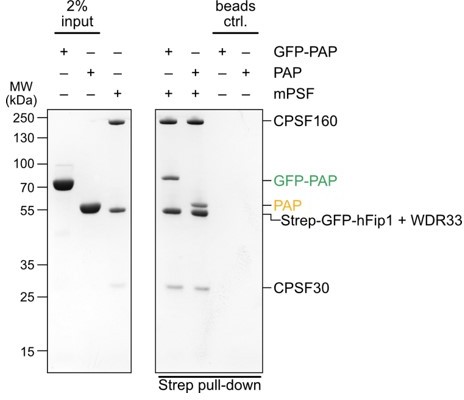
Fip1 is a multivalent interaction scaffold for processing factors in human mRNA 3′ end biogenesis
Figures
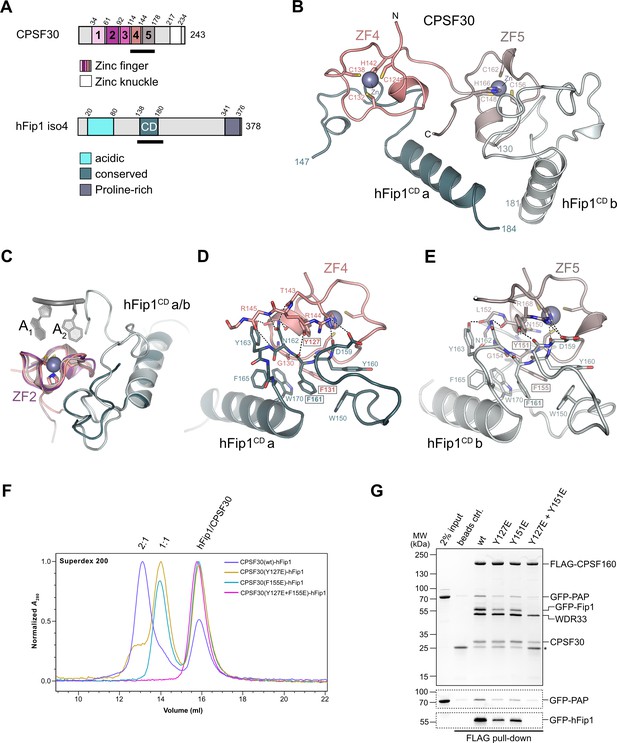
hFip1 interacts with CPSF30 with 2:1 stoichiometry.
(A) Schematic representation of the domain architecture of CPSF30 and hFip1. CPSF30 consists of five zinc finger (ZF) domains and a zinc knuckle domain. hFip1 isoform 4 comprises acidic, conserved, and proline-rich regions but lacks the RE/D region interacting with CF Im, as well as the R-rich region, which has been shown to bind U-rich RNA in hFip1 isoform 1 (Kaufmann et al., 2004). (B) Cartoon representation of the crystal structure of CPSF30ZF4–ZF5 in complex with two hFip1 fragments comprising the conserved domain (CD). (C) Superposition of CPSF30 ZF2 domain in complex with PAS RNA onto ZF4 and ZF5. (D) Detailed interaction interface of hFip1CD with CPSF30 ZF4. (E) Detailed interaction interface of hFip1CD with CPSF30 ZF5. (F) Size-exclusion chromatography coupled to multiangle static light scattering (SEC-MALS) chromatogram of MBP-CPSF30ZF4–ZF5 selective hFip1-binding mutants for stoichiometry analysis with GFP-hFip1. (G) In vitro pull-down analysis of FLAG-epitope-tagged mPSF comprising wild-type CPSF30 and its selective hFip1-binding mutants with GFP-PAP. Asterisk indicates anti-FLAG M2 antibody light chain. GFP-hFip1 and GFP-PAP are also visualized with in-gel GFP fluorescence (bottom).
-
Figure 1—source data 1
Raw gel image for Figure 1, panel G.
- https://cdn.elifesciences.org/articles/80332/elife-80332-fig1-data1-v2.zip
-
Figure 1—source data 2
Raw gel image for Figure 1, panel G.
- https://cdn.elifesciences.org/articles/80332/elife-80332-fig1-data2-v2.zip
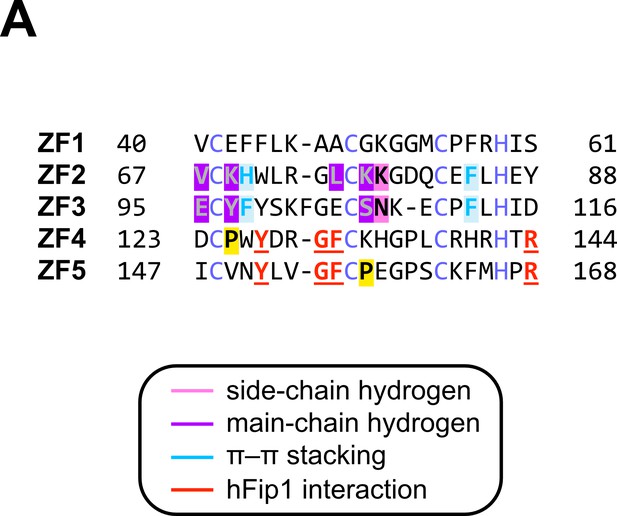
Sequence alignment of CPSF30 zinc finger domains.
