The conserved centrosomin motif, γTuNA, forms a dimer that directly activates microtubule nucleation by the γ-tubulin ring complex (γTuRC)
Figures
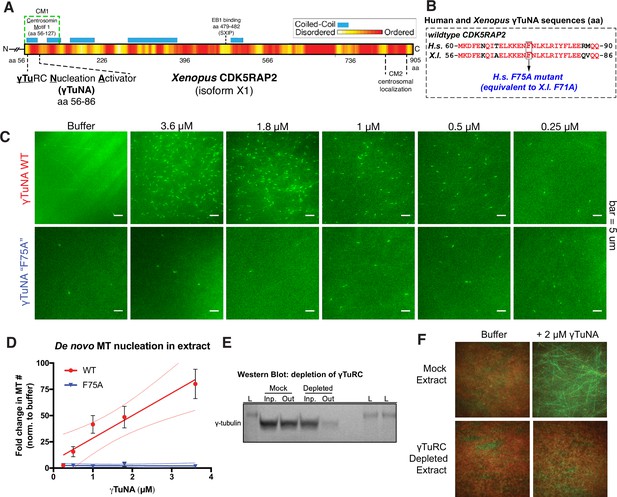
Cdk5rap2’s γTuNA domain increases MT nucleation in Xenopus egg extract and requires the universal MT template, the γ-tubulin ring complex (γTuRC).
(A) Schematic of Xenopus Cdk5rap2’s domains. The γTuRC nucleation activator domain, γTuNA, is located from amino acids 56–86 in Xenopus laevis isoform X1 (905 aa) or 60–90 in human CDK5RAP2 isoform A (1893 aa). Predictions of disorder (PONDR-FIT; Xue et al., 2010) and coiled-coil regions (COILS) are shown as a red/yellow gradient or blue boxes, respectively. (B) Alignment of wildtype human and Xenopus γTuNAs. Identical residues are red. The human F75 residue (first mutated in Fong et al., 2008) is equivalent to residue F71 in Xenopus. In this study, mutations of well-conserved, identical residues are designated according to the human residue number (e.g. human F75A is equivalent to Xenopus F71A; both hereafter referred to as “F75A”). (C) TIRF assay of MT nucleation in Xenopus egg extract. A titration series of wildtype or ‘F75A’ versions of Xenopus γTuNA (Strep-His-Xen. γTuNA-aa 56–89) were added to extract as shown. EB1-mCherry was used to mark growing MT plus-ends (pseudo-colored green in images). Bar = 5 µm. (D) Quantification of the number of EB1 spots in C. The data were normalized by the buffer controls, and are shown as fold-changes. Black error bars are the standard error of the mean (SEM) for three independent extracts. Thin colored lines on either side of the central trendline represent 95% confidence intervals. (E) Western blot of γ-tubulin levels before and after mock-treatment or incubation with Strep-His-Halo-Xenopus γTuNA-coupled beads. After a single pulldown, the majority of γ-tubulin signal is lost. (F) TIRF assay of mock- and γTuRC-depleted extract. Alexa-488 labeled tubulin (green) and EB1-mCherry (red) were used to visualize microtubules in extract with or without 2 µM Strep-His-Xenopus γTuNA. See “Figure 1—source data 1” and “Figure 1—source data 2” for numerical data and raw blot.
-
Figure 1—source data 1
Numerical data for Figure 1.
- https://cdn.elifesciences.org/articles/80053/elife-80053-fig1-data1-v2.zip
-
Figure 1—source data 2
Labeled and raw blots used in Figure 1.
- https://cdn.elifesciences.org/articles/80053/elife-80053-fig1-data2-v2.zip
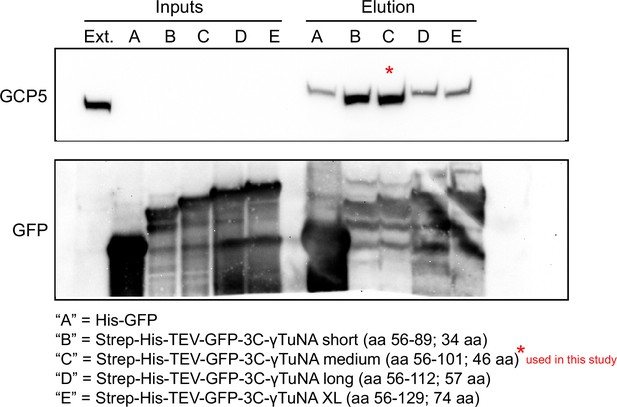
Different sizes of Xenopus γTuNA bind γTuRC with different affinities.
Western blots of Xenopus egg extract pulldowns with GFP-trap beads (cat # gtma-400, Chromotek) coupled to either GFP control or different lengths of GFP-tagged Xenopus γTuNA. Western blots of GCP5 and GFP were used to assess optimal γTuNA length relative to γTuRC binding ability. From these pulldowns, we determined that N-terminally GFP-tagged Xenopus γTuNAs longer than 46aa appeared to lose γTuRC binding ability. For all subsequent pulldowns in this study, the 46 aa version of wildtype Xenopus γTuNA (aa 56–101) was used fused to an N-terminal Halo-tag. Raw blots are in “Figure 1—figure supplement 1—source data 1.”
-
Figure 1—figure supplement 1—source data 1
Labeled and raw blots used in Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/80053/elife-80053-fig1-figsupp1-data1-v2.zip
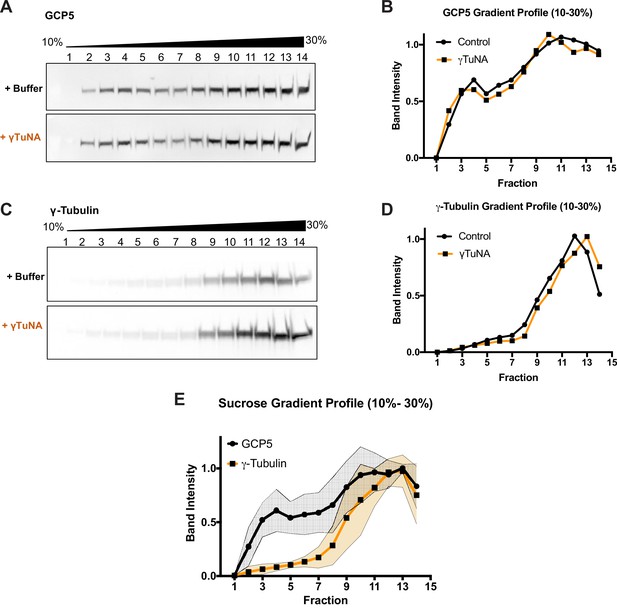
Addition of γTuNA to Xenopus egg extract does not affect γTuRC assembly or stability.
