BRCA1/BRC-1 and SMC-5/6 regulate DNA repair pathway engagement during Caenorhabditis elegans meiosis
Figures
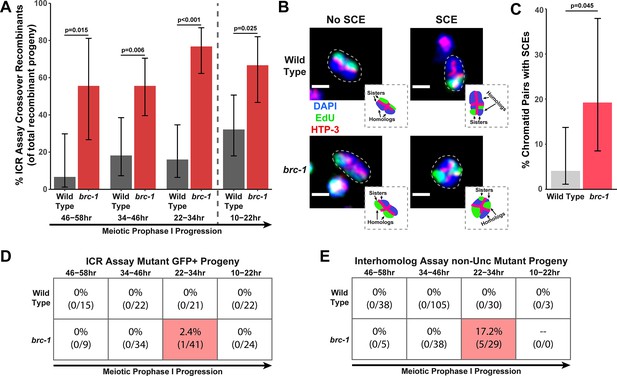
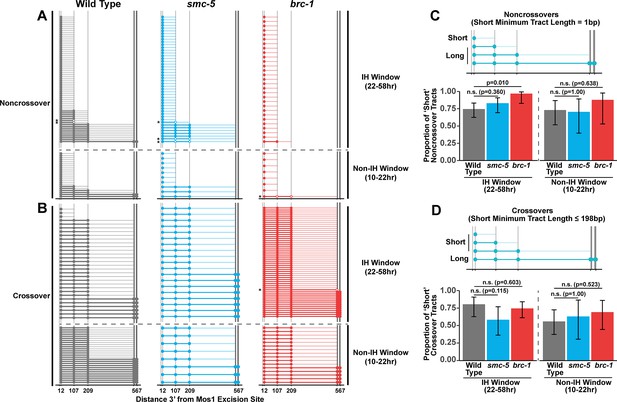
BRC-1 represses intersister crossovers and error-prone repair.
(A) Bar plot displaying the percent of crossover recombinant progeny identified in wild type and brc-1 ICR assays out of all recombinant progeny scored. Frequencies of recombinants identified overall in ICR assays is displayed in Figure 1—figure supplement 2A. Wild type ICR assay data is shared in A and Figures 2A, B, 4A, Figure 1—figure supplement 2, Figure 2—figure supplement 1, and Figure 4—figure supplement 1. brc-1 ICR assay data is shared in A and Figure 4A, Figure 1—figure supplement 2, and Figure 4—figure supplement 1. The total number of recombinant progeny scored in each dataset are (10–22 hr/22–34 hr/34–46 hr/46–58 hr timepoints) wild type: 28/25/22/15, brc-1(xoe4) 24/43/36/9. Specific progeny counts separated by experimental replicate are presented in Figure 1—source data 1. (B) Images of wild type and brc-1(xoe4) mutant bivalent chromosomes displaying an absence or presence of sister chromatid exchanges (SCEs). Scale bars represent 1 μm. Dashed bordered insets contain cartoon depictions of the SCE and non-SCE bivalents that are outlined with dashed lines in the images to aid in visualizing exchange events. (C) Frequency of SCEs identified among wild type (n=49) or brc-1 mutant (n=26) chromatid pairs scored. (D–E) Tables displaying the percent of sequenced GFP+ progeny in wild type and brc-1 ICR assays (D) or non-Unc progeny IH assays (E) that showed signatures of mutagenic repair. Numbers in parentheses indicate the number of mutant worms out of the total number of sequenced progeny. Shaded boxes indicate timepoints in which mutant progeny were identified. The overall frequency of interhomolog assay non-Unc progeny is displayed in Figure 1—figure supplement 3. In panels (D) and (E), note that only mutations that allow for translation of functional GFP (panel D) or UNC-5 (panel E) protein can be detected (see Methods). In all panels, error bars represent 95% Binomial confidence intervals, dashed vertical lines delineate between timepoints within the interhomolog window (22–58 hr post heat shock) and non-interhomolog window (10–22 hr post heat shock), and p values were calculated using Fisher’s Exact Test.
-
Figure 1—source data 1
The source data for Figure 1A is provided.