(A) Sequence alignment of human CPSF30 zinc finger domains. Residues responsible for RNA interactions (in ZF2/ZF3) or hFip1 interaction (in ZF4/ZF5) are highlighted and the nature of their interaction color-coded. ZF4/ZF5 domains contain proline residues (yellow) at positions corresponding to critical main-chain hydrogen bonding interactions in ZF2/ZF3.
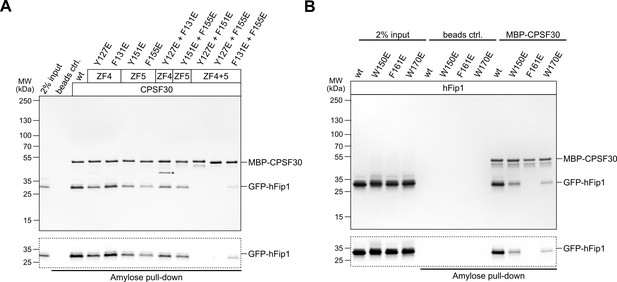
Analysis of the hFip1–CPSF30 interaction using structure-guided point mutants.
(A) Pull-down assay of immobilized MBP-tagged wild-type (wt) or mutant CPSF30 proteins with GFP-hFip1. GFP-hFip1 is visualized by in-gel GFP fluorescence (bottom). Asterisk indicates contaminating free MBP protein. (B) Pull-down assay of immobilized MBP-tagged wt CPSF30 and GFP-hFip1 mutants.
-
Figure 1—figure supplement 2—source data 1
Raw gel image for Figure 1—figure supplement 2, panel A.
- https://cdn.elifesciences.org/articles/80332/elife-80332-fig1-figsupp2-data1-v2.zip
-
Figure 1—figure supplement 2—source data 2
Raw gel image for Figure 1—figure supplement 2, panel A.
- https://cdn.elifesciences.org/articles/80332/elife-80332-fig1-figsupp2-data2-v2.zip
-
Figure 1—figure supplement 2—source data 3
Raw gel image for Figure 1—figure supplement 2, panel B.
- https://cdn.elifesciences.org/articles/80332/elife-80332-fig1-figsupp2-data3-v2.zip
-
Figure 1—figure supplement 2—source data 4
Raw gel image for Figure 1—figure supplement 2, panel B.
- https://cdn.elifesciences.org/articles/80332/elife-80332-fig1-figsupp2-data4-v2.zip
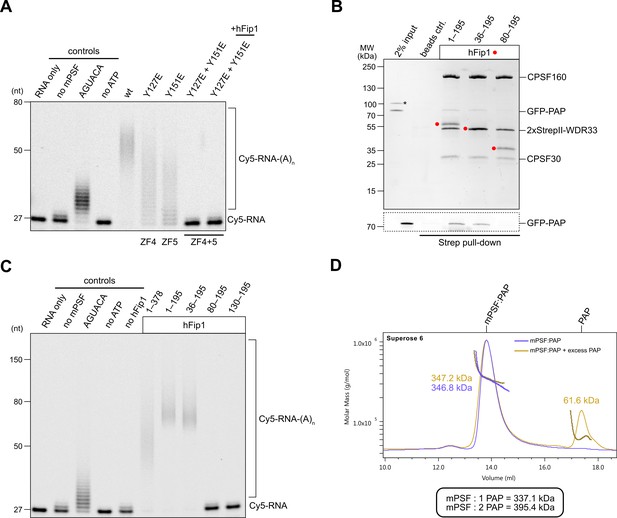
hFip1 directly recruits poly(A) polymerase.
(A) Polyadenylation activity assay of mPSF complexes containing wild-type and mutant CPSF30 proteins as well as hFip1 added in trans (rightmost lane) using a Cy5-labeled PAS-containing RNA substrate. An RNA substrate lacking the canonical AAUAAA PAS hexanucleotide is denoted by its substitute sequence, AGUACA. Polyadenylated RNA products are indicated as RNA-(A)n. (B) Pull-down analysis of immobilized StrepII-tagged mPSF complexes comprising N-terminal truncations of hFip1 with GFP-PAP. GFP-PAP is visualized by in-gel GFP fluorescence (bottom). Asterisk denotes contaminating protein. (C) Polyadenylation activity assay of mPSF complexes containing hFip1 truncations. (D) Size-exclusion chromatography coupled to multiangle static light scattering (SEC-MALS) analysis of reconstituted mPSF:PAP:RNA complexes and in the absence (purple) or presence of excess PAP (yellow). Theoretical molecular masses of 1:1 and 1:2 mPSF:PAP complexes are indicated.