High-speed centrifuged Xenopus egg extracts were run on 10–30% w/w sucrose gradients and fractionated. Western blots of GCP5 and γ-tubulin were quantified to generate a sucrose gradient profile for each target, as shown in A. GCP5 appeared to be in two separate populations: uncomplexed around fraction 3, and another in the γTuRC complex (around fraction 10–11). (A and C) Western blots of GCP5 and γ-tubulin in gradient fractionated extracts treated with either buffer or 6 µM Strep-His-Xenopus γTuNA for 1 h on ice. The presence of γTuNA did not shift intensity from the soluble peak of GCP5 (fraction 3) to the γTuRC fraction (fraction 11), nor did it have any effect on the distribution of γ-tubulin. (B and D) Normalized band intensity profiles for GCP5 and γ-tubulin. (E) Mean gradient profile for GCP5 and γ-tubulin with standard deviation (N=2). The traces for extracts treated with 6 µM γTuNA do not fall outside the colored regions shown in E. Raw blots are in “Figure 1—figure supplement 2—source data 1.”
-
Figure 1—figure supplement 2—source data 1
Labeled and raw blots used in Figure 1—figure supplement 2A and C.
- https://cdn.elifesciences.org/articles/80053/elife-80053-fig1-figsupp2-data1-v2.zip
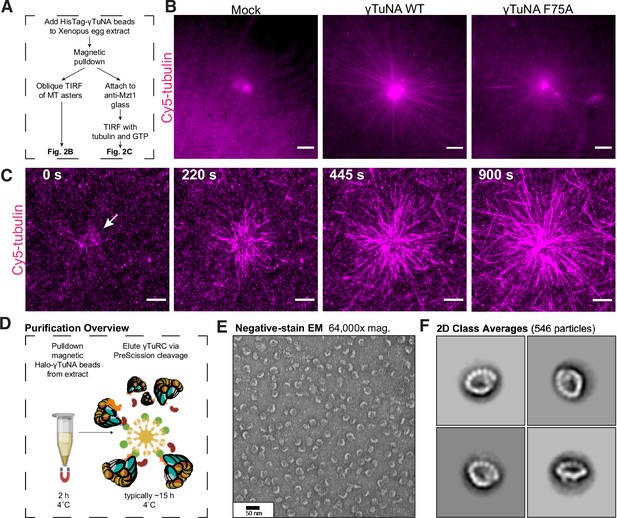
The γTuNA domain strongly recruits MT nucleation factors (including γTuRC) from Xenopus egg extract.
(A) Schematic of experiments for B and C. (B) Oblique TIRF images of MT asters from beads in vitro after 10 min. HisPur magnetic beads coated with either bovine serum albumin (mock), Strep-His-Xenopus γTuNA wildtype (WT), or ‘F75A’ mutant were incubated with extract, pulled-down, and washed. These were then diluted 1/1000 with polymerization mix containing 15 µM tubulin and 1 mM GTP, before imaging with TIRF. 5% Cy5-tubulin was used to label MTs. Bar = 5 µm. (C) Time-lapse imaging of MT aster growth from wildtype γTuNA beads in vitro. As in part B, wildtype γTuNA beads were pulled-down from extract and washed. These were then incubated on DDS-surface treated coverslips coated in anti-Mzt1 antibody to attach beads containing γTuRC. After a wash step, polymerization mix was added prior to time-lapse TIRF imaging. Frames are shown over the course of 15 min (900 s). Bar = 5 µm. (D) Diagram showing purification of endogenous Xenopus γTuRC using magnetic beads coupled to Strep-His-HaloTag-3C-human γTuNA. Made partly with Biorender. (E) Representative image of purified γTuRCs via negative-stain electron microscopy. Magnification is 64,000 x, taken at 80 kV with a Philips CM100 transmission electron microscope. Bar = 50 nm. (F) 2D class averages of 546 γTuRC particles picked from negative-stain EM images like in E. Each image represents one of four top classes. See “Figure 2—source data 1” for uncropped images in B and E.
-
Figure 2—source data 1
Uncropped images for Figure 2.
- https://cdn.elifesciences.org/articles/80053/elife-80053-fig2-data1-v2.zip
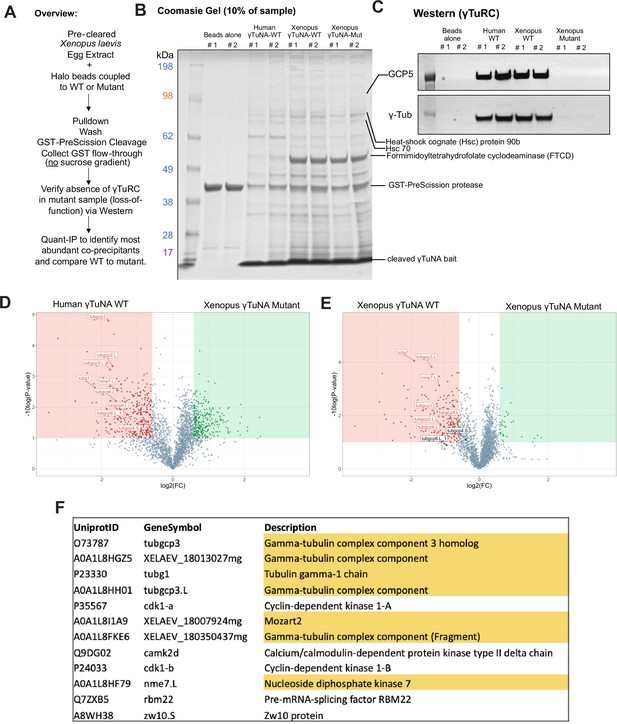
Mass spectrometry (Quant-IP) reveals γTuRC is the dominant factor present after extract pulldown of Halo-γTuNA beads.
