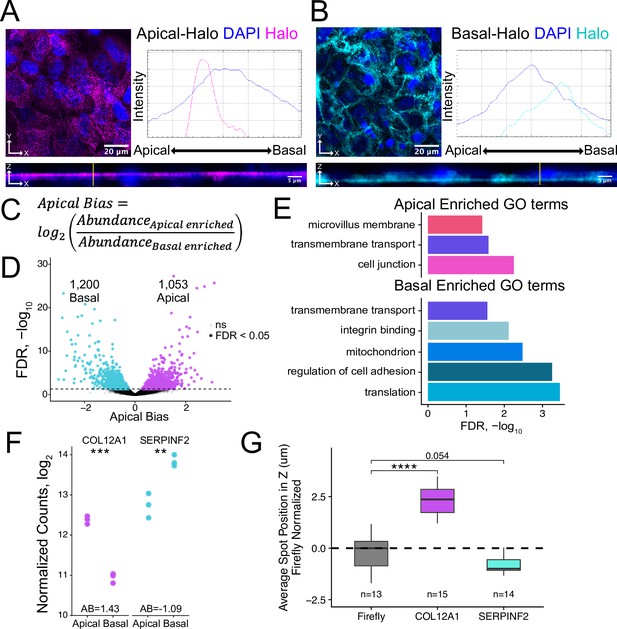
Halo-seq enriches RNAs at the apical and basal poles of enterocyte monolayers.
(A) HaloTag protein fusions to PODXL and (B) ATP1A1 are localized to the apical and basal compartments respectively. HaloTag domains are visualized in fixed cells through the addition of a Halo ligand fluorophore shown in magenta (apical) and cyan (basal). DAPI stain marks the nuclei in blue. Profile lines across the XZ image demonstrate their apical or basal bias. Images are an average projection through the XZ axis of approximately 30 cells. (C) Equation for Apical Bias. RNA localization across the apicobasal axis is calculated with a metric termed Apical Bias as the log2 of a transcript’s abundance in the apical enriched RNA fraction divided by its abundance in the basal enriched fraction. (D) Genes with differing Apical Bias following Halo-seq RNA labeling using the apical and basal-Halo fusions. FDR values calculated by DESeq2. (E) Enriched gene ontology (GO) terms associated with proteins encoded by RNAs identified as localized to the apical or basal pole of enterocytes. FDR calculated by topGO. (F) DESeq2-calculated normalized counts of candidate apical (COL12A1) and basal (SERPINF2) localized RNAs in the apical and basal enriched sequencing samples. p-Values calculated by DESeq2. (G) smFISH puncta position in Z. Firefly luciferase RNA included as a non-localized control. p-Values were calculated using a Wilcoxon rank-sum test. ns (not significant) represents p>0.05, * p<0.05, ** p<0.01, *** p<0.001 and **** represents p<0.0001.
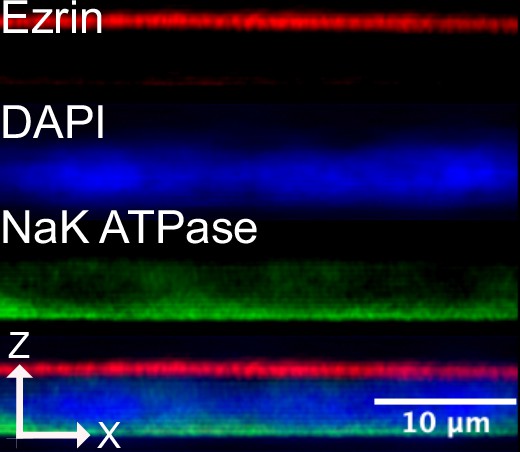
C2bbe1 monolayers differentiated for 7 days on transwell inserts are highly polarized as demonstrated by immunofluorescence for Ezrin (apical) and NaK-ATPase (Basal) endogenous polarity protein markers.
Images are an average projection through the XZ axis of roughly 20 cells.
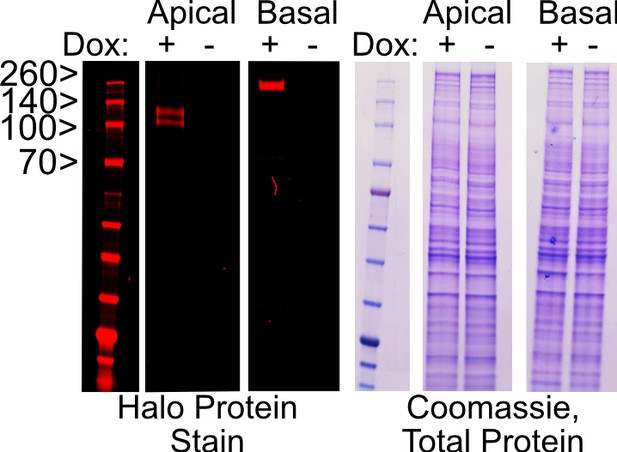
SDS-PAGE gel of doxycycline-induced apical-Halo and basal-Halo constructs (95 and 150 kDa respectively) visualized with Halo ligand fluorophore in red (JF 646).
Total protein is visualized with a Coomassie stain.
-
Figure 2—figure supplement 2—source data 1
GoPAGE Bis-Tris Precast Gel, 4–12% with Halo Ligand JF646 visualizing Halo-tagged proteins in doxycycline induced cell lysates.
The ladder is a broad-spectrum protein ladder (Fisher Scientific PI26623).
- https://cdn.elifesciences.org/articles/80040/elife-80040-fig2-figsupp2-data1-v1.zip
-
Figure 2—figure supplement 2—source data 2
GoPAGE Bis-Tris Precast Gel, 4–12% with coomassie visualizing total protein extracted from the halo tag expressing cell lysates.
The ladder is a broad-spectrum protein ladder (Fisher Scientific PI26623).
- https://cdn.elifesciences.org/articles/80040/elife-80040-fig2-figsupp2-data2-v1.zip
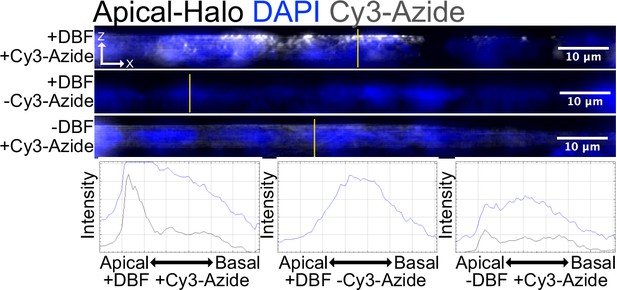
Cy3-Azide fluorophores attached to alkynylated biomolecules by Click chemistry visualize labeled molecules in fixed cells.
Alkynylated molecules are restricted to the apical pole in cells containing apical-Halo and basal-Halo, respectively. This localization is dependent on both addition of DBF and Cy3-azide. Images are an average projection through the XZ axis of roughly 50 cells.
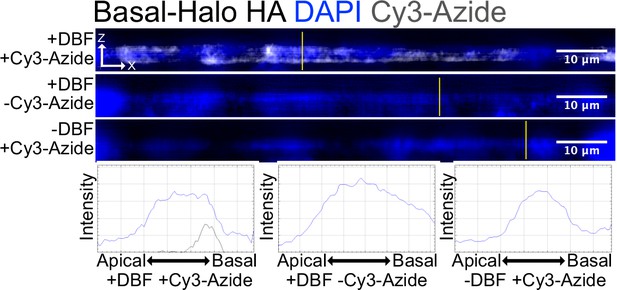
Cy3-Azide fluorophores attached to alkynylated biomolecules by Click chemistry visualize labeled molecules in situ.
Alkynylated molecules are restricted to the basal pole in cells containing apical-Halo and basal-Halo, respectively. This localization is dependent on both addition of DBF and Cy3-azide. Images are an average projection through the XZ axis of roughly 50 cells.

in vitro biotinylation of RNA labeled in apical-Halo and basal-Halo expressing cells is dependent on DBF addition as visualized by streptavidin-HRP on an RNA dot blot.
Biotinylated input (In) RNA are cleared from the supernatant (Sn) by streptavidin pull-downs but efficiently eluted (El) off the beads. Methylene Blue stains total RNA.
-
Figure 2—figure supplement 5—source data 1
RNA is spotted and crosslinked onto an Amersham Hybond-N +western blotting membrane.
Biotinylated RNA is visualized with Streptavidin HRP.
- https://cdn.elifesciences.org/articles/80040/elife-80040-fig2-figsupp5-data1-v1.zip
-
Figure 2—figure supplement 5—source data 2
RNA is spotted and crosslinked onto an Amersham Hybond-N +western blotting membrane.
Total RNA is visualized with 1% Methylene Blue.
- https://cdn.elifesciences.org/articles/80040/elife-80040-fig2-figsupp5-data2-v1.zip
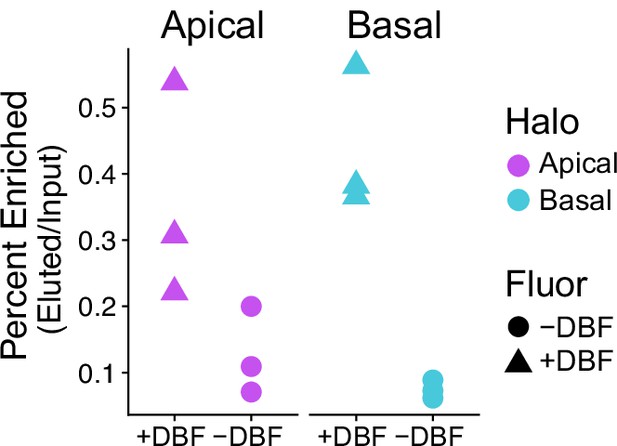
Percent enriched RNA after streptavidin pull-down of Apical-Halo and Basal-Halo-labeled RNAs.
Input and eluted RNA were quantified by Qubit.
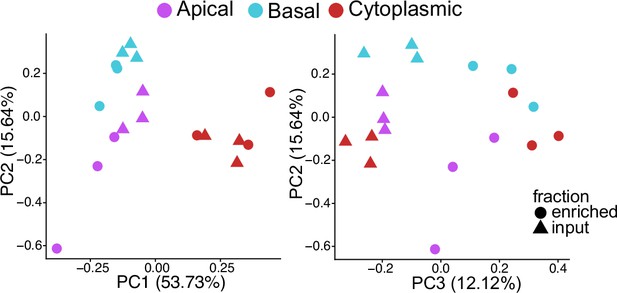
Principal component analysis of gene expression values from the cytoplasmic-Halo, apical-Halo, and basal-Halo experiments.
PC1 and 2 separate Halo-construct lines while PC2 and 3 separate input and enriched fractions.