-
Figure 2—source data 1
Raw gel image for Figure 2, panel A.
- https://cdn.elifesciences.org/articles/80332/elife-80332-fig2-data1-v2.zip
-
Figure 2—source data 2
Raw gel image for Figure 2, panel A.
- https://cdn.elifesciences.org/articles/80332/elife-80332-fig2-data2-v2.zip
-
Figure 2—source data 3
Raw gel image for Figure 2, panel B.
- https://cdn.elifesciences.org/articles/80332/elife-80332-fig2-data3-v2.zip
-
Figure 2—source data 4
Raw gel image for Figure 2, panel B.
- https://cdn.elifesciences.org/articles/80332/elife-80332-fig2-data4-v2.zip
-
Figure 2—source data 5
Raw gel image for Figure 2, panel C.
- https://cdn.elifesciences.org/articles/80332/elife-80332-fig2-data5-v2.zip
-
Figure 2—source data 6
Raw gel image for Figure 2, panel C.
- https://cdn.elifesciences.org/articles/80332/elife-80332-fig2-data6-v2.zip
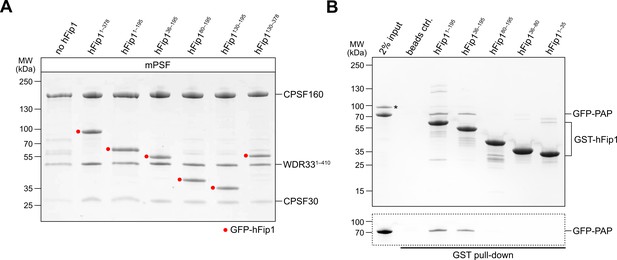
Analysis of hFip1 regions required for PAP recruitment.
(A) Purified mPSF complexes containing hFip1 isoform 4 and truncations thereof (indicidated with red dots) used in polyadenylation activity assay (Figure 2C). (B) Pull-down assay of immobilized glutathione-S-transferase (GST)-tagged hFip1 fragments with GFP-PAP. GFP-PAP is visualized by in-gel GFP fluorescence (bottom). Asterisk denotes protein contaminant.
-
Figure 2—figure supplement 1—source data 1
Raw gel image for Figure 2—figure supplement 1, panel A.
- https://cdn.elifesciences.org/articles/80332/elife-80332-fig2-figsupp1-data1-v2.zip
-
Figure 2—figure supplement 1—source data 2
Raw gel image for Figure 2—figure supplement 1, panel A.
- https://cdn.elifesciences.org/articles/80332/elife-80332-fig2-figsupp1-data2-v2.zip
-
Figure 2—figure supplement 1—source data 3
Raw gel image for Figure 2—figure supplement 1, panel B.
- https://cdn.elifesciences.org/articles/80332/elife-80332-fig2-figsupp1-data3-v2.zip
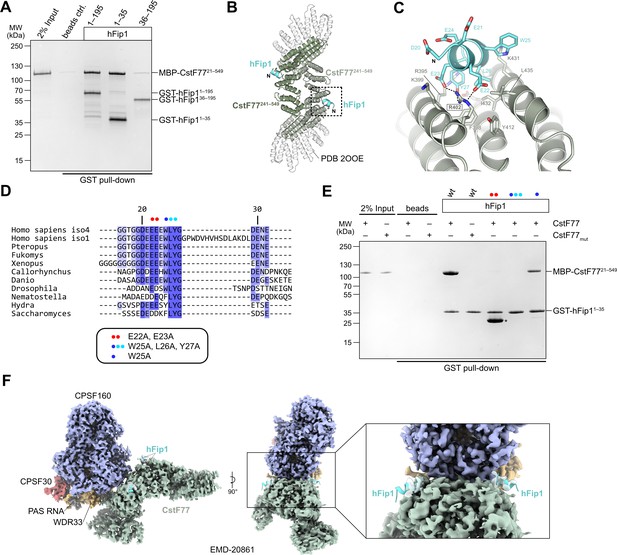
hFip1 interacts with CstF77 through a conserved motif within its N-terminal acidic domain.