(A) Experiment overview of extract pulldowns with beads coupled to mock, human wildtype, Xenopus wildtype, and Xenopus F75A mutant versions of γTuNA. Bound factors were eluted using PreScission cleaveage of Halo-3C- γTuNA bait. (B) SDS-PAGE and Coomassie stain of 10% of elution sample. The band at ~40 kDa corresponds to GST-PreScission. The band at ~7 kDa (dominant band <17 kDa) corresponds to cleaved γTuNA bait. The dominant band at ~50 kDa was identified as Xenopus formimidoyltransferase cyclodeaminase (FTCD). (C) Western blot of γTuRC components, GCP5 and γ-tubulin, in the elution samples. Note that only the wildtype γTuNA baits had detectable γTuRC signal. (D and E) Volcano plots of proteins specifically enriched in the human or Xenopus γTuNA baits, as compared to the Xenopus F75A γTuNA mutant bait. (F) Table displays the top 12 proteins specifically enriched by wildtype γTuNA bait, after subtraction of proteins present in F75A mutant bait. If a protein was not present in both human and Xenopus wildtype conditions or if it was identified with fewer than 5 unique peptides, it was not included in our analysis. Raw blots are in “Figure 2—figure supplement 1—source data 1.” Raw mass spectrometry data is in “Figure 2—figure supplement 1—source data 2.
-
Figure 2—figure supplement 1—source data 1
Labeled and raw blots used in Figure 2—figure supplement 1B and C.
- https://cdn.elifesciences.org/articles/80053/elife-80053-fig2-figsupp1-data1-v2.zip
-
Figure 2—figure supplement 1—source data 2
Raw mass spectrometry data for pulldowns of Halo-γTuNA from Xenopus egg extract (TCMP- ProQuant).
- https://cdn.elifesciences.org/articles/80053/elife-80053-fig2-figsupp1-data2-v2.zip
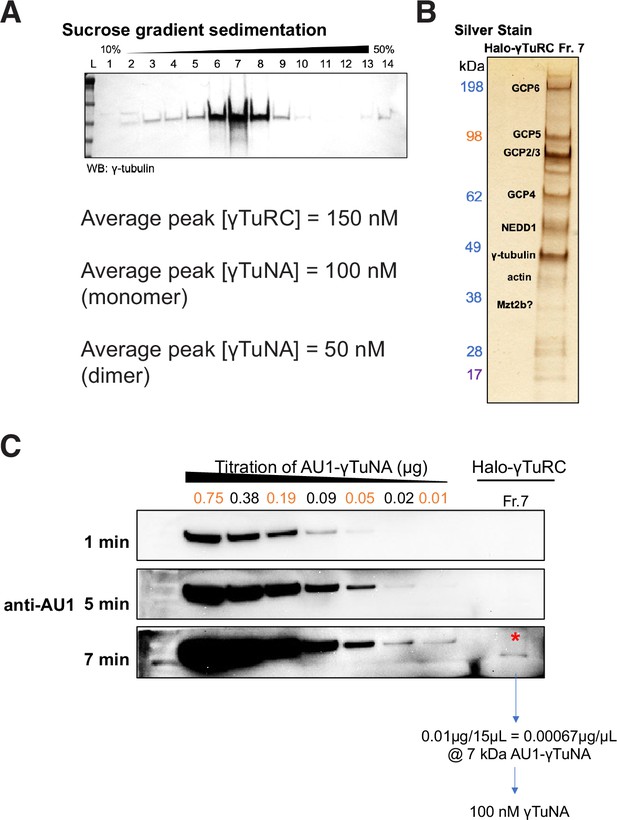
Purity and concentration assessment of γTuRCs purified via Halo-γTuNA pulldown.
(A) Western blot of γ-tubulin in fractions from a 10% to 50% w/w sucrose gradient. The γTuRC peak was routinely located in fraction 7, as shown in A. Peak γTuRC concentration refers to the peak fraction (usually Fraction 7). This was compared in a western blot against a known titration of recombinant GCP4 to determine [GCP4]. As there are two copies of GCP4 per γTuRC, this meant final [γTuRC] = [GCP4]*0.5. (B) SDS-PAGE and silver stain of the peak γTuRC fraction. Dominant bands are known γTuRC components, as verified by mass spectrometry. (C) Western blot-based determination of γTuNA concentration remaining in γTuRC peak fraction. For this, Halo-γTuRC preps were done with Halo-3C-AU1 reporter tagged versions of human γTuNA. After elution of target protein via PreScission (3 C) cleavage, any remaining γTuNA will still be fused to the AU1 epitope. Probing of the γTuRC peak fraction with a sensitive anti-AU1 antibody (against a titration of known amounts of AU1) allowed us to determine that 100 nM of AU1 remained. This corresponded to 50 nM γTuNA dimer. All raw blots are available in “Figure 2—figure supplement 1—source data 1.”
-
Figure 2—figure supplement 2—source data 1
Labeled and raw blots used in Figure 2—figure supplement 2A, B, C.
- https://cdn.elifesciences.org/articles/80053/elife-80053-fig2-figsupp2-data1-v2.zip
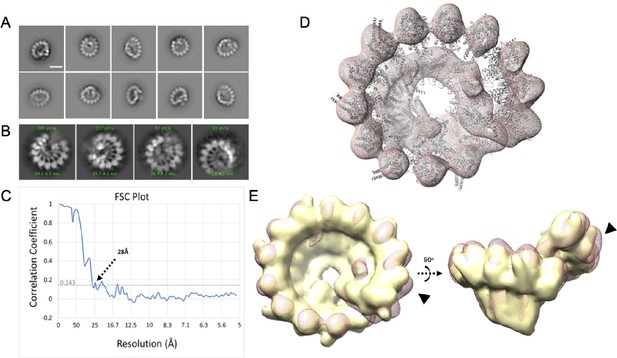
γTuRCs purified via Halo-γTuNA pulldown are fully assembled rings.
