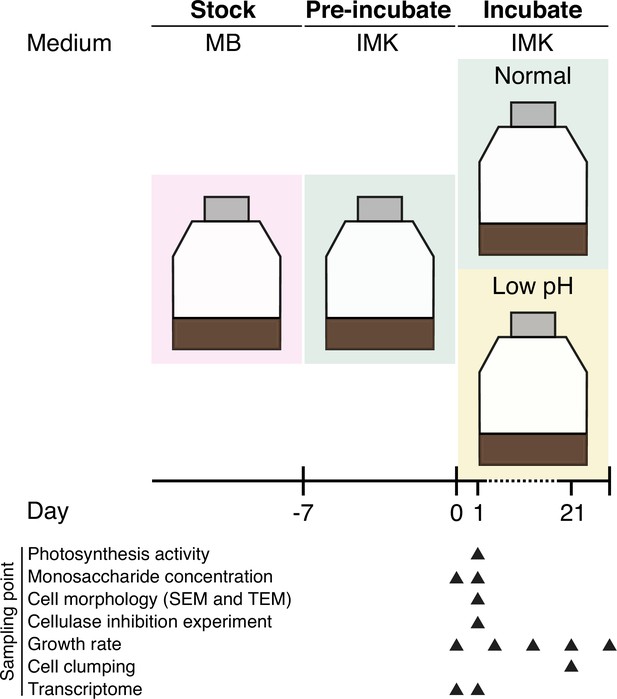
Environmental pH signals the release of monosaccharides from cell wall in coral symbiotic alga
Figures
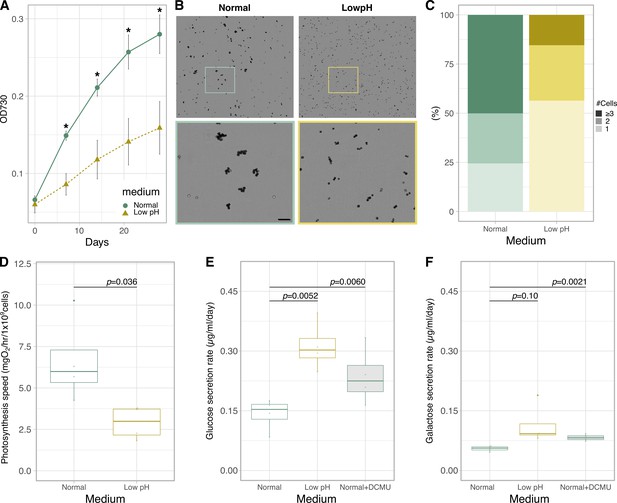
Physiological characterization and monosaccharide secretion of cultured Breviolum.
(A) Growth rate (n = 6 per treatment, t-test). Asterisks indicate statistically significant differences (t-test, p < 0.005). (B) Bright field images of the cells under different conditions. The lower panels show high-magnification views of boxed areas in the upper panels. Scale bar = 50 μm. (C) Quantification of the number of cells forming clusters (Fisher’s exact test for “1 or 2” vs “3 or more”, p = 1.727 × 10-7). (D) Photosynthesis activity (n = 4 per treatment, t-test) (E, F) Quantification of glucose (E) and galactose (F) secreted in normal, low pH and normal+DCMU media during incubation for 1 day using ion chromatography (n = 4 per treatment, t-test).
-
Figure 1—source data 1
Raw data of growth rates.
- https://cdn.elifesciences.org/articles/80628/elife-80628-fig1-data1-v1.txt
-
Figure 1—source data 2
Raw data of photosynthesis activities.
- https://cdn.elifesciences.org/articles/80628/elife-80628-fig1-data2-v1.txt
-
Figure 1—source data 3
Raw data of glucose and galactose concentrations.
- https://cdn.elifesciences.org/articles/80628/elife-80628-fig1-data3-v1.txt
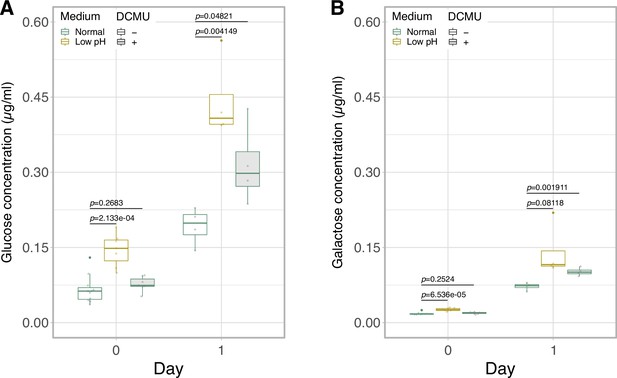
Effect of photosynthesis inhibitor on glucose and galactose secretion.