(A) Pull-down analysis of immobilized GST-hFip1 fragments with MBP-CstF7721–549. (B) Cartoon representation of the crystal structure of the CstF77241–549–hFip11–35 complex, superimposed onto the structure of murine CstF77 (white, PDB ID: 2OOE). (C) Zoomed-in view of the hFip1–CstF interaction interface. (D) Multiple sequence alignment of the N-terminal region of Fip1 orthologs. (E) Pull-down analysis of immobilized wild-type and mutant GST-hFip11–35 proteins with MBP-CstF7721–549 and MBP-CstF77mut (R395A/R402A/K431A). Asterisk indicates contaminating free GST protein. (F) 3D cryo-EM density map (EMD-20861) of the human CPSF160–WDR33–CPSF30–PAS RNA–CstF77 complex (Zhang et al., 2019), displayed at contour level 0.015 and color coded according to the corresponding atomic protein model (PDB ID 6URO). The hFip1–CstF77 crystal structure from this study was superimposed onto the atomic model of CstF77, and atomic model of hFip1 is shown (cyan). Inset shows a zoomed-in view of unassigned density that matches hFip1.
-
Figure 3—source data 1
Raw gel image for Figure 3, panel A.
- https://cdn.elifesciences.org/articles/80332/elife-80332-fig3-data1-v2.zip
-
Figure 3—source data 2
Raw gel image for Figure 3, panel E.
- https://cdn.elifesciences.org/articles/80332/elife-80332-fig3-data2-v2.zip
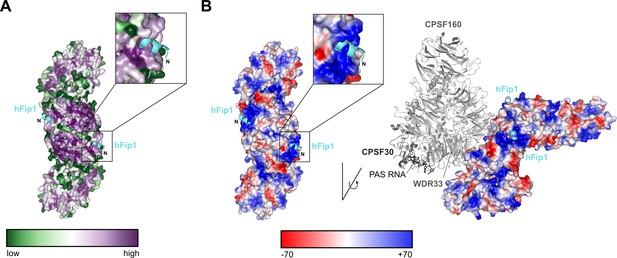
hFip1 binds to a conserved positively charged patch on CstF77.
(A) Color-coded representation of surface conservation of CstF77. Cartoon representation of bound hFip11–35 (cyan). Inset shows a zoomed-in view of the sequence conservation of CstF77 surrounding at the Fip1-binding site. (B) Color-coded electrostatic surface representation of CstF77 HAT homodimer alone (left) or bound to mPSF (right), both with hFip1 (cyan) shown in cartoon representation. Inset shows a zoomed-in view of the CstF77 surface electrostatics at the hFip1-binding site.
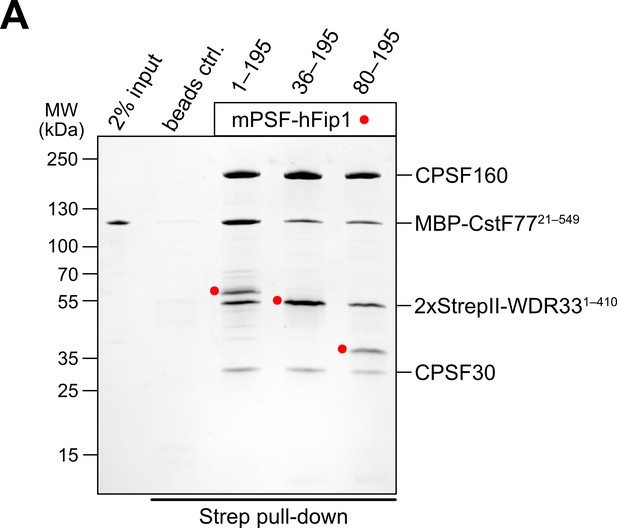
The N-terminal region of hFip1 contributes to mPSF–CstF77 interaction.
(A) Pull-down analysis of immobilized StrepII-tagged mPSF complexes containing N-terminally truncated hFip1 proteins with MBP-CstF77 HAT domain.
-
Figure 3—figure supplement 2—source data 1
Raw gel image for Figure 3—figure supplement 2, panel A.
- https://cdn.elifesciences.org/articles/80332/elife-80332-fig3-figsupp2-data1-v2.zip
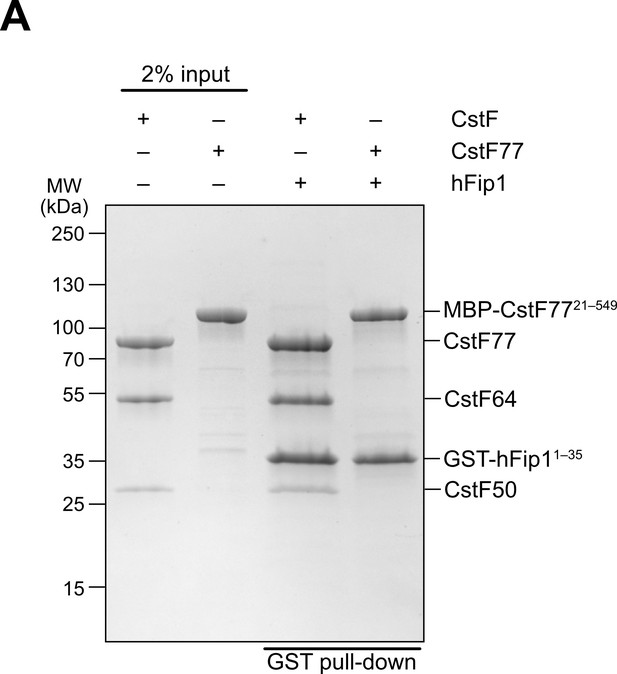
The N-terminal region of hFip1 interacts with CstF complex.