Electron-microscopy data of γTuNA-prepped γTuRC. (A) 2D class averages from 2692 particles isolated from negative-stain data. Scale bar = 20 nm. (B) Selected 2D class averages from 529 particles selected from cryoEM data. (C) Fourier shell correlation (FSC) plot from 3D reconstructions and final refinements of particles of γTuRC from negative-stain EM data, showing a final resolution of 28 Å. (D) Map of γTuNA-prepped γTuRC (red; mesh) with previously published holo-γTuRC structure from Xenopus laevis docked. (E) Overlay of γTuNA-prepped γTuRC with a 28 Å simulated map from the published Xenopus laevis structure of γTuRC (yellow), showing a high correlation between the two (0.89). Highlighted is the area where γTuNA is predicted to bind γTuRC (arrows). Figure made using Chimera. (D and E) Both parts use structure PDB 6TF9 for comparison.
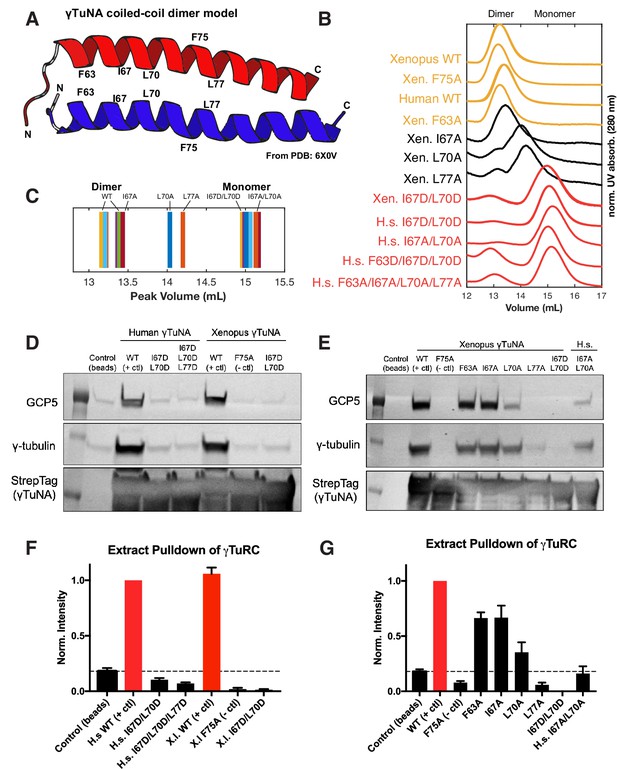
γTuNA requires both dimerization and the F75 residue to bind γTuRC in extract.
(A) Model of dimerized, coiled-coil γTuNA with labeled side-chains for residues F63, I67, L70, F75, and L77. Made using PyMOL (RRID:SCR_000305) and chains C/G (red color) and D/H (blue color) from PDB: 6X0 V (Wieczorek et al., 2020a). (B) Size-exclusion chromatograms for human (aa 53–98) and Xenopus (aa 56–101) Halo-γTuNA wildtype and mutant constructs. Proteins were run at 50 µM (monomer) on a Superdex 200 increase 10/300 GL column (Cytiva) on an Äkta Pure system. Absorbance traces (A280 nm) were normalized by their peaks and plotted stacked as shown. (C) Diagram of peak retention volumes for each construct tested. (D) Western blots for γTuRC components, GCP5 and γ-tubulin, pulled down by beads coupled to human and Xenopus Halo-γTuNAs incubated in egg extract. The Strep-tag blot is shown as a bead loading control. (E) Western blots as in D, except comparing pulldowns done with Halo-Xenopus γTuNA alanine point mutants, with wildtype and F75A mutants as positive and negative controls. (F) Quantification of γTuRC pulldowns shown in D, normalized to the band intensity for human wildtype Halo-γTuNA beads. N=3. Error bars are SEM. (G) Same quantification of γTuRC pulldowns as in F, except for pulldowns as done in E. Normalized to the band intensity of wildtype Xenopus γTuNA. N=2. Error bars are SEM. See “Figure 3—source data 1” for numerical data and “Figure 3—source data 2” for raw blots.
-
Figure 3—source data 1
Numerical data for Figure 3, includes normalized size-exclusion chromatography traces for Figure 3B, quantified pulldowns in Figure 3F, and quantified pulldowns in Figure 3G.
- https://cdn.elifesciences.org/articles/80053/elife-80053-fig3-data1-v2.zip
-
Figure 3—source data 2
Labeled and raw blots used in Figure 3D and E.
- https://cdn.elifesciences.org/articles/80053/elife-80053-fig3-data2-v2.zip
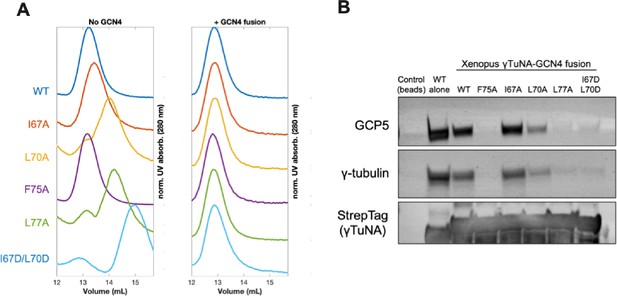
Rescuing γTuNA dimerization is not enough to rescue γTuRC binding.
(A) Size-exclusion chromatography traces for Xenopus Strep-His-Halo-3C-γTuNA (Halo-γTuNA) or Strep-His-Halo-γTuNA-GCN4 fusion constructs. These constructs were either wildtype or single alanine mutations of residues I67, L70, F75, and L77. Traces were normalized by their maximum peak absorbance and plotted as shown. Proteins were run at 50 µM in a Superdex 200 increase 10/300 GL column (Cytiva) on an Äkta Pure system. (B) Western blot of γTuRC components, GCP5 and γ-tubulin, pulled down from extract by beads coupled to Halo-Xenopus γTuNA wildtype (as a positive control) or Halo-Xenopus γTuNA-GCN4 fusion constructs (wildtype, alanine point mutants, or an aspartate double mutant). StrepTag blot is shown as a bait loading control. Raw blots available in “Figure 3—figure supplement 1—source data 1.”
-
Figure 3—figure supplement 1—source data 1
Labeled and raw blots used in Figure 3—figure supplement 1B.