The total number of ICR assay progeny with GFP+ or non-GFP+ phenotypes are listed. Wild type data is shared with Figure 1—figure supplement 2, Figure 2, Figure 2—figure supplement 1, Figure 4, and Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/80687/elife-80687-fig1-data1-v2.xlsx
-
Figure 1—source data 2
The source data for Figure 1C is provided.
The number of scorable chromatid pairs with SCE or no SCE events (no_SCE) are listed for each image assessed in generating this dataset. Wild type data is shared with Figure 2.
- https://cdn.elifesciences.org/articles/80687/elife-80687-fig1-data2-v2.xlsx
-
Figure 1—source data 3
The source data for Figure 1E is provided.
The total number of IH assay progeny with recombinant or mutant nonUnc phenotypes or Unc nonrecombinant phenotypes are listed. Wild type data is shared with Figure 1—figure supplement 3, Figure 2, and Figure 2—figure supplement 2.
- https://cdn.elifesciences.org/articles/80687/elife-80687-fig1-data3-v2.xlsx
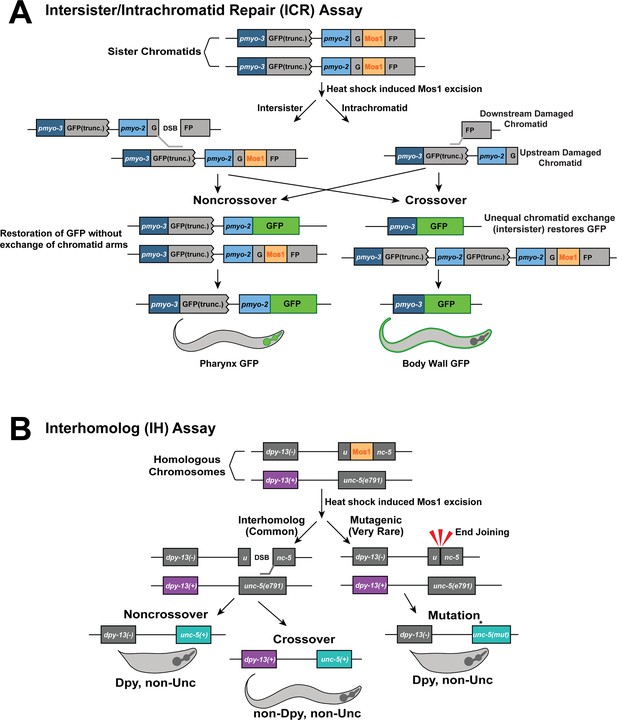
Diagram of the Intersister/Intrachromatid and Interhomolog assays.
Cartoon depictions of how noncrossover and crossover events can be generated in the Intersister/Intrachromatid Repair assay (A) (Toraason et al., 2021c) and Interhomolog assay (B) (Rosu et al., 2011). Panel A is adapted from Toraason et al., 2021c.
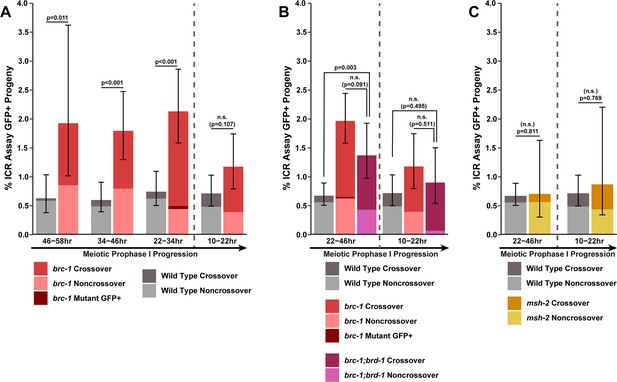
Intersister/intrachromatid repair (ICR) assay GFP+ progeny are elevated in brc-1 and brc-1;brd-1 mutants.