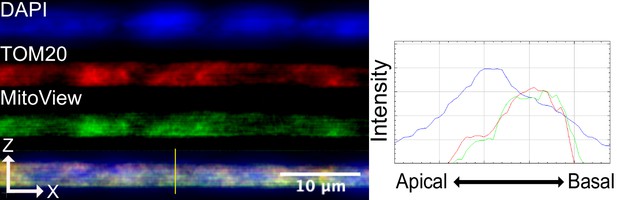
Mitochondria are visualized with both MitoView Green and immunofluorescence for TOM20 protein in fixed cells.
Profile lines across the XZ axis show basal localization of mitochondria. Images are an average projection through the XZ axis of roughly 30 cells.
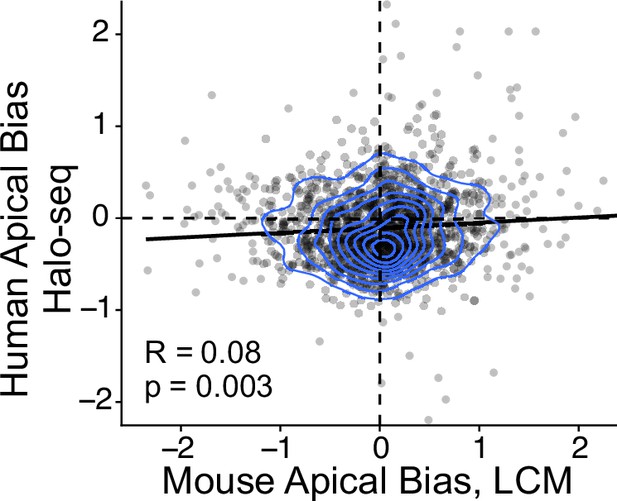
Direct comparison of Halo-seq calculated Apical Bias in Human C2bbe1 monolayers and Laser Capture Microdissection (LCM) calculated Apical Bias in adult Mouse enterocytes (Moor et al., 2017).
Correlation and p-values calculated with Spearman’s correlation test.
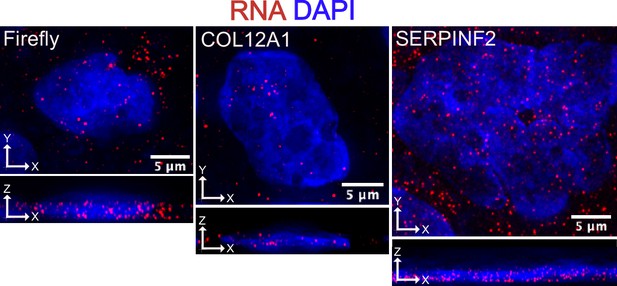
Representative smFISH images in fixed cells.
Images are a max projection through the XZ axis of a single cell. RNA signal is in red and DAPI nuclear stain is in blue.
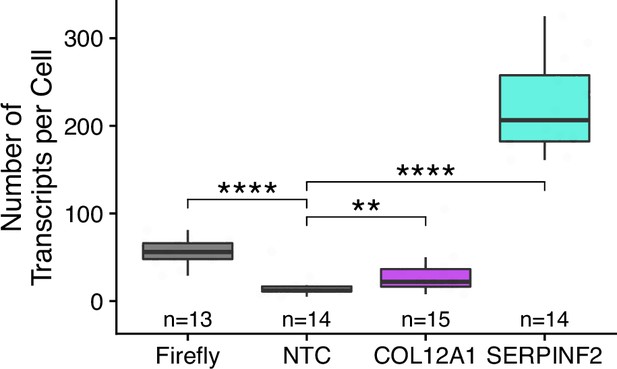
Number of smFISH puncta per cell.
p-Values were calculated using a Wilcoxon rank-sum test. No template control (Firefly luciferase smFISH probes in cells not expressing the Firefly luciferase transgene) included to show noise level in smFISH. ns (not significant) represents p>0.05, * p<0.05, ** p<0.01, *** p<0.001 and **** represents p<0.0001.
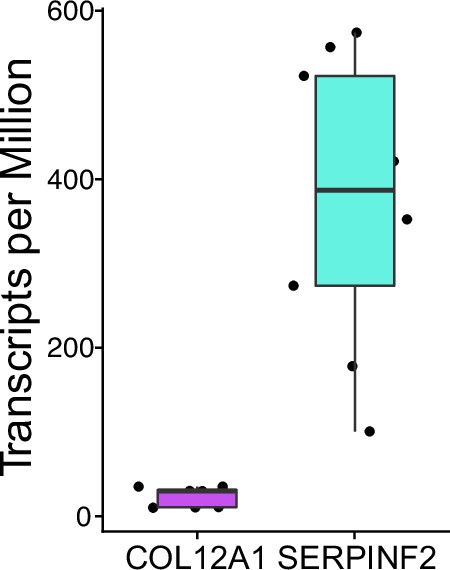
Transcripts per million for each candidate localized RNA in each sequenced input sample (n=9).
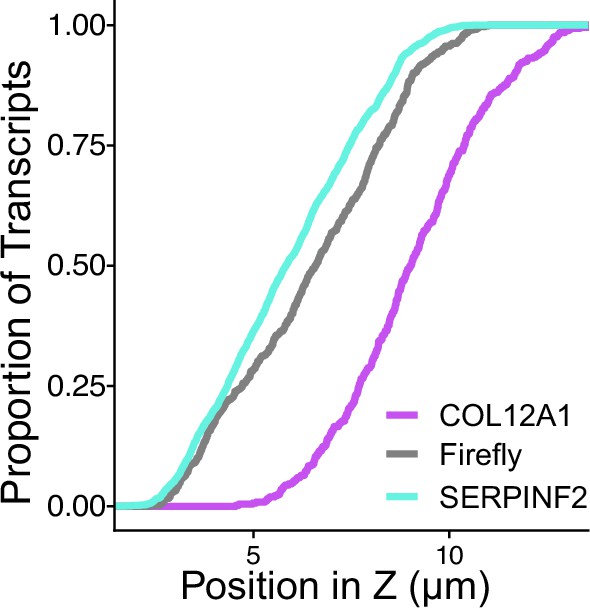
Cumulative distribution of smFISH spots across their position in Z as calculated by FISH-quant.
p-Values were all significant (<0.05) compared to Firefly luciferase as calculated by Wilcoxon rank-sum test and printed on Figure 2G.
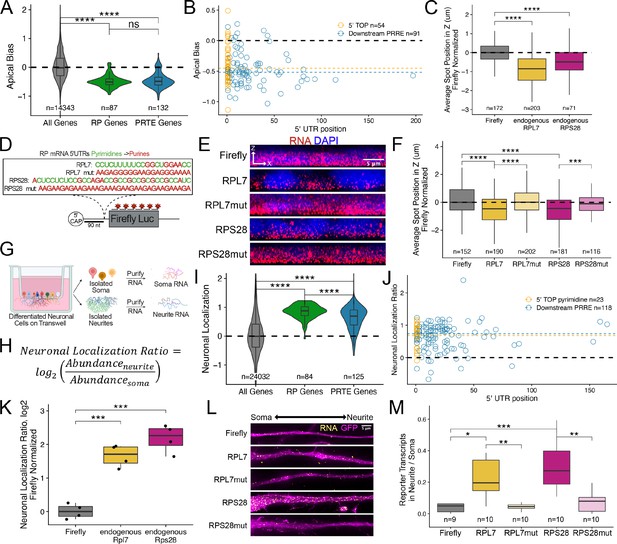
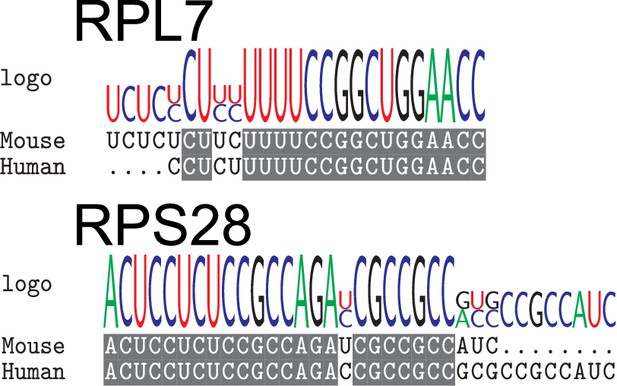
Ribosomal protein mRNAs are localized in a PRRE-dependent manner in multiple cell types.
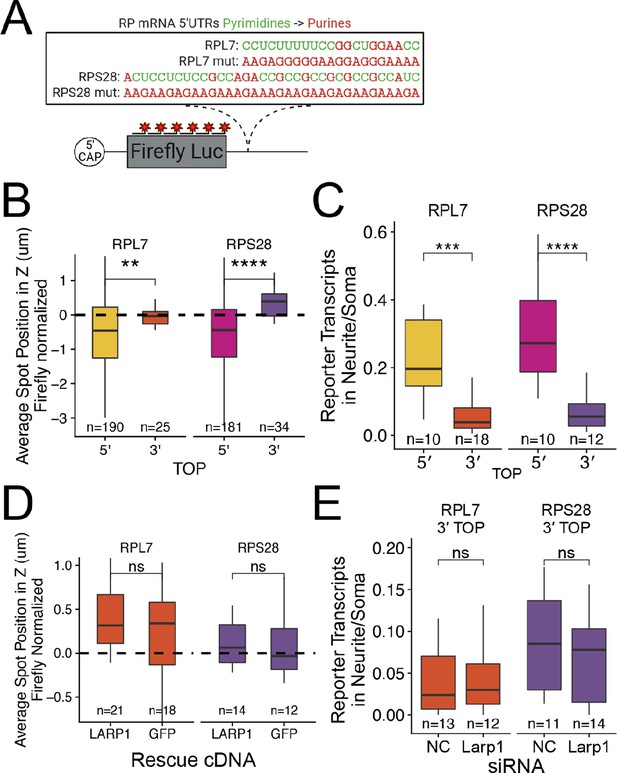
(A) Apical Bias for ribosomal protein (RP) genes and PRTE-containing genes as compared to all genes. P-values were calculated using a Wilcoxon rank-sum test. (B) Apical Bias for transcripts containing pyrimidine-rich 5′ UTR elements as a function of the position of the element within the 5′ UTR. Dotted lines represent medians for each class. (C) Localization of endogenous RPL7 and RPS28 transcripts in C2bbe1 monolayers as determined by smFISH. Values represent the average position per cell. Numbers of cells interrogated for each transcript are indicated. (D) Schematic of ribosomal protein PRRE reporter constructs. Mutant versions were created by swapping each pyrimidine for a purine. Motifs were inserted roughly 90 nucleotides downstream of the transcription start site. (E) Representative images of smFISH spots for each PRRE reporter construct. RNA signal shown in red while DAPI stains mark the nuclei blue. Images are a max projection through the XZ axis of roughly 20 cells. (F) smFISH puncta position in Z normalized to median untagged Firefly luciferase transcript position. P-values were calculated using a Wilcoxon rank-sum test. Values represent the average position per cell. Numbers of cells interrogated for each construct are indicated. (G) Schematic of neuronal subcellular fractionation and localized RNA isolation. (H) Equation for Neuronal Localization Ratio. Neuronal RNA localization is calculated with a metric termed Neuronal Localization Ratio as the log2 of a transcript’s abundance in the neurite fraction divided by its abundance in the soma fraction. (I) Neuronal Localization Ratio (LR) for ribosomal protein genes and PRTE-containing genes as compared to all genes. p-Values were calculated using a Wilcoxon rank-sum test. (J) Neurite localization ratio for transcripts containing pyrimidine-rich 5′ UTR elements as a function of the position of the element within the 5′ UTR. Dotted lines represent medians for each class. (K) Neurite-enrichment of endogenous Rpl7 and Rps28 transcripts as quantified by RT-qPCR following neuronal fractionation. Endogenous gene expression normalized to the endogenous RNA Tsc1. (L) Representative images of smFISH for each PRRE reporter construct. Images are a max projection through the XY axis of a single neurite positioned with the soma to the left. RNA signal shown in yellow while cell outlines marked by GFP signal shown as magenta. (M) Number of smFISH puncta in neurites normalized to soma. For all panels, p-values were calculated using a Wilcoxon rank-sum test. Numbers of cells interrogated for each construct are indicated. ns (not significant) represents p>0.05, * p<0.05, ** p<0.01, *** p<0.001 and **** represents p<0.0001.