- https://cdn.elifesciences.org/articles/80053/elife-80053-fig3-figsupp1-data1-v2.zip
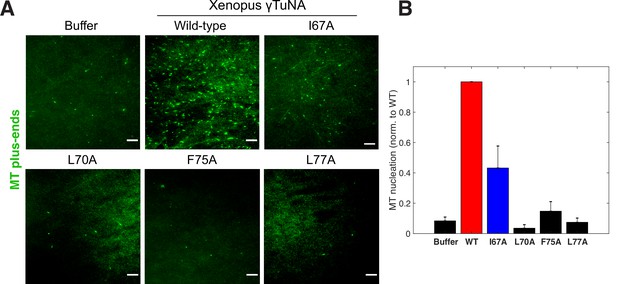
Complete γTuNA dimerization is required to maximally increase MT nucleation in extract.
(A) TIRF assay of MT nucleation in extract after addition of 2 µM (1 µM dimer) wildtype or single alanine mutants of Strep-His-Xenopus γTuNA. EB1-mCherry was used to count MTs (MT nucleation) and is shown pseudo-colored green. Images were taken after 5 min at 18–20°C. Bar = 5 µm. (B) Quantification of MT nucleation (MT number) normalized by the wildtype condition across four independent experiments. Red bar denotes wildtype level, while the blue bar denotes the effect of the I67A mutant. Error bars are SEM. See “Figure 4—source data 1” for numerical data.
-
Figure 4—source data 1
Numerical data used in Figure 4.
- https://cdn.elifesciences.org/articles/80053/elife-80053-fig4-data1-v2.zip
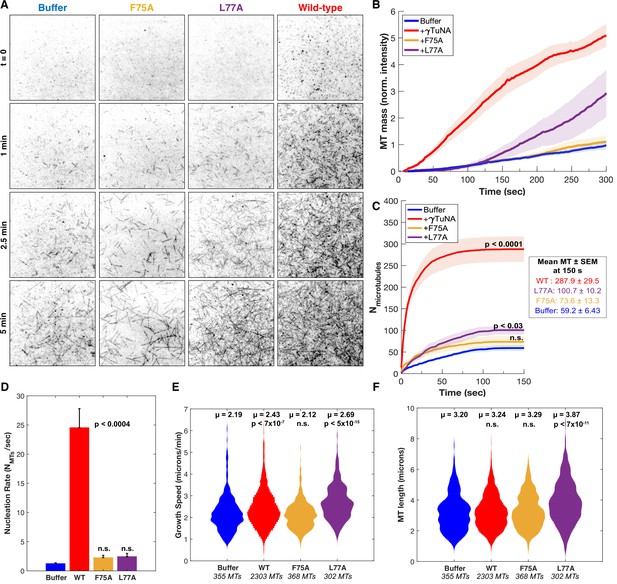
γTuNA dimers directly activate γTuRC MT nucleation ability in vitro.
(A) Single molecule TIRF assays of γTuRC-mediated MT nucleation in vitro. Purified Xenopus γTuRCs were biotinylated and attached to passivated coverslips via surface-bound Neutravidin molecules. Polymerization mix containing 15 μM tubulin,1 mM GTP, and either control buffer or 3.3 μM (1.7 μM dimer) Strep-His-Xenopus γTuNA was then added. Wildtype, F75A, and L77A versions of γTuNA were tested. 5% Alexa 568-tubulin was used to visualize MTs. Images were taken every 2 s, for 5 min total, at 33.5 °C. Wildtype γTuNA (n=8), buffer control (n=6), γTuNA-F75A (n=5), and γTuNA-L77A (n=3). (B) Mean MT signal (MT mass) over time, normalized to the buffer condition at 300 s. (C) Mean MT number over time (measured for the first 150 s). The box shows the mean MT number ± SEM at 150 s for each condition. (B and C) Solid lines are the mean over time, with error clouds representing SEM. (D) Initial nucleation rates (Mts nucleated per sec) for each condition (± SEM). The curves shown in part C were fit to an exponential function to determine k (the nucleation rate). Each k was then averaged; see Materials and methods. The following are mean nucleation rate ± SEM. Buffer: 1.2±0.15 MTs/s, WT: 24.5±3.27 MTs/s, F75A: 2.3 ± 0.41 MTs/s, L77A: 2.4±0.55 MTs/s. (E and F) Violin plots of MT growth speeds (in E) or MT lengths (in F) for each condition. Means (μ) are shown alongside p-values. Wildtype γTuNA (n=2303 MTs), buffer control (n=355 MTs), γTuNA-F75A (n=368 MTs), and γTuNA-L77A (n=302 MTs). (C-F) Two-sample unpaired t-tests were used to compare the buffer control to the experimental values. Significance is p<0.05. See “Figure 5—source data 1” for all numerical data presented here.
-
Figure 5—source data 1
Numerical data from Figure 5’s in vitro TIRF assays with purified γTuRC and γTuNA: including MT mass measurements, MT number, MT growth speed, and MT lengths.
- https://cdn.elifesciences.org/articles/80053/elife-80053-fig5-data1-v2.zip
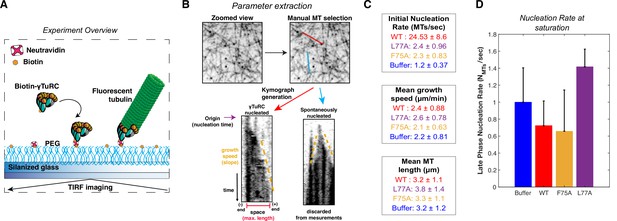
Overview and additional single molecule TIRF data.
(A) Diagram of single molecule TIRF assay for γTuRC MT nucleation. (B) Process diagram of parameter extraction from single molecule TIRF data: individual MTs were manually selected in FIJI (ImageJ, NIH). Next, time-lapse data (stack) was resliced for each MT to generate space vs time plots (better known as kymographs). From these kymographs, we determined whether a MT nucleated from a γTuRC (no growth at one end) or spontaneously (growth from both ends). If a MT was nucleated by a γTuRC, then we manually extracted the time that MT nucleated (origin), the growth speed, and the max length of that MT in the kymograph. If spontaneously nucleated, we did not extract these parameters. (C) Numerical representations of data presented in Figure 5C–F. Mean ± standard deviation. (D) Nucleation rate measured at saturation (30–150 s), normalized to the buffer mean. Error bars are standard deviation. Source data is available in “Figure 5—figure supplement 1—source data 1” file.