The quantification of glucose (A) and galactose (B) in the media of the Breviolum cultures on 0- and 1-day incubation using ion chromatography (n = 4-11 per treatment, t-test).
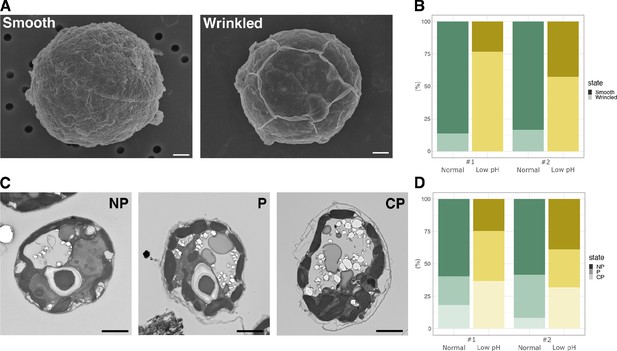
Cell structures under different pH condition.
(A) SEM images of the representative cells. Scale bar = 1 μm. (B) Quantification of the cell surface structures of the SEM images (Fisher’s exact test, #1; p < 2.2 × 10-16, #2; p < 2.2 × 10-16). (C) TEM images of the representative cells. NP, ‘non-peeled’ where the outer struc- ture of the cell wall is not shed from the cell surface; P, ‘peeled’ at some parts of the cell surface; CP, ‘completely peeled’. Scale bar = 2 μm. (D) Quantification of the cell surface structures of the TEM images (Fisher’s exact test for “P or CP” vs “NP”, #1; p = 4.621 × 10-11, #2; p = 3.525 × 10-4).
-
Figure 2—source data 1
Raw data of bright field cell counts.
- https://cdn.elifesciences.org/articles/80628/elife-80628-fig2-data1-v1.txt
-
Figure 2—source data 2
Raw data of SEM cell counts.
- https://cdn.elifesciences.org/articles/80628/elife-80628-fig2-data2-v1.txt
-
Figure 2—source data 3
Raw data of TEM cell counts.
- https://cdn.elifesciences.org/articles/80628/elife-80628-fig2-data3-v1.txt
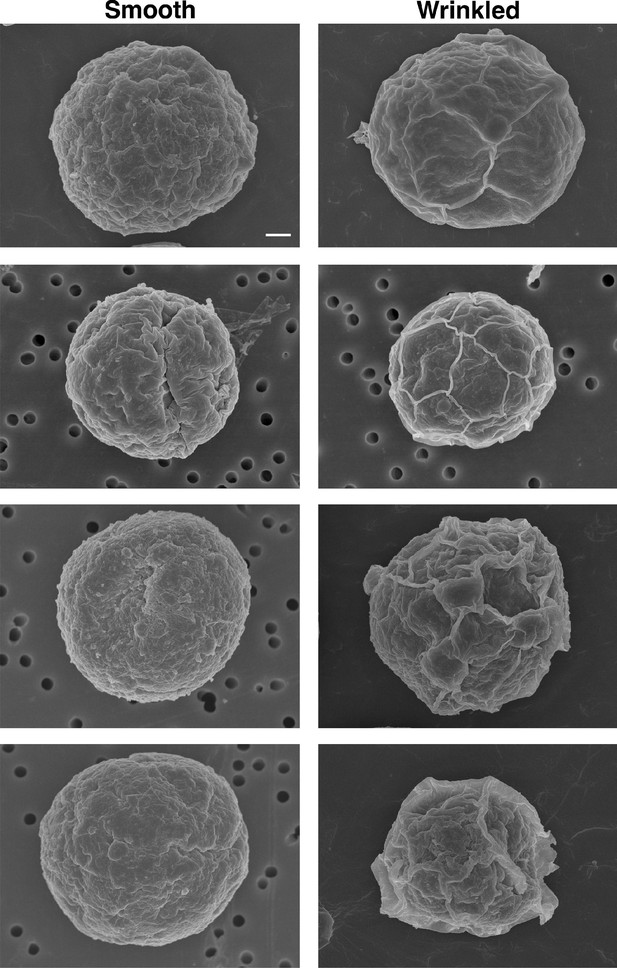
SEM images of cell surface structures.
Representative examples of Smooth and Wrinkled. Scale bar = 1 μm.