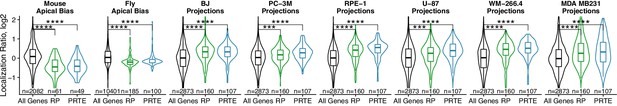
Localization of ribosomal protein mRNAs across the apicobasal axis of adult mouse enterocytes (Moor et al., 2017), the apicobasal axis of Drosophila follicular cells (Cassella and Ephrussi, 2022) and in migrating cell projections (Dermit et al., 2020) according to various sequencing experiments.
p-Values were calculated using a Wilcoxon rank-sum test. ns (not significant) represents p>0.05, * p<0.05, ** p<0.01, *** p<0.001 and **** represents p<0.0001.

Genes with differing Apical Bias with candidate RP mRNAs highlighted.
FDR values were calculated by DESeq2.

Representative smFISH images for endogenous RPL7 and RPS28 transcripts in an orthogonal, XZ projection of roughly 20 cells.
The apical side of the cell is toward the top while the basal side is toward the bottom. Large, non-spherical puncta toward the apical side of cells are background fluorescence (perhaps associated with probes getting stuck in the mucus found on the apical side of C2bbe1 cells), and are computationally removed prior to the calculation of puncta positions.
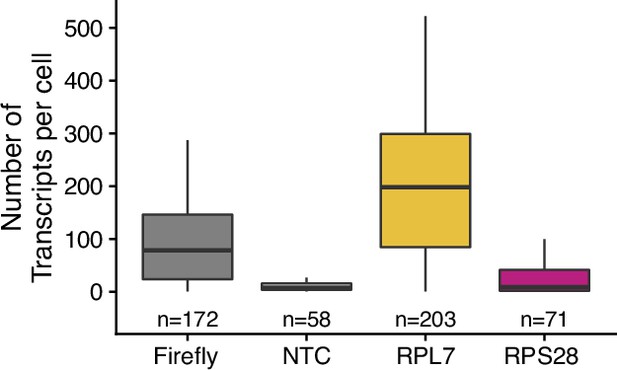
Transcript abundance per cell for endogenous RPL7 and RPS28 transcripts as measured by smFISH.
Numbers of cells interrogated for each construct are indicated. No template control (Firefly luciferase smFISH probes in cells not expressing the Firefly luciferase transgene) included to show noise level in smFISH.
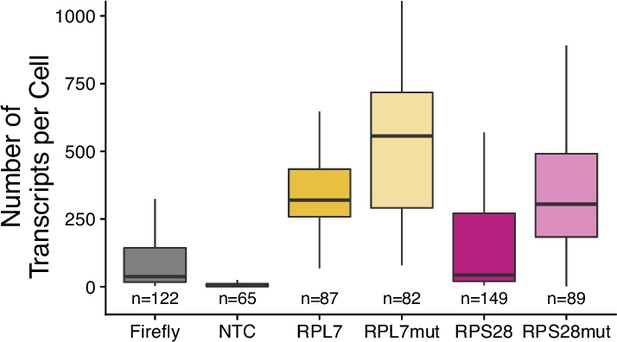
Transcript abundance per cell for PRRE-containing reporters in C2bbe1 cells as measured by smFISH.
Numbers of cells interrogated for each construct are indicated. No template control (Firefly luciferase smFISH probes in cells not expressing the Firefly luciferase transgene) included to show noise level in smFISH.
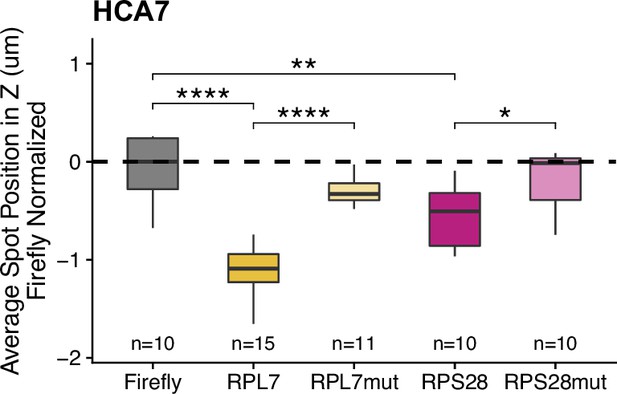
PRRE reporter transcript localization in HCA-7 monolayers.
Reporter RNA smFISH spot position in Z normalized to median untagged Firefly luciferase transcript position. p-Values were calculated using a Wilcoxon rank-sum test. Values represent the average value per cell. Numbers of cells interrogated for each construct are indicated. ns (not significant) represents p>0.05, * p<0.05, ** p<0.01, *** p<0.001 and **** represents p<0.0001.
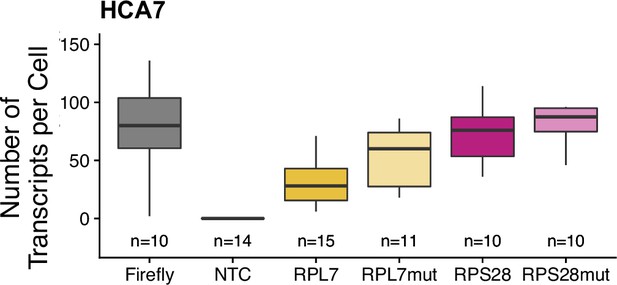
Transcript abundance per cell for PRRE-containing reporters in HCA7 cells as measured by smFISH.
Numbers of cells interrogated for each construct are indicated. No template control (Firefly luciferase smFISH probes in cells not expressing the Firefly luciferase transgene) included to show noise level in smFISH.
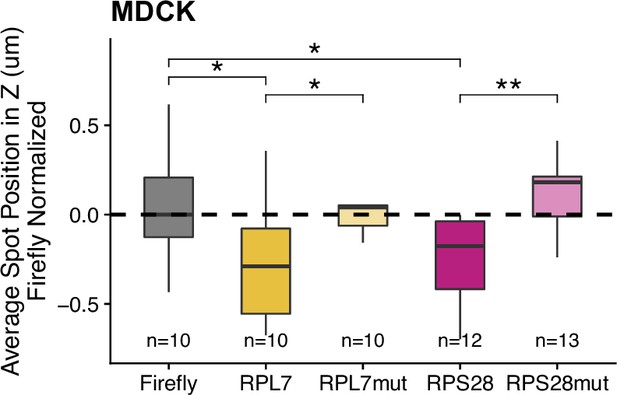
PRRE reporter construct localization in MDCK monolayers.
Reporter RNA smFISH spot position in Z normalized to median untagged Firefly transcript position. p-Values were calculated using a Wilcoxon rank-sum test. Values represent the average value per cell. Numbers of cells interrogated for each construct are indicated. ns (not significant) represents p>0.05, * p<0.05, ** p<0.01, *** p<0.001 and **** represents p<0.0001.
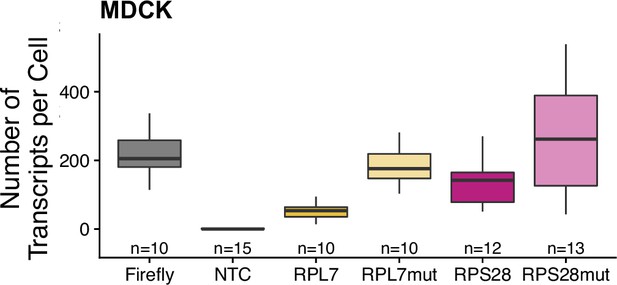
Transcript abundance per cell for PRRE-containing reporters in MDCK cells as measured by smFISH.
Numbers of cells interrogated for each construct are indicated. No template control (Firefly luciferase smFISH probes in cells not expressing the Firefly luciferase transgene) included to show noise level in smFISH.
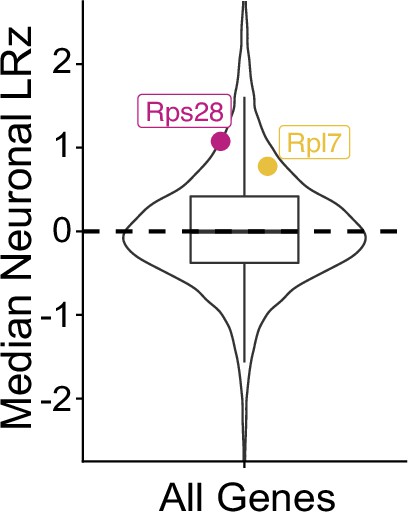
Neuronal localization ratio of all genes with candidate RP mRNAs highlighted.
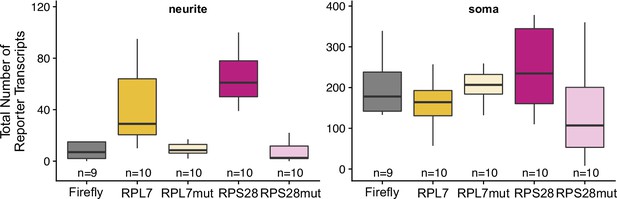
Total smFISH reporter construct puncta detected in neurites and soma by FISH-quant.
Numbers of cells interrogated for each construct are indicated.
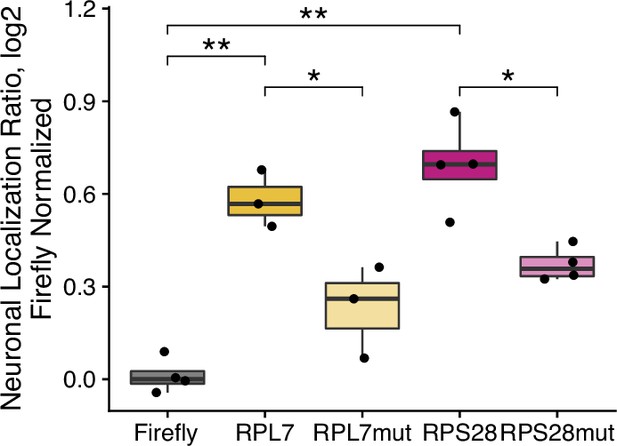
Enrichment of PRRE-containing reporter transcripts in neurite RNA fractions as determined by RT-qPCR.