-
Figure 5—figure supplement 1—source data 1
Numerical data used in Figure 5—figure supplement 1; late-stage nucleation rates for experiments from Figure 5.
- https://cdn.elifesciences.org/articles/80053/elife-80053-fig5-figsupp1-data1-v2.zip
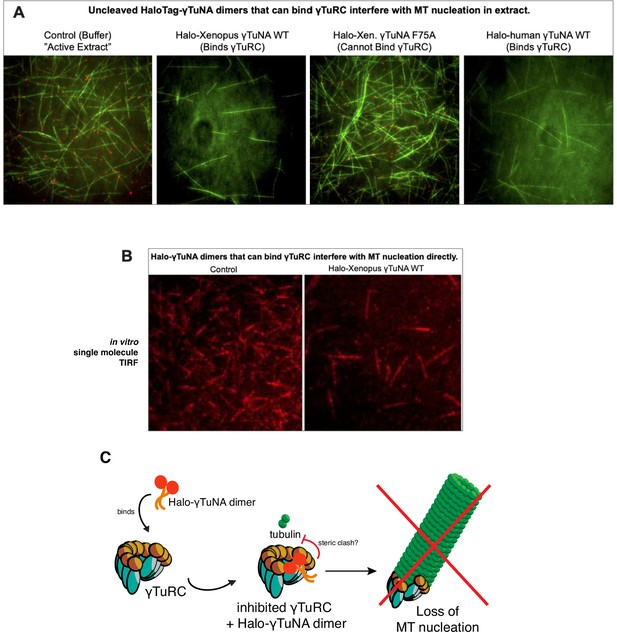
The presence of large, bulky N-terminal tags on γTuNA directly inhibits γTuRC activity in extract and in vitro.
(A) TIRF assay of Xenopus egg extract in the presence of N-terminally tagged, Halo-γTuNA constructs (wildtype and F75A) after 15 min. Alexa488-tubulin and EB1-mCherry were used to visualize MTs and MT plus-ends. Halo-γTuNA constructs that can bind γTuRC (as determined in Figure 3) drastically decreased the amount of MTs nucleated (human and Xenopus wildtype). Halo-γTuNA F75A mutant, which cannot bind γTuRC, had no effect on MT levels. Images were taken at 18–20°C. Halo-γTuNA was added at 2 µM final. (B) Single molecule TIRF assay of γTuRC-mediated MT nucleation in vitro. After 5 min, γTuRCs alone efficiently nucleate MTs. In the presence of 3.3 µM wildtype Xenopus Halo-γTuNA, few γTuRCs nucleate MTs, indicating a direct inhibition of γTuRC activity. (C) Model for inhibition of γTuRC activity by N-terminal Halo-tagged γTuNA dimers.
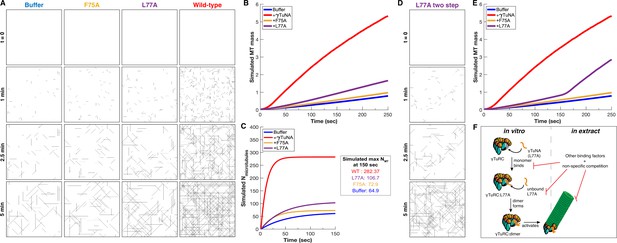
Simulation of γTuNA’s effect on γTuRC MT nucleation activity.
(A) Simulated frames of MT nucleation and growth (simulated 40x40 µm2 plane) over 5 minutes. Based on just the initial nucleation rate and a single constant growth speed, the simulated data correlates well with the observed data in Figure 5 (see Video 3 for side-by-side comparison). (B) Simulated MT mass over time, measured from simulated movies using the same FIJI (ImageJ) pipeline as used with real data in Figure 5. (C) Simulated number of nucleated MTs over time, with simulated maximum number of MTs at 150 s. (D) Simulated frames of MT nucleation and growth of a two-step model for the L77A mutant. (E) Simulated MT mass over time for the two-step L77A model, now reflecting a late stage increase in MT mass, similar to what was observed in Figure 5B. (F) Model for L77A’s two stage behavior: involving sequential binding of two separate L77A monomers that form a dimer on γTuRC before triggering its activation. In extract, this behavior is not observed, suggesting the presence of other extract factors compete against this interaction. Code used in this figure is available as Source code 1 and Source code 2.
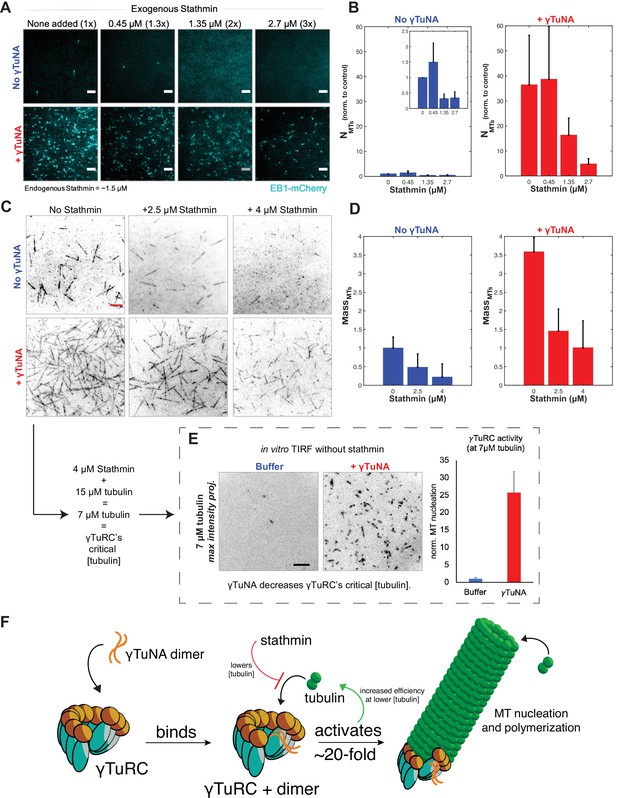
γTuNA enhances γTuRC activity at low tubulin concentrations in extract and in vitro.