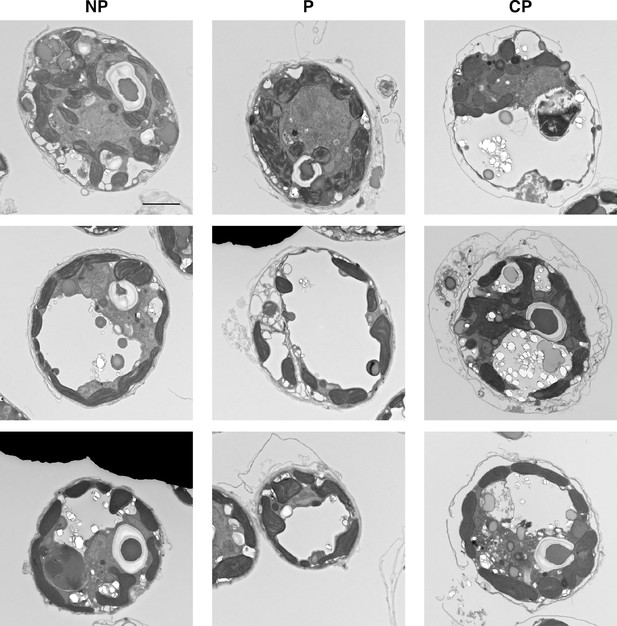
TEM images of cell surface structures.
Representative examples of NP (non-peeled), P (peeled) and CP (completely peeled). Scale bar = 2 μm.
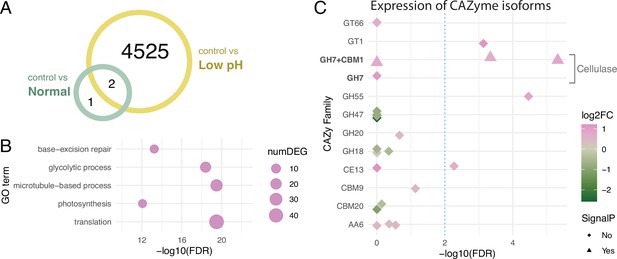
Differentially expressed genes under different pH conditions.
(A) Venn diagram showing the numbers of DEGs under different conditions. (B) GO term enrichment analysis. Circles indicate the statistical significance (FDR) of the enriched GO terms, with the numbers of DEGs (numDEG) associated with each GO term. (C) Isoform-level expression analysis of genes encoding Carbohydrate-Active enZymes (CAZymes). Symbols indicate isoforms associated with the DEGs, with the presence (triangle) or absence (rhombus) of signal peptide predicted in the amino acid sequence. Symbol colors represent the log2 fold-changes (log2FC) of the expression levels of each isoform (low pH/control). Dashed line indicates a threshold for differential expression (FDR = 0.01).
-
Figure 3—source data 1
Expression levels of the annotated low pH DEGs.
- https://cdn.elifesciences.org/articles/80628/elife-80628-fig3-data1-v1.txt
-
Figure 3—source data 2
GO enrichment analysis results.
- https://cdn.elifesciences.org/articles/80628/elife-80628-fig3-data2-v1.xlsx
-
Figure 3—source data 3
CAZome analysis results.
- https://cdn.elifesciences.org/articles/80628/elife-80628-fig3-data3-v1.xlsx
-
Figure 3—source data 4
Multiple alignment of the cellulase proteins.
- https://cdn.elifesciences.org/articles/80628/elife-80628-fig3-data4-v1.txt
-
Figure 3—source data 5
Newick format file for phylogenetic tree of the cellulase proteins.
- https://cdn.elifesciences.org/articles/80628/elife-80628-fig3-data5-v1.txt

Mapping of DEGs between the low pH and control groups on KEGG pathways.
The KEGG pathways for glycolysis/gluconeogenesis are shown. The colours indicate gene expression levels.

Phylogenetic tree of the cellulase proteins.
A maximum likelihood tree of cellulase proteins labeled with species names and GenBank IDs is shown. Support values using an SH-like approximate likelihood ratio test (left) and ultrafast boot strap approximation (right) are shown for each branch. For Breviolum sp. SSB01, the Trinity assembly contig IDs are shown.
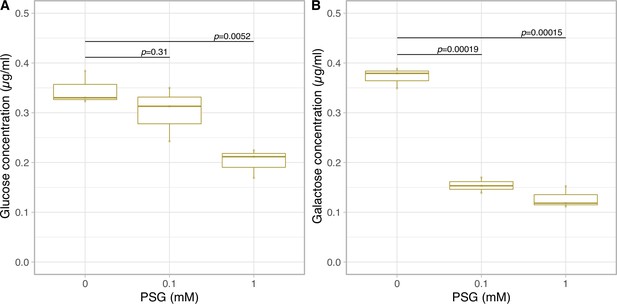
Glucose and galactose secretion in cellulase inhibitor treatment.
The quantification of glucose (A) and galactose (B) in the medium on 1 day incubation with PSG using LC-MS/MS (n = 3 per treatment, t-test).
-
Figure 4—source data 1
Raw data of glucose and galactose concentrations with PSG.
- https://cdn.elifesciences.org/articles/80628/elife-80628-fig4-data1-v1.txt
-
Figure 4—source data 2
Raw data of cellulase activity in vitro.
- https://cdn.elifesciences.org/articles/80628/elife-80628-fig4-data2-v1.txt