In each sample both the PRRE-containing firefly luciferase transcript and an unfused, control renilla luciferase transcript are quantified. The ratio of firefly to renilla transcript levels is then calculated, and this ratio is compared in neurite and soma fractions. Values were normalized to those obtained for an unfused, control firefly luciferase transcript. Triplicate technical replicates are averaged to represent a single biological replicate. ns (not significant) represents p>0.05, * p<0.05, ** p<0.01, *** p<0.001 and **** represents p<0.0001.
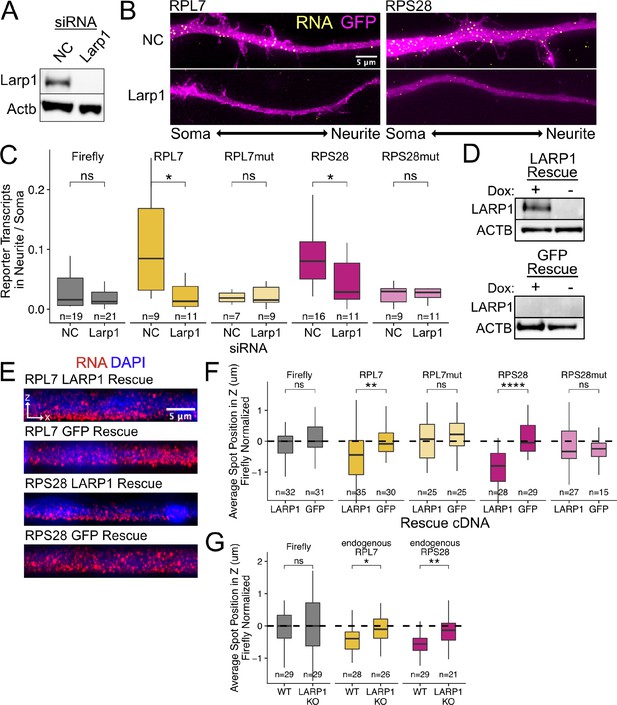
PRRE localization is mediated by LARP1 in enterocyte monolayers and neurons.
(A) Immunoblot for Larp1 and Actb showing knockdown of Larp1 protein in neuronal cells when treated with Larp1 siRNAs as compared to a negative control (NC) siRNA. (B) Representative images of smFISH for each wildtype PRRE reporter construct treated with Larp1 siRNAs or NC siRNAs. Images are a max projection through the XY axis of a single neurite positioned with the soma to the left. RNA signal shown in yellow while cell outlines marked by GFP signal shown as magenta. (C) Number of smFISH reporter transcript puncta in neurites normalized to soma. p-Values were calculated using a Wilcoxon rank-sum test. Numbers of cells interrogated for each construct are indicated. (D) Immunoblot for LARP1 and ACTB showing knockout and doxycycline-induced rescue of LARP1 levels in C2bbe1 LARP1 knockout cells. (E) Representative images of smFISH for each wildtype PRRE reporter construct. RNA signal shown in red while DAPI marks the nuclei in blue. Images are a max projection through the XZ axis of many cells. (F) smFISH puncta position in Z of the indicated Firefly luciferase reporters normalized to median untagged Firefly luciferase transcript position. p-Values were calculated using a Wilcoxon rank-sum test. Values represent the average value per cell. Numbers of cells interrogated for each construct are indicated. (G) smFISH puncta position in Z of the indicated endogenous transcripts normalized to median untagged Firefly luciferase transcript position. Vp-alues were calculated using a Wilcoxon rank-sum test. Values represent the average value per cell. Numbers of cells interrogated for each transcript are indicated. ns (not significant) represents p>0.05, * p<0.05, ** p<0.01, *** p<0.001 and **** represents p<0.0001.
-
Figure 4—source data 1
Protein lysates from siRNA treated CAD cells were ran on a GoPAGE Bis-Tris Precast Gel, 4–12% before being transferred to a PVDF membrane via the iBlot dry transfer device.
LARP1 expression was visualized with a mouse monoclonal anti LARP1 antibody.
- https://cdn.elifesciences.org/articles/80040/elife-80040-fig4-data1-v1.zip
-
Figure 4—source data 2
The ladder from the same blot was imaged.
The ladder is a broad-spectrum protein ladder (Fisher Scientific PI26623).
- https://cdn.elifesciences.org/articles/80040/elife-80040-fig4-data2-v1.zip
-
Figure 4—source data 3
Protein lysates from siRNA-treated CAD cells were ran on a GoPAGE Bis-Tris Precast Gel, 4–12% before being transferred to a PVDF membrane via the iBlot dry transfer device.
Beta-actin expression was used as a loading control and visualized with a mouse monoclonal anti ACTB antibody.
- https://cdn.elifesciences.org/articles/80040/elife-80040-fig4-data3-v1.zip
-
Figure 4—source data 4
The ladder from the same blot was imaged.
The ladder is a broad-spectrum protein ladder (Fisher Scientific PI26623).
- https://cdn.elifesciences.org/articles/80040/elife-80040-fig4-data4-v1.zip
-
Figure 4—source data 5
Protein lysates from C2bbe1 LARP1 Knockout cells rescued with doxycycline-inducible cDNAs were ran on a GoPAGE Bis-Tris Precast Gel, 4–12% before being transferred to a PVDF membrane via the iBlot dry transfer device.
LARP1 expression was visualized with a mouse monoclonal anti-LARP1 antibody.
- https://cdn.elifesciences.org/articles/80040/elife-80040-fig4-data5-v1.zip
-
Figure 4—source data 6
Protein lysates from C2bbe1 LARP1 Knockout cells rescued with doxycycline-inducible cDNAs were ran on a GoPAGE Bis-Tris Precast Gel, 4–12% before being transferred to a PVDF membrane via the iBlot dry transfer device.
Beta-actin expression was used as a loading control and visualized with a mouse monoclonal anti-ACTB antibody.
- https://cdn.elifesciences.org/articles/80040/elife-80040-fig4-data6-v1.zip
-
Figure 4—source data 7
The ladder from the same blot was imaged.
The ladder is a broad-spectrum protein ladder (Fisher Scientific PI26623).
- https://cdn.elifesciences.org/articles/80040/elife-80040-fig4-data7-v1.zip
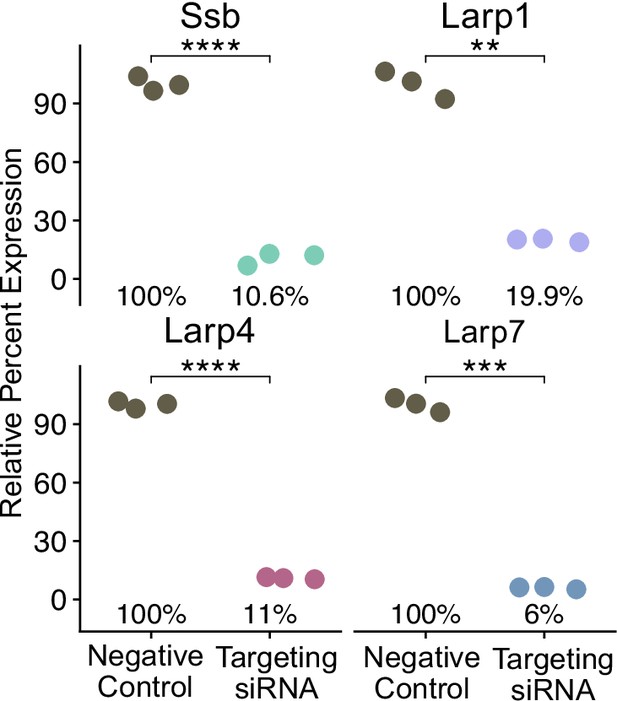
Percent knockdown of Larp RNAs by targeting and negative control siRNAs as measured by qPCR normalized to HPRT.
p-Values were calculated using a student’s t test. Triplicate technical replicates are averaged to represent a single biological replicate. ns (not significant) represents p>0.05, * p<0.05, ** p<0.01, *** p<0.001 and **** represents p<0.0001.
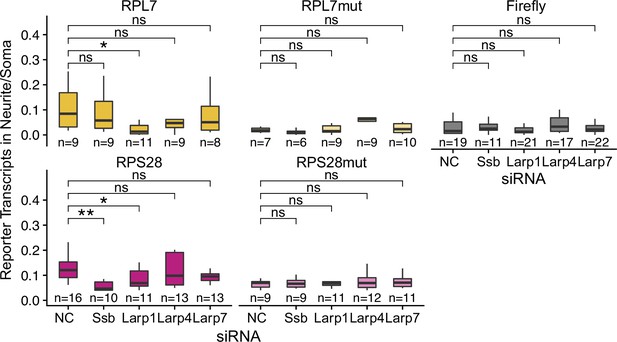
Reporter localization in neurons treated with different Larp siRNAs.
Reporter RNA smFISH puncta in neurites normalized to soma. p-Values were calculated using a Wilcoxon rank-sum test. Numbers of cells interrogated for each construct are indicated. ns (not significant) represents p>0.05, * p<0.05, ** p<0.01, *** p<0.001 and **** represents p<0.0001.

Representative brightfield images of Larp1 and negative control siRNA-treated differentiated CAD cells.
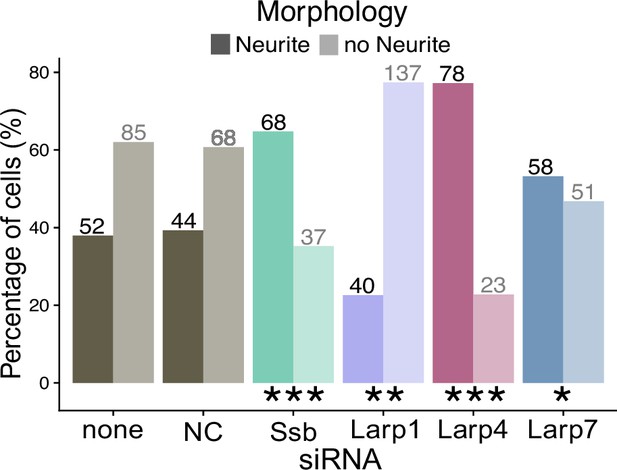
Proportion of cells with or without defined neurites when treated with different Larp siRNAs.
Raw count data is printed. p-Values were calculated using a Fisher’s exact test. ns (not significant) represents p>0.05, * p<0.05, ** p<0.01, *** p<0.001 and **** represents p<0.0001.