(A) TIRF assay of extract MT nucleation (Strep-His-γTuNA vs stathmin). His-SNAP-tag-Xenopus stathmin (isoform 1 A) was added at 0.45, 1.35, and 2.7 µM final concentration to extract. Either buffer or 2.3 µM (1.15 µM dimer) Strep-His-Xenopus wildtype γTuNA was also added. EB1-mCherry was used to visualize MT plus-ends (MT number). Images were taken after 5 min at 18–20°C and pseudo-colored cyan. Bars = 5 µm. (B) Normalized MT number for each concentration of stathmin tested, n=4; bars are SEM. (C) Single molecule TIRF assay of γTuRC MT nucleation in vitro. Purified γTuRCs were assayed at 33.5 °C with 15 µM tubulin, 1 mM GTP, either alone, in the presence of 2.5 µM or 4 µM stathmin, or with 3.3 µM (1.65 µM dimer) Strep-His-Xenopus γTuNA. 5% Alexa-568-tubulin was used to visualize MTs. Red bar = 5 µm. (D) Normalized MT mass at 300 seconds (intensity normalized to buffer control) from C. Error bars are SEM. N=3 minimum for all conditions, except: n=4 for “4 µM stathmin” and, n=5 for “4 µM stathmin + γTuNA”. (E) TIRF assay of γTuRC activity in vitro at its critical tubulin concentration (7 µM) with or without 3.3 µM (1.65 µM dimer) Strep-His-Xenopus γTuNA. Images are max intensity projections of 5 min time-series. Normalized to buffer control: n=3 (γTuNA); n=2 (buffer). (F) Model of γTuNA’s activation of γTuRC-mediated microtubule nucleation. See “Figure 6—source data 1” for all numerical data.
-
Figure 6—source data 1
Numerical data used in Figure 6: including raw EB1 counts for stathmin/γTuNA experiments in extract, and MT mass measurement for in vitro TIRF assays with purified γTuRC, γTuNA, and stathmin.
- https://cdn.elifesciences.org/articles/80053/elife-80053-fig6-data1-v2.zip
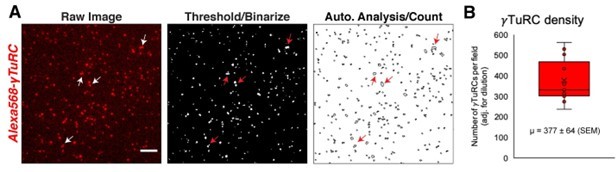
Analysis of the number of γTuRCs present in the in vitro TIRF assays.
(A) Image analysis of fluorescent γTuRC (labeled with NHS-Alexa 568) in an in vitro TIRF assay. Unlike in assays shown in Figure 5, γTuRC was further diluted two-fold with buffer to aid in accurate counting. Images were binarized and counted via ImageJ’s “Analyze Particles” function. Outlines of counted complexes are shown in “A” rightmost panel. Arrows mark complexes that the automatic analysis undercounted as a single γTuRC. Without dilution of γTuRC, this undercounting is worsened and leads to significant error. (B) Quantification of the number of γTuRC spots, corrected for dilution factor. The mean, marked by an “X”, is 377 γTuRCs (±64 SEM). The first to third quartile range (red box) is 302 to 469 γTuRCs.
Videos
Post-pulldown wildtype γTuNA beads nucleate asters in vitro.
Wildtype γTuNA directly stimulates γTuRC in vitro.
Video of simulated γTuNA-dependent activation of γTuRC.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Xenopus laevis) | cdk5rap2.L (Xenopus laevis) | NCBI | XP_018085184.1; isoform X1 | |
| Strain, strain background (Escherichia coli) | DH5-alpha (High Efficiency) | New England Biolabs | C2987I | Chemically competent; cloning strain |
| Strain, strain background (Escherichia coli) | BL21(DE3) | Sigma-Aldrich | 71402 | Chemically competent; expression strain |
| Biological sample (Xenopus laevis) | Xenopus laevis eggs and egg extract | This study | Method previously described; see Good and Heald, 2018 | |
| Antibody | anti-MZT1, (Rabbit polyclonal) | Abcam | ab178359 | 375 µg/mL, used in bead attachment assay |
| Antibody | Anti-gamma-tubulin, GTU-88 clone (Mouse, monoclonal) | Sigma | T6557; RRID:AB_2863751 | 1:1000 dilution |
| Antibody | anti-GCP5, E-1 clone (Mouse monoclonal) | Santa Cruz Biotechnology | sc-365837; RRID:AB_10847352 | 1:250 dilution |
| Antibody | anti-GFP, ChIP grade, (Rabbit polyclonal) | Abcam | Ab290 | 1:1000 dilution |
| Antibody | anti-StrepTagII, (Mouse monoclonal) | Qiagen | 34850; RRID:AB_2810987 | 1:1000 dilution |
| Antibody | anti-AU1, (Mouse monoclonal) | Biolegend | 901905 | 1:1000 dilution |
| Antibody | Mouse IgG, HRP-linked whole Ab, secondary (Sheep, clonality not reported by manufacturer) | Amersham | NA931-1ML | 1:3000 dilution |
| Antibody | Rabbit IgG, HRP-linked whole Ab, secondary (Donkey, clonality not reported by manufacturer) | Amersham | NA934-1ML | 1:3000 dilution |
| Recombinant DNA reagent | pET28a-Hook3 aa 1–160-GCN4 (plasmid) | Addgene | 74608; RRID:Addgene_74608 | Ron Vale; Schroeder and Vale, 2016 |
| Recombinant DNA reagent | Modified pST50Trc (StrepTagII-6xHis-PreScission cleavage site) with human or X. laevis γTuNA for bacterial expression (plasmids) | This study | See Table 1 for all constructs | |
| Commercial assay or kit | 2 x Gibson Assembly Master Mix | New England Biolabs | E2611L | |
| Commercial assay or kit | Strep-Tactin Superflow | IBA | 2-1206-025 | |
| Commercial assay or kit | Halo Magne beads | Promega | G7287 | |
| Commercial assay or kit | HisPur Ni-NTA magnetic beads | ThermoFisher | 88831 | |
| Commercial assay or kit | Quick Start Bradford 1 x Dye Reagent | BioRad | 5000205 | |
| Commercial assay or kit | Akta Püre System with Superdex 200 increase 10/300 GL column | Cytiva (formerly GE Healthcare) | 28-9909-44 | |
| Commercial assay or kit | SNAP i.d. 2.0 rapid Western blotting system | EMD-Millipore | SNAP2MM | |
| Chemical compound, drug | Pluronic-F127 | ThermoFisher | P6866 | |
| Chemical compound, drug | NHS-PEG4-Biotin | ThermoFisher | A39259 | |
| Chemical compound, drug | Dichlorodimethylsilane | Sigma | 440272–100 ML | |
| Software, algorithm | Fiji (ImageJ) | NIH | RRID:SCR_002285 | |
| Software, algorithm | MATLAB | MathWorks | ver. R2019a; RRID:SCR_001622 | |
| Software, algorithm | Prism 7 | GraphPad Software | RRID:SCR_002798 | |
| Software, algorithm | Relion 3.1 | Zivanov et al., 2018 | ||
| Software, algorithm | CryoSparc 3.2 | Punjani et al., 2017 | ||
| Other | Nikon Ti-E inverted scope system | Nikon | RRID:SCR_021242 | See Materials and Methods. |
| Other | Optima MAX-XP ultracentrifuge | Beckman Coulter | 393315 | See Materials and Methods. |
γTuNA constructs generated in this study.