(A) Pull-down analysis of immobilized GST-tagged hFip11–35 with holo-CstF complex.
-
Figure 3—figure supplement 3—source data 1
Raw gel image for Figure 3—figure supplement 3, panel A.
- https://cdn.elifesciences.org/articles/80332/elife-80332-fig3-figsupp3-data1-v2.zip
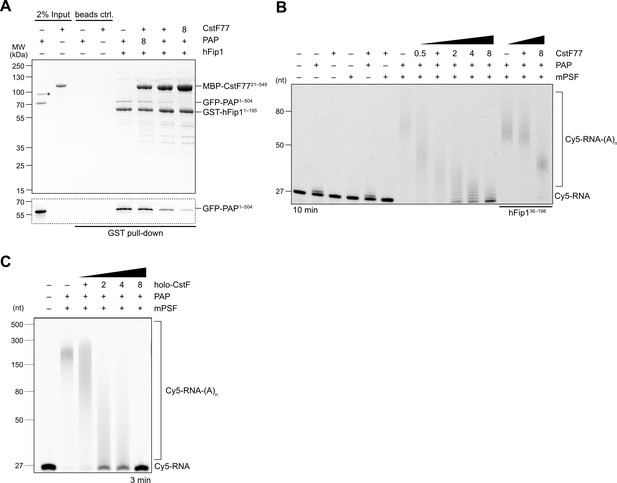
CstF77 competitively inhibits 3′ polyadenylation.
(A) Pull-down analysis of immobilized GST-hFip11–195 with varying molar ratios of GFP-PAP and MBP-CstF7721–549. GFP-PAP is visualized by in-gel GFP fluorescence (bottom). Asterisk denotes contaminating protein. (B) Polyadenylation activity assay of mPSF complexes containing full-length hFip1 and N-terminally truncated hFip1 (hFip136–195) in the presence of varying molar ratios of CstF77. Polyadenylated RNA products are indicated as RNA-(A)n. (C) Polyadenylation activity assay of mPSF in the presence of varying molar ratios of holo-CstF complex.
-
Figure 4—source data 1
Raw gel image for Figure 4, panel A.
- https://cdn.elifesciences.org/articles/80332/elife-80332-fig4-data1-v2.zip
-
Figure 4—source data 2
Raw gel image for Figure 4, panel A.
- https://cdn.elifesciences.org/articles/80332/elife-80332-fig4-data2-v2.zip
-
Figure 4—source data 3
Raw gel image for Figure 4, panel B.
Same as Figure 2—source data 5.
- https://cdn.elifesciences.org/articles/80332/elife-80332-fig4-data3-v2.zip
-
Figure 4—source data 4
Raw gel image for Figure 4, panel B.
- https://cdn.elifesciences.org/articles/80332/elife-80332-fig4-data4-v2.zip
-
Figure 4—source data 5
Raw gel image for Figure 4, panel C; same as Figure 2—source data 5.
- https://cdn.elifesciences.org/articles/80332/elife-80332-fig4-data5-v2.zip
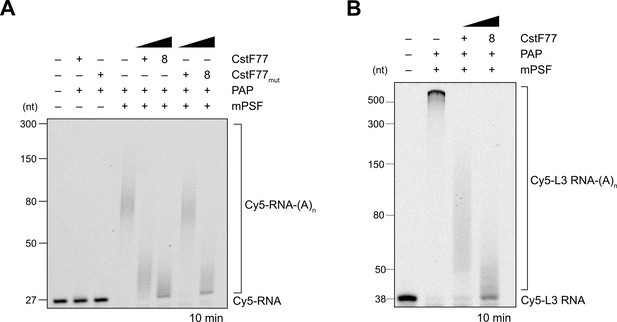
CstF77 reduces RNA 3′ polyadenylation rate.
(A) Polyadenylation activity assay of mPSF in the presence of wild-type CstF77 or a CstF77 mutant (CstFmut) incapable of binding hFip1 at mPSF:CstF77 molar ratios of 1:1 and 1:8. Polyadenylated RNA is indicated as RNA-(A)n. (B) Polyadenylation activity assay of mPSF with a 38-nt L3 RNA substrate, in the presence of wild-type CstF77 (1:1 and 1:8 molar ratios).
-
Figure 4—figure supplement 1—source data 1
Raw gel image for Figure 4—figure supplement 1, panel A.
Same as Figure 2—source data 5.