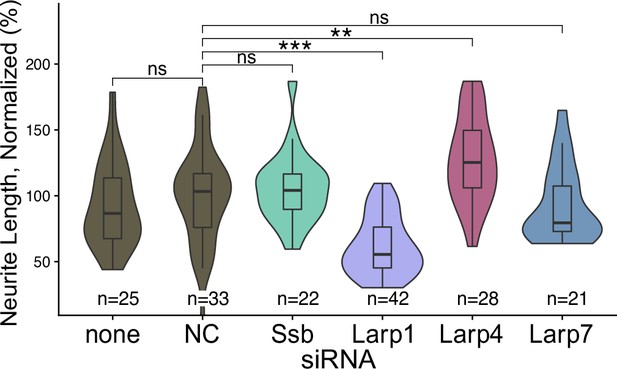
Neurite length of multiple Larp siRNA-treated cells normalized to negative control (NC) siRNA-treated cells.
p-Values were calculated using a student’s t test. ns (not significant) represents p>0.05, * p<0.05, ** p<0.01, *** p<0.001 and **** represents p<0.0001.
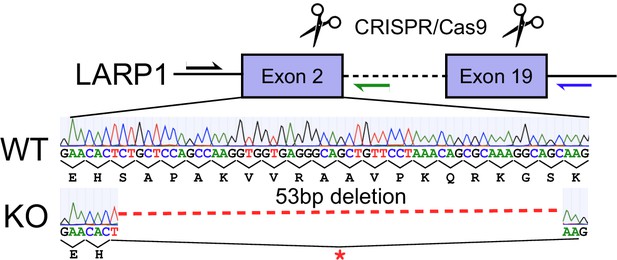
LARP1 CRISPR/Cas9 knockout Strategy.
Guide RNAs targeting exon 2 and exon 19 facilitated loss of the intervening sequence and frameshifting deletions in Exon 2. Traces from sanger sequencing show a 53 base pair deletion in exon 2 that creates an early stop codon.
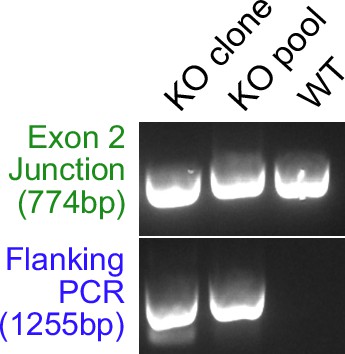
PCR amplification of the locus using primers that flank both gRNA cut sites shows loss of the sequence between exons 2 and 19.
This PCR product is roughly 1255 bp when drop out occurs and 24 kb when drop out does not occur. Exon 2 PCR shows decreased size due to deletions. Its expected size is 774 bp.
-
Figure 4—figure supplement 7—source data 1
1% Agarose gel of PCR products using primers flanking Exon2 of LARP1 as well as the complete deletion of LARP1 by CRISPR/Cas9 targeting gRNAs in Exon2 and 19.
DNA from several knockout clones, a heterogeneous pool of knockout cells and wildtype cells were assayed. The ladder used is the 1 kb Plus ladder (NEB N3200).
- https://cdn.elifesciences.org/articles/80040/elife-80040-fig4-figsupp7-data1-v1.zip
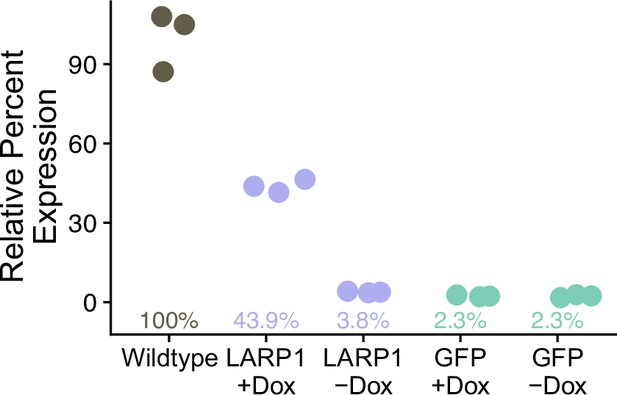
Relative expression of LARP1 rescue lines as normalized to HPRT then normalized to wildtype C2bbe1 LARP1 expression.
Triplicate technical replicates are averaged to represent a single biological replicate.
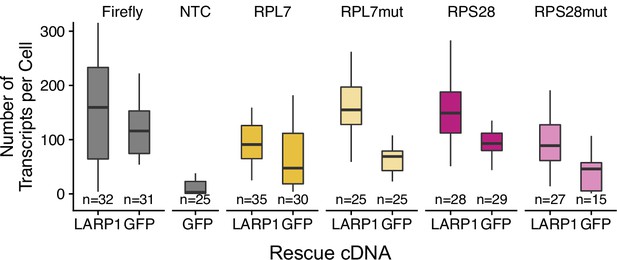
Expression of PRRE-containing reporter transcripts in LARP1 knockout and rescue cells as assayed by smFISH.
Numbers of cells interrogated for each construct are indicated. No template control (Firefly luciferase smFISH probes in cells not expressing the Firefly luciferase transgene) included to show noise level in smFISH.
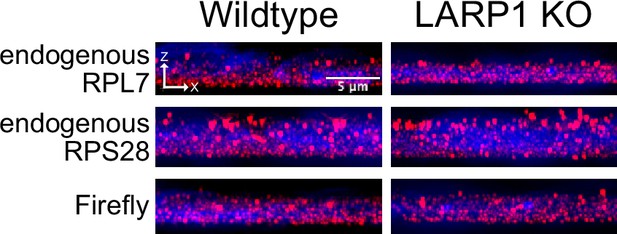
Localization of endogenous RPL7 and RPS28 transcripts in wildtype and LARP1 knockout C2bbe1 monolayers as assayed by smFISH.
Representative images are shown in an orthogonal, XZ projection of roughly 20 cells. The apical side of the cell is toward the top while the basal side is toward the bottom. Large, non-spherical puncta toward the apical side of cells are background fluorescence (perhaps associated with probes getting stuck in the mucus found on the apical side of C2bbe1 cells), and are computationally removed prior to the calculation of puncta positions.
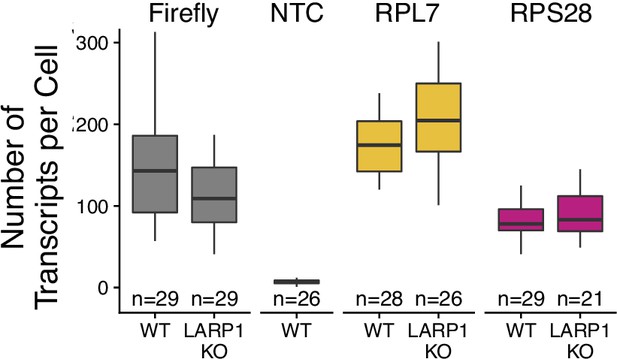
Transcript abundance of endogenous RPL7 and RPL28 transcripts in wildtype and LARP1 knockout C2bbe1 monolayers as assayed by smFISH.
Values represent the average value per cell. Numbers of cells interrogated for each construct are indicated. No template control (Firefly luciferase smFISH probes in cells not expressing the Firefly luciferase transgene) included to show noise level in smFISH.
PRRE motifs must be in the 5′ UTR for efficient transcript localization.
(A) Schematic of ribosomal protein 3′ PRRE reporter constructs. Mutant versions were created by swapping each pyrimidine for a purine. PRRE motif sequences are identical to 5′ PRRE reporter constructs. (B) 3′ PRRE reporter transcript localization in C2bbe1 monolayers. smFISH puncta position of the indicated Firefly luciferase reporters in Z normalized to median untagged Firefly transcript position. p-Values were calculated using a Wilcoxon rank-sum test. Values represent the average value per cell. (C) 3′ PRRE construct localization in neurons. Number of reporter RNA smFISH puncta in neurites normalized to soma. p-Values were calculated using a Wilcoxon rank-sum test. (D) 3′ PRRE construct localization in LARP1 knockout and rescued C2bbe1 monolayers. smFISH puncta position of the indicated Firefly luciferase reporters in Z normalized to median untagged Firefly luciferase transcript position. p-Values calculated using a Wilcoxon rank-sum test. Values represent the average value per cell. (E) 3′ PRRE construct localization in neurons depleted of Larp1 by siRNA. Number of reporter RNA smFISH puncta in neurites normalized to soma. p-Values calculated using a Wilcoxon rank-sum test. ns (not significant) represents p>0.05, * p<0.05, ** p<0.01, *** p<0.001 and **** represents p<0.0001. For B-E, Numbers of cells interrogated for each construct are indicated.
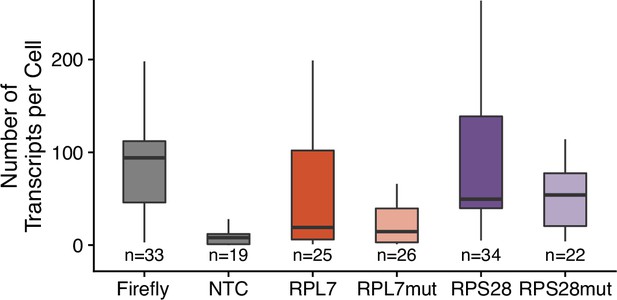
Transcript abundance of 3′ PRRE reporter transcripts in C2bbe1 monolayers as calculated by smFISH.
Numbers of cells interrogated for each construct are indicated. No template control (Firefly luciferase smFISH probes in cells not expressing the Firefly luciferase transgene) included to show noise level in smFISH.
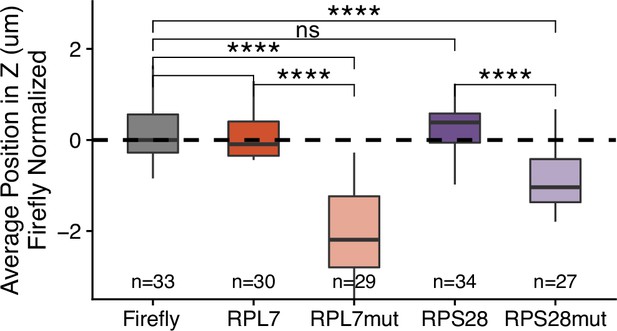
3′ PRRE reporter transcript localization in C2bbe1 monolayers.
Reporter RNA smFISH spot position in Z normalized to median untagged Firefly luciferase transcript position. p-Values were calculated by Wilcoxon rank-sum test.Values represent the average value per cell. Numbers of cells interrogated for each construct are indicated. ns (not significant) represents p>0.05, * p<0.05, ** p<0.01, *** p<0.001 and **** represents p<0.0001.