Key: S = StrepTagII, H = 6xHis-tag, 3C = human rhinovirus 3C (PreScission) protease cleavage site, TEV = tobacco etch virus protease cleavage site, GCN4 = yeast transcription factor GCN4 dimerization domain. Mutated residues in the “Sequence” column are designated using { } brackets. Primer sequences and a copy of this table are included in Supplementary file 1.
| Name | Species | N-term tag versions | Fused GCN4 version? | Sequence | Mutation Type |
|---|---|---|---|---|---|
| Wildtype | H. sapiens (H.s) | SH-3C; SH-Halo-3C-; SH-Halo-3C-AU1 | No | SPTRARNMKDFENQITELKKENFNLKLRIYFLEERMQQEFHGPTEH | None |
| Wildtype | X. laevis (X.l.) | SH-TEV; SH-3C; SH-TEV-GFP; SH-Halo-3C- | Yes, SH-Halo-3C-(γTuNA)-GCN4 | ...MKDFEKQIAELKKENFNLKLRIYFLEEQVQQKCDNSSEDLYRMNIE | None |
| F63A | H.s./X.l. | SH-3C; SH-Halo-3C | No | ...MKD{A}EKQIAELKKENFNLKLRI… | Weakened hydrophobic |
| I67A | H.s./X.l. | SH-3C; SH-Halo-3C | Yes, SH-Halo-3C-(γTuNA)-GCN4 | ...MKDFEKQ{A}AELKKENFNLKLR... | Weakened hydrophobic |
| L70A | H.s./X.l. | SH-3C; SH-Halo-3C | Yes, SH-Halo-3C-(γTuNA)-GCN4 | ...MKDFEKQIAE{A}KKENFNLKLR... | Weakened hydrophobic |
| F75A | H.s./X.l. | SH-3C; SH-Halo-3C | Yes, SH-Halo-3C-(γTuNA)-GCN4 | ...MKDFEKQIAELKKEN{A}NLKLR... | Weakened hydrophobic |
| L77A | H.s./X.l. | SH-3C; SH-Halo-3C | Yes, SH-Halo-3C-(γTuNA)-GCN4 | ...MKDFEKQIAELKKENFN{A}KLR... | Weakened hydrophobic |
| I67A/L70A | H.s./X.l. | SH-3C; SH-Halo-3C | No | ...MKDFEKQ{A}AE{A}KKENFNLKLR... | Weakened hydrophobic (2 x) |
| F63D | X.l. | SH-Halo-3C | No | ...MKD{D}EKQIAELKKENFNLKLR… | Flip to hydrophilic |
| I67D | X.l. | SH-Halo-3C | No | ...MKDFEKQ{D}AELKKENFNLKLR... | Flip to hydrophilic |
| L70D | X.l. | SH-Halo-3C | No | ...MKDFEKQIAE{D}KKENFNLKLR... | Flip to hydrophilic |
| F75D | X.l. | SH-Halo-3C | No | ...MKDFEKQIAELKKEN{D}NLKLR... | Flip to hydrophilic |
| L77D | X.l. | SH-Halo-3C | No | ...MKDFEKQIAELKKENFN{D}KLR... | Flip to hydrophilic |
| I67D/L70D | H.s./X.l. | SH-Halo-3C | Yes, SH-Halo-3C-(γTuNA)-GCN4 | ...MKDFEKQ{D}AE{D}KKENFNLKLR... | Flip to hydrophilic (2 x) |
| I67D/L70D/L77D | H.s. | SH-Halo-3C | No | SPTRARNMKDFENQ{D}TE{D}KKENFN{D}KLRIYFLEERMQQEFHGPTEH | Flip to hydrophilic (3 x) |
Additional files
-
Supplementary file 1
Primers used to generate γTuNA constructs used in this study; pertains to Table 1.
- https://cdn.elifesciences.org/articles/80053/elife-80053-supp1-v2.zip
-
MDAR checklist
- https://cdn.elifesciences.org/articles/80053/elife-80053-mdarchecklist1-v2.pdf
-
Source code 1
MATLAB source code for numerical simulation of MT nucleation from purified γTuRCs in the presence of γTuNA constructs (used to generate Figure 5—figure supplement 3 and Video 3).
- https://cdn.elifesciences.org/articles/80053/elife-80053-code1-v2.zip
-
Source code 2
MATLAB source code for graphical simulation of MT nucleation from purified γTuRCs in the presence of γTuNA constructs (uses Source code 1 as input; used to generate Figure 5—figure supplement 3 and Video 3).
- https://cdn.elifesciences.org/articles/80053/elife-80053-code2-v2.zip