- https://cdn.elifesciences.org/articles/80332/elife-80332-fig4-figsupp1-data1-v2.zip
-
Figure 4—figure supplement 1—source data 2
Raw gel image for Figure 4—figure supplement 1, panel A.
- https://cdn.elifesciences.org/articles/80332/elife-80332-fig4-figsupp1-data2-v2.zip
-
Figure 4—figure supplement 1—source data 3
Raw gel image for Figure 4—figure supplement 1, panel B.
- https://cdn.elifesciences.org/articles/80332/elife-80332-fig4-figsupp1-data3-v2.zip
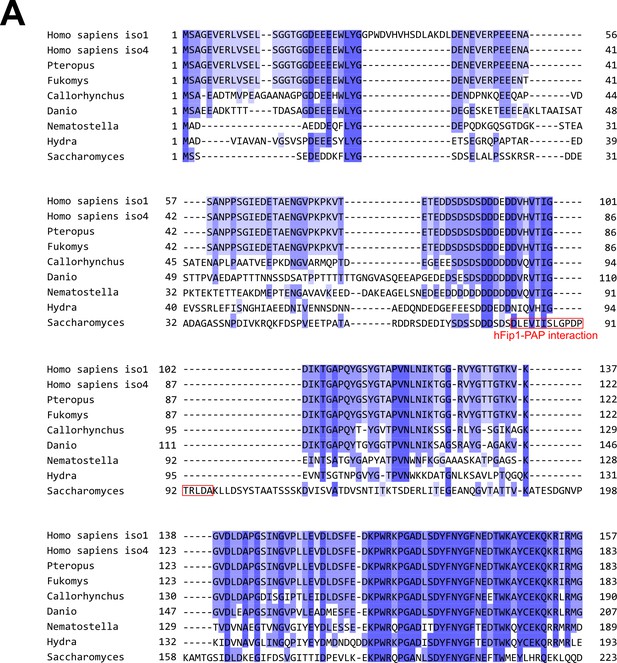
Pap1 interaction motif in Fip1 orthologs is poorly conserved.
(A) Multiple sequence alignment of hFip1 isoform 1, hFip1 isoform 4, and selected orthologs, colored by sequence similarity. Residues of yeast Fip1 that directly interact with Pap1 (Meinke et al., 2008, PDB ID: 3C66) are indicated in red.
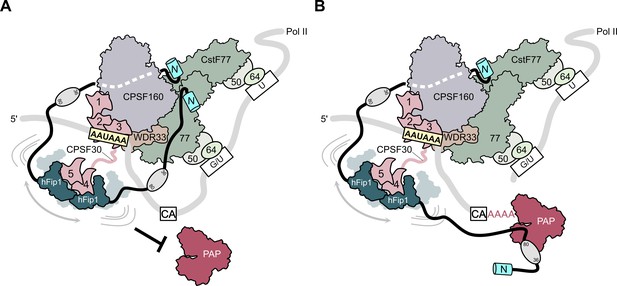
Model of CPSF-mediated pre-mRNA cleavage and polyadenylation and CstF77-dependent inhibition of polyadenylation.
(A) Prior to pre-mRNA cleavage, PAP recruitment is inhibited by CStF, in part due to competitive interactions of CstF77 and hFip1 (left). (B) Upon pre-mRNA cleavage, structural remodeling of the CPSF–CstF complex enables hFip1 to recruit PAP to the nascent 3′ end of the mRNA and consequently stimulates polyadenylation. Figure 1—figure supplement 1: sequence alignment of CPSF30 zinc finger domains. (A) Sequence alignment of human CPSF30 zinc finger domains. Residues responsible for RNA interactions (in ZF2/ZF3) or hFip1 interaction (in ZF4/ZF5) are highlighted and the nature of their interaction color coded. ZF4/ZF5 domains contain proline residues (yellow) at positions corresponding to critical main chain hydrogen bonding interactions in ZF2/ZF3.
Tables
Crystallographic data collection and refinement statistics.