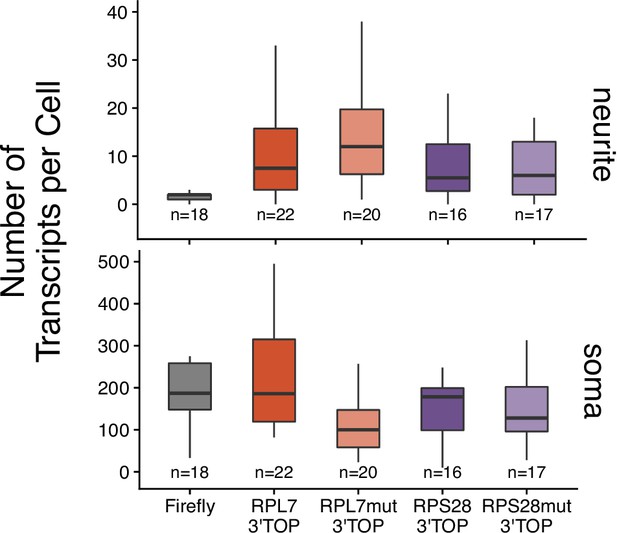
Transcript abundance of 3′ PRRE reporter transcripts in the neurite and soma compartments of CAD cells as calculated by smFISH.
Numbers of cells interrogated for each construct are indicated.
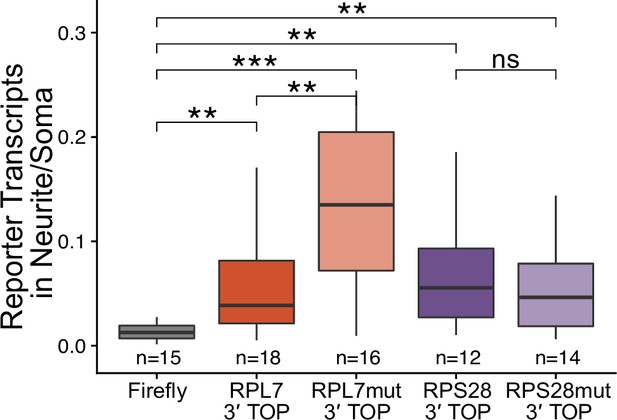
Number of 3′ PRRE reporter transcript puncta detected in neurites normalized to soma.
ns (not significant) represents p>0.05, * p<0.05, ** p<0.01, *** p<0.001 and **** represents p<0.0001.
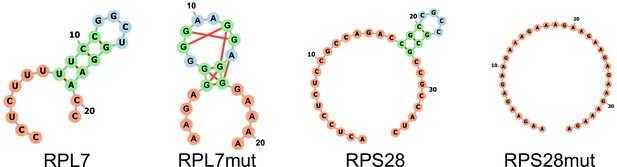
PRRE motif structural conformations as calculated by RNAfold (Lorenz et al., 2011).
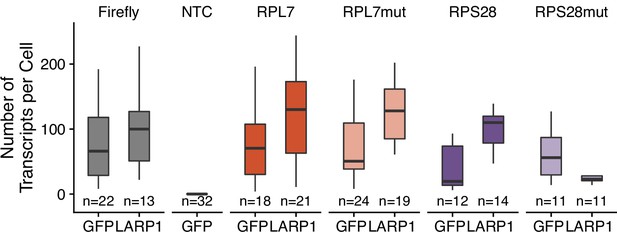
Transcript abundance as calculated by smFISH of 3′ PRRE reporter transcripts in LARP1 knockout C2bbe1 cells that have been rescued with either LARP1 or GFP.
Numbers of cells interrogated for each construct are indicated. No template control (Firefly luciferase smFISH probes in cells not expressing the Firefly luciferase transgene) included to show noise level in smFISH.
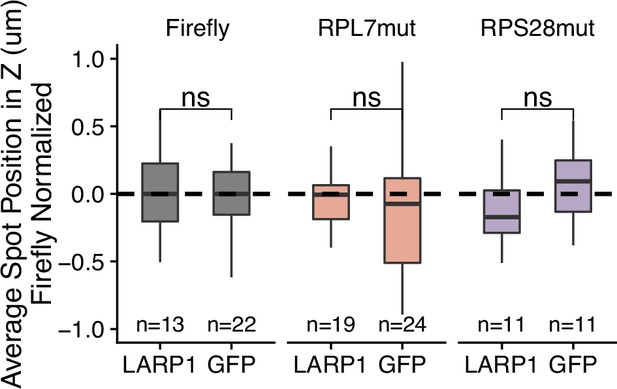
3′ PRRE reporter localization in LARP1 knockout C2bbe1 cells that have been rescued with either LARP1 or GFP.
Reporter RNA smFISH puncta position in Z normalized to median untagged Firefly luciferase transcript position. p-Values were calculated using a Wilcoxon rank-sum test. Values represent the average value per cell. Numbers of cells interrogated for each construct are indicated. ns (not significant) represents p>0.05, * p<0.05, ** p<0.01, *** p<0.001 and **** represents p<0.0001.
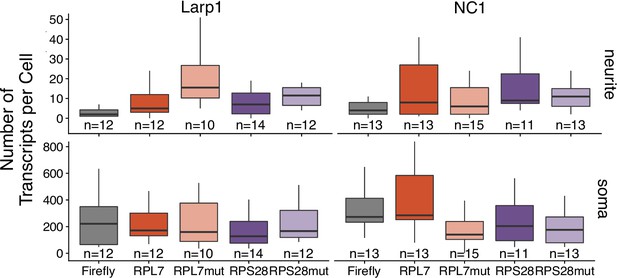
Transcript abundance of reporter transcripts as calculated by smFISH of 3′ PRRE reporter transcripts in the soma and neurite compartments of CAD cells that have been treated with either siRNA against LARP1 or a control, nontargeting siRNA.
Numbers of cells interrogated for each construct are indicated. No template control (Firefly luciferase smFISH probes in cells not expressing the Firefly luciferase transgene) included to show noise level in smFISH.
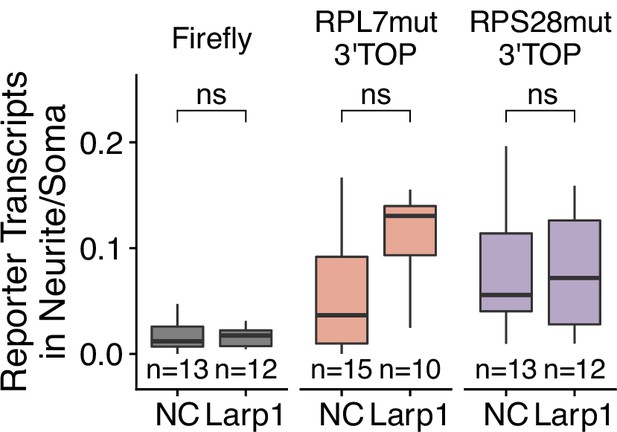
Number of 3′ PRRE reporter transcript puncta detected in neurites normalized to soma in Larp1 siRNA-treated CAD cells.
p-Values were calculated using a Wilcoxon rank-sum test. Numbers of cells interrogated for each construct are indicated. ns (not significant) represents p>0.05, * p<0.05, ** p<0.01, *** p<0.001 and **** represents p<0.0001.
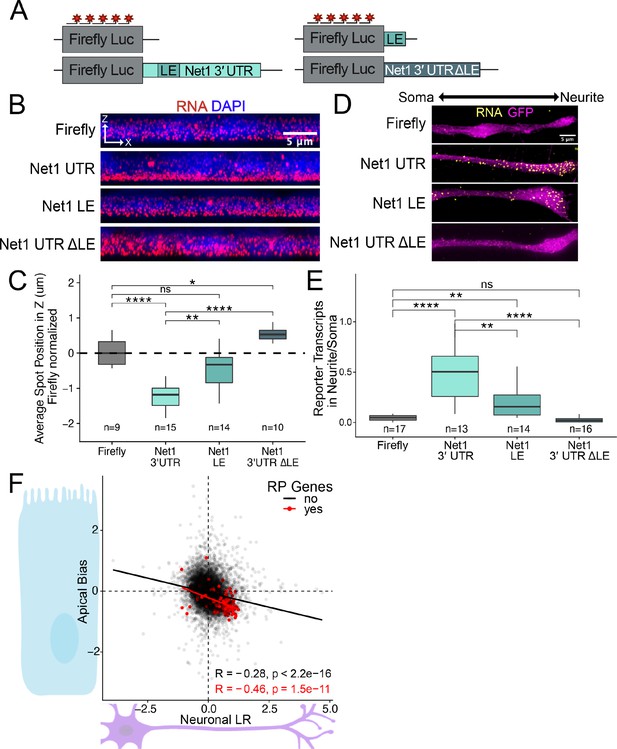
RNA localization mechanisms beyond PRRE motifs are conserved across species and cell morphology.
(A) Schematic of Net1 reporter constructs used to investigate necessity and sufficiency of Net1’s localization element (LE). (B) Representative images of smFISH puncta for each Net1 reporter construct in C2bbe1 monolayers. RNA signal shown in red while DAPI marks the nuclei in blue. Images are a max projection through the XZ axis of roughly 50 cells. (C) smFISH puncta position in Z of the indicated reporter RNAs normalized to median untagged Firefly luciferase transcript position. p-Values were calculated using a Wilcoxon rank-sum test. Values represent the average value per cell. Numbers of cells interrogated for each construct are indicated. (D) Representative images of smFISH puncta for Net1 constructs in CAD cells. Images are a max projection through the XY axis of a single neurite positioned with the soma to the left. RNA signal shown in yellow while cell outlines marked by GFP signal shown as magenta. (E) Number of reporter RNA smFISH puncta in neurites normalized to soma. p-Values were calculated using a Wilcoxon rank-sum test. Numbers of cells interrogated for each construct are indicated. (F) Direct comparison of the Halo-seq-derived Apical Bias values in human C2bbe1 monolayers and mechanical fractionation-derived neuronal Localization Ratio (LR) values in neuronal cells for all genes. Ribosomal protein mRNAs are highlighted in red. Correlation and p-values calculated using a Spearman’s correlation test. ns (not significant) represents p>0.05, * p<0.05, ** p<0.01, *** p<0.001 and **** represents p<0.0001.
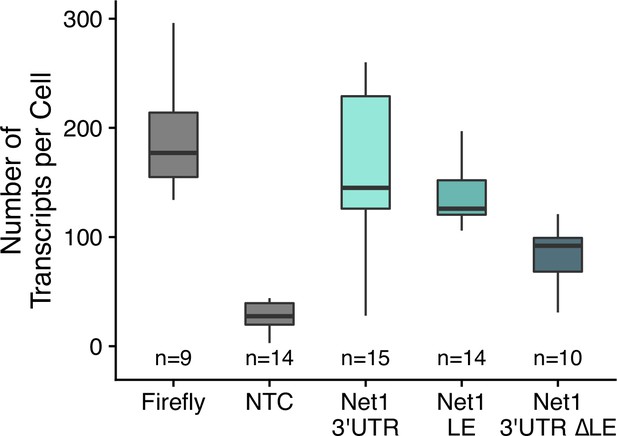
Transcript abundance of reporter transcripts in C2bbe1 monolayers as calculated by smFISH.
Numbers of cells interrogated for each construct are indicated. No template control (Firefly luciferase smFISH probes in cells not expressing the Firefly luciferase transgene) included to show noise level in smFISH.
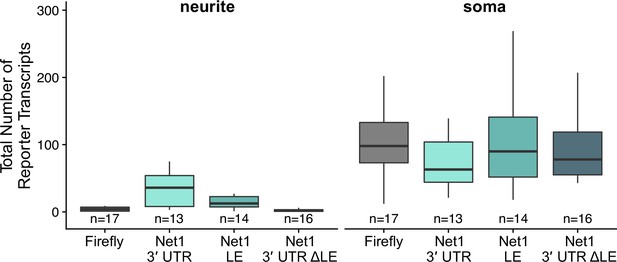
Transcript abundance in CAD cell neurites and soma as calculated by smFISH.
Numbers of cells interrogated for each construct are indicated.
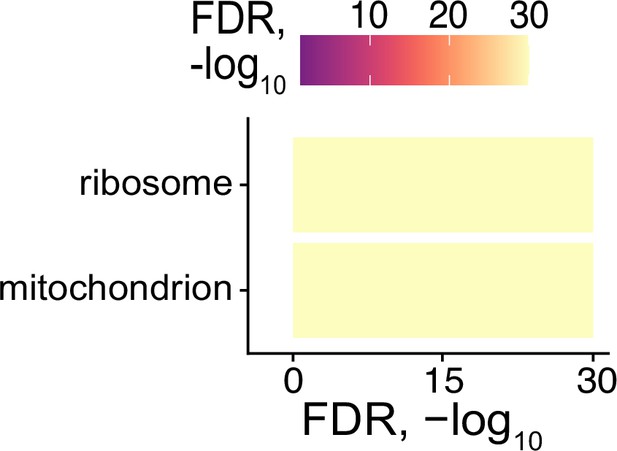
Enriched gene ontology terms derived from RNAs identified as localized to the basal pole of enterocytes and to neurites of neurons.
FDR calculated by topGO.
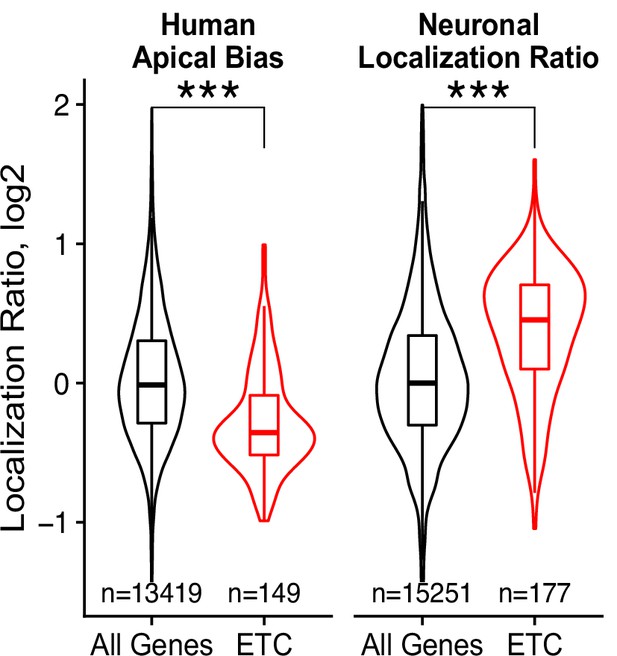
Localization of transcripts encoding electron transport chain (ETC) associated proteins in apicobasal and neuronal localization datasets.
p-Values were calculated using a Wilcoxon rank-sum test. ns (not significant) represents p>0.05, * p<0.05, ** p<0.01, *** p<0.001 and **** represents p<0.0001.
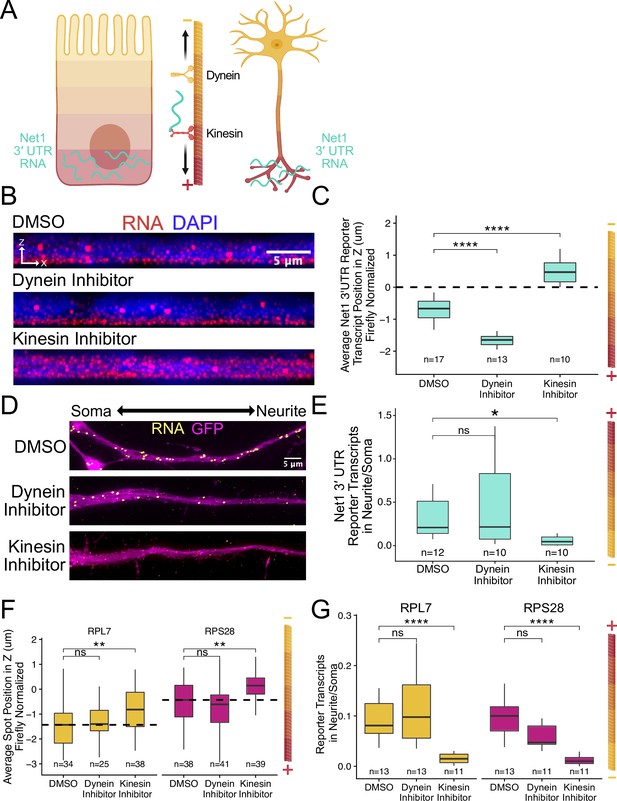
RNA localization to neurites and the basal pole of epithelial cells is dependent on kinesin.
(A) Schematic of microtubule organization in both enterocytes and neurons where the plus ends are oriented basally and out to neurites. Net1 3′ UTR Reporter constructs are teal, hypothesized to reach their destination via kinesin transport. (B) Representative images of Net1 3′ UTR reporter transcript localization with each inhibitor treatment or DMSO control. RNA signal shown in red while DAPI marks the nuclei in blue. Images are a max projection through the XZ axis of roughly 30 cells. (C) smFISH puncta position in Z of Firefly luciferase-Net1 3′ UTR constructs normalized to median untagged Firefly luciferase transcript position in Z. p-Values were calculated using a Wilcoxon rank-sum test. Values represent the average value per cell. (D) Representative images of smFISH for Net1 3′ UTR transcripts in CAD cells. Images are a max projection through the XY axis of a single neurite positioned with the soma to the left. RNA signal shown in yellow while cell outlines marked by GFP signal shown as magenta. (E) Number of reporter RNA smFISH puncta in neurites normalized to soma. p-Values were calculated using a Wilcoxon rank-sum test. (F) smFISH puncta position in Z of RPL7 and RPS28 PRRE constructs normalized to median untagged Firefly luciferase transcript position in Z. p-Values were calculated using a Wilcoxon rank-sum test. Values represent the average value per cell. (G) Number of reporter RNA smFISH puncta in neurites normalized to soma. p-Values were calculated using a Wilcoxon rank-sum test. ns (not significant) represents p>0.05, * p<0.05, ** p<0.01, *** p<0.001 and **** represents p<0.0001. For panels C, E, F, and G, numbers of cells interrogated for each construct are indicated.
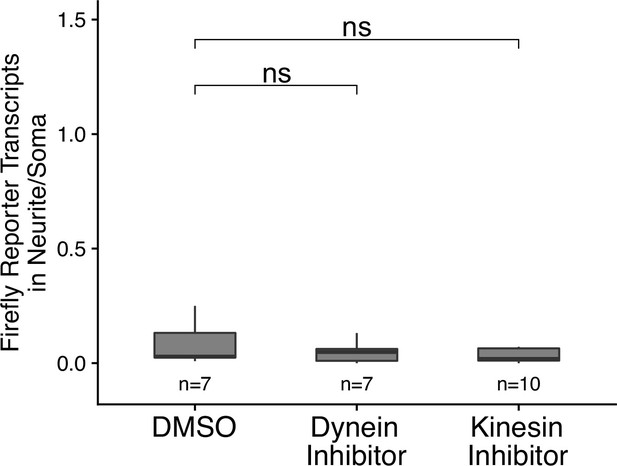
Number of Firefly reporter transcript puncta detected in neurites normalized to soma with different drug treatments in CAD cells.
p-Values were calculated using a Wilcoxon rank-sum test. Numbers of cells interrogated for each construct are indicated. ns (not significant) represents p>0.05, * p<0.05, ** p<0.01, *** p<0.001 and **** represents p<0.0001.
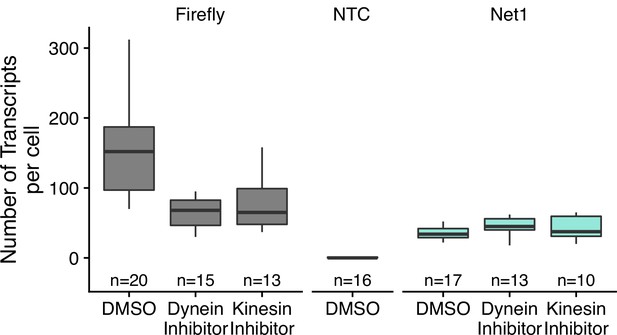
Transcript abundance of reporter transcripts in C2bbe1 monolayers treated with the indicated drugs as calculated by smFISH.
Numbers of cells interrogated for each construct are indicated. No template control (Firefly luciferase smFISH probes in cells not expressing the Firefly luciferase transgene) included to show noise level in smFISH.
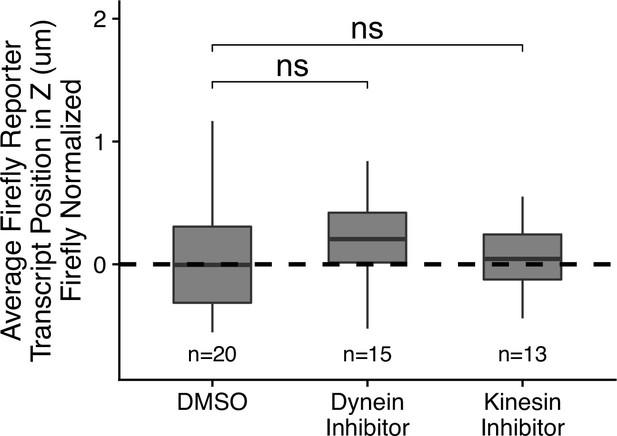
Firefly luciferase reporter transcript localization with different drug treatments normalized to median transcript position in DMSO treatment.
p-Values calculated by Wilcoxon rank-sum test. Values represent the average value per cell. Numbers of cells interrogated for each construct are indicated. ns (not significant) represents p>0.05, * p<0.05, ** p<0.01, *** p<0.001 and **** represents p<0.0001.