| hFip1–CPSF30 | hFip1–CstF77 | |
|---|---|---|
| Data collection | ||
| Space group | P21 | P6122 |
| Cell dimensions | ||
| a, b, c (Å) | 60.127, 115.125, 66.444 | 157.612, 157.612, 161.005 |
| α, β, γ (°) | 90, 116.781, 90 | 90, 90, 120 |
| Wavelength (Å) | 1.2809 | 1.0000 |
| Resolution (Å) | 48.65–2.201 (2.28–2.201) | 56.31–2.55 (2.641–2.55) |
| Total reflections | 226,720 (15,294) | 1,577,004 (162,259) |
| Unique reflections | 37,698 (3244) | 38,981 (3836) |
| Rmerge (%) | 7.5 (95.9) | 9.2 (186.1) |
| Rpim (%) | 3.2 (46.9) | 1.5 (28.8) |
| I/σI | 13.5 (1.1) | 36.0 (2.6) |
| Cc(1/2) | 0.998 (0.557) | 1 (0.836) |
| Completeness (%) | 92.3 (80.22) | 99.96 (100.00) |
| Redundancy | 6.0 (4.7) | 40.5 (42.3) |
| Refinement | ||
| Resolution (Å) | 48.65–2.201 | 56.31–2.55 |
| No. reflections | 37,698 | 38,975 |
| Rwork / Rfree | 0.2406/0.2622 | 0.2410/0.2647 |
| No. non-hydrogen atoms | ||
| Protein | 4607 | 5188 |
| Ligand/ion | 8 | 98 |
| Water | 67 | 25 |
| B-factors (Å2) | ||
| Protein | 56.53 | 65.34 |
| Ligand/ion | 63.69 | 69.46 |
| Water | 49.83 | 55.9 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.008 | 0.009 |
| Bond angles (°) | 1.03 | 1.1 |
| Ramachandran plot | ||
| % favored | 95.83 | 97.9 |
| % allowed | 4.17 | 2.1 |
| % outliers | 0 | 0 |
-
Values in parentheses are for highest resolution shell.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | BL21 star (DE3) | Thermo Fisher scientific | BL21 star (DE3) | Chemically competent cells |
| Strain, strain background (Escherichia coli) | BL21(DE3)-AI | Thermo Fisher scientific | BL21(DE3)-AI | Chemically competent cells |
| Strain, strain background (Escherichia coli) | Rosetta2 (DE3) | Novagen | Rosetta2 (DE3) | Chemically competent cells |
| Cell line (Spodoptera frugiperda) | Sf9 | Thermo Fisher Scientific | Cat. #11496015 | |
| Recombinant DNA reagent | pLM B042; pLM B043 (plasmid) | This paper | Holo-CstF | |
| Recombinant DNA reagent | pLM B092 (plasmid) | This paper | MBP-CstF77 | |
| Recombinant DNA reagent | pLM B123 (plasmid) | This paper | CstF77 | |
| Recombinant DNA reagent | pLM B142 (plasmid) | This paper | PAP | |
| Recombinant DNA reagent | pLM B156 (plasmid) | This paper | MBP-PAP | |
| Recombinant DNA reagent | pLM B157 (plasmid) | This paper | GFP-PAP | |
| Recombinant DNA reagent | pLM B164 (plasmid) | This paper | MBP-CstF7721–549 or MBP-CstF77 | |
| Recombinant DNA reagent | pLM B168 (plasmid) | This paper | MBP-CstF77mut | |
| Recombinant DNA reagent | pLM B170 (plasmid) | This paper | CstF77mut | |
| Recombinant DNA reagent | pMC B051 (plasmid) | This paper | CPSF30ZF4–ZF5 | |
| Recombinant DNA reagent | pMC B054 (plasmid) | This paper | MBP-CPSF30ZF4–ZF5 | |
| Recombinant DNA reagent | pMC B055 (plasmid) | This paper | CPSF30 ZF4 mutant | |
| Recombinant DNA reagent | pMC B056 (plasmid) | This paper | CPSF30 ZF5 mutant | |
| Recombinant DNA reagent | pMC B057 (plasmid) | This paper | CPSF30 ZF4 and ZF5 mutant | |
| Recombinant DNA reagent | pMC B058 (plasmid) | This paper | CPSF30 ZF4 mutant | |
| Recombinant DNA reagent | pMC B059 (plasmid) | This paper | CPSF30 ZF4 mutant | |
| Recombinant DNA reagent | pMC B060 (plasmid) | This paper | CPSF30 ZF5 mutant | |
| Recombinant DNA reagent | pMC B061 (plasmid) | This paper | CPSF30 ZF5 mutant | |
| Recombinant DNA reagent | pMC B062 (plasmid) | This paper | CPSF30 ZF4 and ZF5 mutant | |
| Recombinant DNA reagent | pMC B063 (plasmid) | This paper | CPSF30 ZF4 and ZF5 mutant | |
| Recombinant DNA reagent | pMC C011 (plasmid) | This paper | hFip1CD | |
| Recombinant DNA reagent | pMC C015 (plasmid) | This paper | GST-hFip1 fragment or hFip180–195 | |
| Recombinant DNA reagent | pMC C030 (plasmid) | This paper | hFip1CD | |
| Recombinant DNA reagent | pMC C049 (plasmid) | This paper | GFP-hFip1 | |