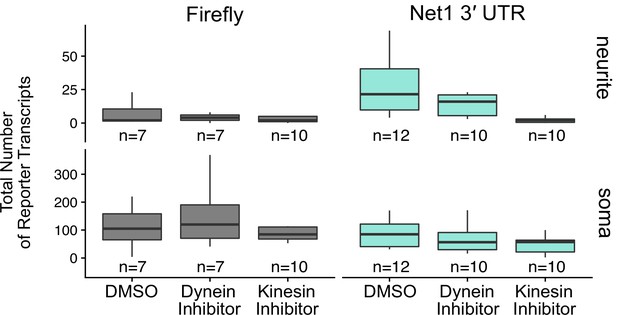
Abundance of Net1 3′ UTR reporter transcripts as assayed by smFISH in the soma and neurite compartments of CAD cells treated with the indicated drugs.
Numbers of cells interrogated for each construct are indicated.
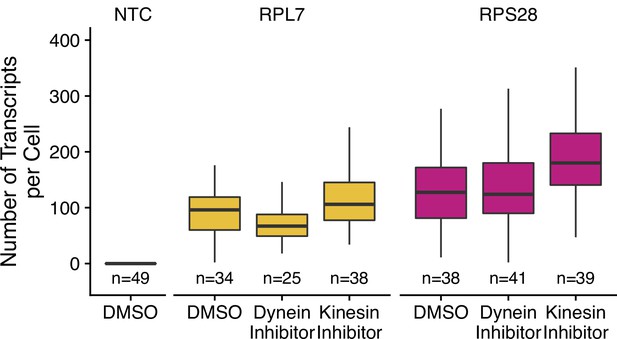
Abundance of PRRE-containing reporter transcripts as assayed by smFISH in C2bbe1 monolayers treated with the indicated drugs.
Numbers of cells interrogated for each construct are indicated. No template control (Firefly luciferase smFISH probes in cells not expressing the Firefly luciferase transgene) included to show noise level in smFISH.
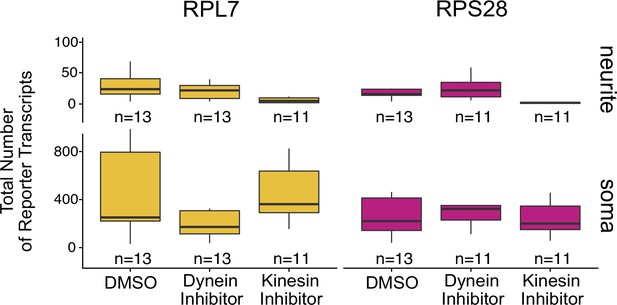
Abundance of PRRE-containing reporter transcripts as assayed by smFISH in the soma and neurite compartments of CAD cells treated with the indicated drugs.
Numbers of cells interrogated for each construct are indicated.
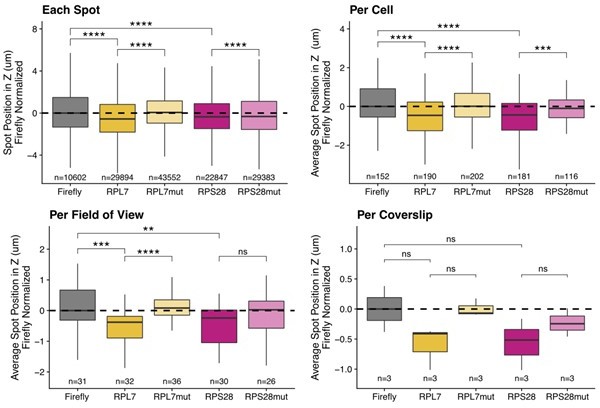
C2bbe1 monolayer smFISH spot position analysis.
RNA localization across the apicobasal axis is measured by smFISH spot position in the Z axis. This can be plotted for each spot, where thousands of spots over-power the statistics. Spot position can be averaged per cell as outlined manually within the FISH-quant software. This reduces sample size and allows for more accurate statistical analysis. When spot position is averaged per field of view, sample size further decreases, statistics are less powered but the localization trends are still robust. Finally, we can average spot position per coverslip, which represents biological replicates. We lose almost all statistical power as sample size is limited to 3 coverslips. Despite this, the localization trends are still recognizable..
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | P65 | hg38 | ENSG00000173039 | |
| Gene (Homo sapiens) | PODXL | hg38 | ENSG00000128567 | |
| Gene (Homo sapiens) | ATP1A1 | hg38 | ENSG00000163399 | |
| Gene (Homo sapiens, Mus musculus) | RPL7 | hg38, mm10 | ENSG00000147604, ENSMUSG00000043716 | |
| Gene (Homo sapiens, Mus musculus) | RPS28 | hg38, mm10 | ENSG00000233927, ENSMUSG00000067288 | |
| Gene (Homo sapiens, Mus musculus) | LARP1 | hg38, mm10 | ENSG00000155506, ENSMUSG00000037331 | |
| Gene (Homo sapiens, Mus musculus) | NET1 | hg38, mm10 | ENSG00000173848, ENSMUSG00000021215 | |
| Sequence-based reagent | LARP1 guide RNA exon2 | Synthego | GCUGUUCCUAAACAGCGCAA | Used to create human LARP1-null cells |
| Sequence-based reagent | LARP1 guide RNA exon19 | Synthego | CAACCCCCUACACCACCCAC | Used to create human LARP1-null cells |
| Sequence-based reagent | AAVS1 guide RNA | Synthego | UAGUGGCCCCACUGUGGGGU | Used to create human loxP cells |
| Sequence-based reagent | Larp1 siRNA | IDT | mm.RI.Larp1.13.2 | Used as equimolar mix |
| Sequence-based reagent | Larp1 siRNA | IDT | mm.RI.Larp1.13.3 | Used as equimolar mix |
| Sequence-based reagent | SSB siRNA | IDT | mm.RI.Larp1.13.1 | Used alone |
| Sequence-based reagent | Larp4 siRNA | IDT | mm.RI.Larp1.13.1 | Used alone |
| Sequence-based reagent | LARP7 siRNA | IDT | mm.RI.Larp1.13.1 | Used alone |
| Sequence-based reagent | FLAP_Y | IDT | /5Cy3/AA TGC ATG TCG ACG AGG TCC GAG TGT AA/3Cy3Sp/ | hybridized to smiFISH probes |
| Sequence-based reagent | RPL7 5'TOP | IDT | CCTCTTTTTCCGGCTGGAACC | used for cloning into reporter constructs |
| Sequence-based reagent | RPL7 mutant 5'TOP | IDT | AAGAGGGGGAAGGAGGGAAAA | used for cloning into reporter constructs |
| Sequence-based reagent | RPS28 5'TOP | IDT | ACTCCTCTCCGCCAGACCGCCGCCGCGCCGCCATC | used for cloning into reporter constructs |
| Sequence-based reagent | RPS28 mutant 5'TOP | IDT | AAGAAGAGAAGAAAGAAAGAAGAAGAGAAGAAAGA | used for cloning into reporter constructs |
| Cell line (Mus musculus) | CAD | Sigma | 08100805-1VL, RRID:CVCL_0199 | |
| Cell line (Mus musculus) | CAD/loxP | Khandelia et al., 2011 | Contains single integration of loxP cassette | |
| Cell line (Homo sapiens) | C2bbe1 | ATCC | CRL-2102 | |
| Cell line (Homo sapiens) | C2bbe1/loxP | Khandelia et al., 2011 | Contains single integration of loxP cassette | |
| Cell line (Homo sapiens) | HCA-7 | ECACC | 6061902 | |
| Cell line (Homo sapiens) | HCA-7/loxP | Khandelia et al., 2011 | Contains single integration of loxP cassette | |
| Cell line (Canis lupus) | MDCK | ATCC | CRL-2935 | |
| Cell line (Canis lupus) | MDCK/loxP | Khandelia et al., 2011 | Contains random integration of loxP cassette | |
| Transfected construct (Homo sapiens) | PODXL cDNA | Horizon | MHS6278-202858197 | used for cloning into Halo plasmids |
| Transfected construct (Homo sapiens) | ATP1A1 cDNA | Horizon | MHS6278-202759485 | used for cloning into Halo plasmids |
| Transfected construct (Homo sapiens) | P65 cDNA | R.Spitale, UC Irvine | used for cloning into Halo plasmids | |
| Transfected construct (Homo sapiens) | LARP1 cDNA | Horizon | MHS6278-202827213 | used for cloning into Halo plasmids |
| Antibody | mouse monoclonal anti LARP1 | Santa Cruz | sc-515873 | 1:100 dilution for immunoblotting |
| Antibody | mouse monoclonal anti ACTB | Sigma | A5441 | 1:5000 for immunoblotting |
| Antibody | mouse monoclonal anti H3 | Abcam | 10799 | 1:10000 for immunoblotting |
| Antibody | rabbit polyclonal anti TOM20 | ProteinTech | 11802–1-AP | 1:250 for immunofluorescence |
| Antibody | rabbit polyclonal anti EZRIN | Cell Signaling Technology | 3145 S | 1:250 for immunofluorescence |
| Antibody | mouse monoclonal anti NaKATPase | DSHB | A5-s | 1:50 for immunofluorescence |
| Sequence-based reagent | smFISH probes against Firefly luciferase | BioSearch | VSMF-1006–5 | |
| Commercial assay or kit | Zymo Quick-RNA Microprep kit | Zymo Research | R1050 | |
| Commercial assay or kit | Zymo Quick-RNA Miniprep kit | Zymo Research | R1055 | |
| Commercial assay or kit | Roche RNA HyperPrep Kit with HMR depletion | Roche | ||
| Other | Cell culture inserts for differentiation | Corning | 353090 | C2bbe1 are differentiation on 0.4 uM transwell inserts |
| Other | Kinesore | Sigma-Aldrich | SML2361 | Kinesin-1 Inhibitor |
| Other | Ciliobrevin-A | Selleckchem | 302803-72-1 | Dynein Inhibitor |
| Other | MitoView Green | Biotium | 70,054 T | 100 ng/mL for immunofluorescence imaging of mitochondria |
| Other | Halo-Dibromofluoroscein | R.Spitale, UC Irvine | ROS producing small molecule for Halo-seq labeling |
Additional files
-
Supplementary file 1
Localization values in C2bbe1 monolayers.
DESeq2 output of Cytoplasmic Bias (CB) and calculated FDR values (CB_FDR). Cytoplasmic bias is the log2 of a gene’s cytoplasmic enriched abundance divided by the abundance in total RNA. Apical Bias (AB) and AB_FDR values are also included for all observed genes where Apical bias is the log2 of a gene’s apical enriched abundance divided by its basal enriched abundance.
- https://cdn.elifesciences.org/articles/80040/elife-80040-supp1-v1.txt
-
Supplementary file 2
smFISH probe sequences.
Sequences of individual probes used as equimolar pool to visualize endogenous RNAs in fixed cells. Note that the last 28 nt of each probe correspond to the sequence that hybridize to the Cy3-labeled secondary probe.
- https://cdn.elifesciences.org/articles/80040/elife-80040-supp2-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/80040/elife-80040-mdarchecklist1-v1.docx