| Recombinant DNA reagent | pMC C050 (plasmid) | This paper | GST-hFip1 fragment or hFip136–80 | |
| Recombinant DNA reagent | pMC C059 (plasmid) | This paper | GST-hFip1 fragment, GST-hFip11–35, or hFip11–35 | |
| Recombinant DNA reagent | pMC C060 (plasmid) | This paper | GST-hFip1 fragment, GST-hFip11–195, or hFip11–195 | |
| Recombinant DNA reagent | pMC C066 (plasmid) | This paper | His6-GFP-TEV-hFip11–195 point mutant | |
| Recombinant DNA reagent | pMC C067 (plasmid) | This paper | His6-GFP-TEV-hFip11–195 point mutant | |
| Recombinant DNA reagent | pMC C068 (plasmid) | This paper | His6-GFP-TEV-hFip11–195 point mutant | |
| Recombinant DNA reagent | pMC C073 (plasmid) | This paper | GST-hFip1 fragment, GST-hFip136–195, or hFip136–195 | |
| Recombinant DNA reagent | pMC C093 (plasmid) | This paper | mutant GST-hFip11–35 | |
| Recombinant DNA reagent | pMC C094 (plasmid) | This paper | mutant GST-hFip11–35 | |
| Recombinant DNA reagent | pMC C096 (plasmid) | This paper | mutant GST-hFip11–35 | |
| Recombinant DNA reagent | pMC N015 (plasmid) | This paper | mPSF | |
| Recombinant DNA reagent | pMC N018 (plasmid) | This paper | mPSF | |
| Recombinant DNA reagent | pMC N018A (plasmid) | This paper | mPSF | |
| Recombinant DNA reagent | pMC N018C-2 (plasmid) | This paper | mPSF | |
| Recombinant DNA reagent | pMC N018G (plasmid) | This paper | mPSF | |
| Recombinant DNA reagent | pMC N018G-0 (plasmid) | This paper | mPSF | |
| Recombinant DNA reagent | pMC N018G-8 (plasmid) | This paper | mPSF | |
| Recombinant DNA reagent | pMC N018G-10 (plasmid) | This paper | mPSF | |
| Recombinant DNA reagent | pMC N018G-12 (plasmid) | This paper | mPSF | |
| Recombinant DNA reagent | pMC N018G-14 (plasmid) | This paper | mPSF | |
| Recombinant DNA reagent | pMC N018G-15 (plasmid) | This paper | mPSF | |
| Recombinant DNA reagent | pMC N018G-21 (plasmid) | This paper | FLAG-epitope-tagged mPSF | |
| Recombinant DNA reagent | pMC N018G-22 (plasmid) | This paper | FLAG-epitope-tagged mPSF | |
| Recombinant DNA reagent | pMC N018G-23 (plasmid) | This paper | FLAG-epitope-tagged mPSF | |
| Recombinant DNA reagent | pMC N018G-24 (plasmid) | This paper | FLAG-epitope-tagged mPSF | |
| Recombinant DNA reagent | pMC N018H (plasmid) | This paper | mPSF | |
| Recombinant DNA reagent | pMC N018I (plasmid) | This paper | mPSF | |
| Recombinant DNA reagent | pMC N018J (plasmid) | This paper | mPSF | |
| Recombinant DNA reagent | pMC N018K (plasmid) | This paper | mPSF | |
| Sequence-based reagent | rLM 011 | This paper | 27 nt RNA substrate based on SV40 pre-mRNA; CUGCAAUAAACAACUUAACAACAAAAA | |
| Sequence-based reagent | rLM 015 | This paper | 5′ Cy5-labeled 27 nt RNA substrate based on SV40 pre-mRNA; CUGCAAUAAACAACUUAACGUCAAAAA | |
| Sequence-based reagent | rLM 016 | This paper | 5′ Cy5-labeled 27 nt RNA substrate based on SV40 pre-mRNA; CUGCAGUACACAACUUAACGUCAAAAA | |
| Sequence-based reagent | rLM 031 | This paper | 5′ Cy5-labeled 38 nt RNA substrate based on adenoviral L3 pre-mRNA; ACUUUCAAUAAAGGCAAAUGUUUUUAUUUGUACAAAAA |
Quantitation of co-precipitated GFP-hFip1 and GFP-PAP protein levels (GFP fluorescence detected at 473 nm), normalized against WT mPSF.
| wt | ZF4 | ZF5 | |
|---|---|---|---|
| GFP-hFip | 1 | 0.29 | 0.42 |
| GFP-PAP | 1 | 0.28 | 0.43 |
Additional files
-
Supplementary file 1
List of protein expression constructs used in this study.
- https://cdn.elifesciences.org/articles/80332/elife-80332-supp1-v2.docx
-
Supplementary file 2
Sequences of RNA oligonucleotides used in this study.
- https://cdn.elifesciences.org/articles/80332/elife-80332-supp2-v2.docx
-
Supplementary file 3
hFip1 ortholog input sequences used for multiple sequence alignment.
- https://cdn.elifesciences.org/articles/80332/elife-80332-supp3-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/80332/elife-80332-mdarchecklist1-v2.pdf