Stacked bar plots displaying the percent of all progeny scored in wild type and brc-1(xoe4) (A), brc-1(tm1145);brd-1(dw1) (B), and msh-2(ok2410) (C) ICR assays that were determined to be GFP+ noncrossover recombinants, crossover recombinants, or mutants. Error bars represent the 95% Binomial confidence intervals for the frequencies of GFP+ progeny. Wild type ICR assay data is shared in Figures 1A, 2A, B, 4A, Figure 1—figure supplement 2, Figure 2—figure supplement 1, and Figure 4—figure supplement 1. brc-1 ICR assay data is shared in Figures 1A and 4A, Figure 1—figure supplement 1, and Figure 4—figure supplement 1. The total number of progeny scored in each dataset are (10–22 hr/22–34 hr/34–46 hr/46–58 hr timepoints) wild type: 3921/3359/3664/2383, brc-1(xoe4) 2042/2017/2005/467, (10–22 hr/22–46 hr) brc-1(tm1145);brd-1(dw1) 1562/2340, msh-2(ok2410) 460/711. Specific progeny counts separated by experimental replicate are presented in Figure 1—source data 1. p values were calculated by Fisher’s Exact Test comparing the sum totals of GFP+ progeny between groups. Vertical dashed lines demarcate the interhomolog window (≥22 hr post heat shock) and non-interhomolog window (10–22 hr post heat shock) timepoints.
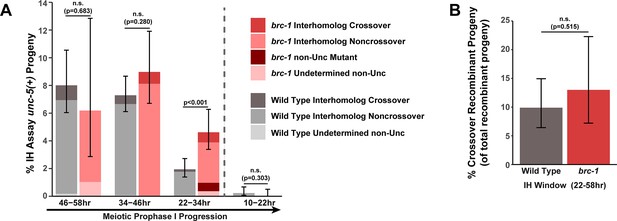
Interhomolog repair is largely unperturbed in brc-1 mutants.
(A) Stacked bar plot displaying the percent of all progeny scored in wild type and brc-1 IH assays that were determined to be noncrossover recombinants, crossover recombinants, non-Unc mutants, or undetermined non-Unc. The vertical dashed line demarcates the interhomolog window (22–58 hr post heat shock) and non-interhomolog window (10–22 hr post heat shock) timepoints. (B) Percent of all recombinant progeny identified within the interhomolog window of wild type and brc-1 IH assays that were crossover recombinants. Error bars represent the 95% Binomial confidence intervals for the frequencies of non-Unc progeny. Wild type data is shared between Figures 1E and 2E, Figure 1—figure supplement 3, and Figure 2—figure supplement 2. The total number of progeny scored in each dataset are (10–22 hr/22–34 hr/34–46 hr/46–58 hr timepoints) wild type: 1308/1695/1592/562, brc-1 758/823/468/97. Specific progeny counts separated by experimental replicate are presented in Figure 1—source data 3. p values were calculated by Fisher’s Exact test comparing the overall frequency of non-Unc progeny (A) or proportion of crossover recombinants of all confirmed recombinant progeny (B, see Materials and Methods).
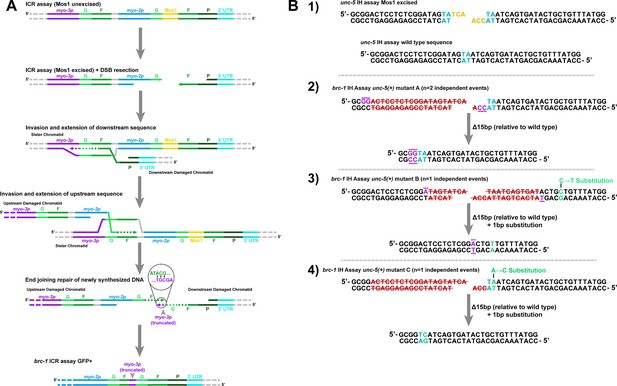
Illustrations of mutants identified in brc-1 ICR and IH assays.
(A) Illustrated depiction of ICR assay GFP+ mutant identified in a brc-1 mutant background (Figure 1D, Figure 1—figure supplement 2). The partial tandem duplication produced (bottom) can best be parsimoniously explained by two independent strand invasion and extension events on either end of the DSB. For simplicity, intersister recombination is depicted in this diagram. However, intrachromatid templates could also have been engaged to produce the final product. (B) Illustrations of unc-5 lesions identified in IH assay non-Unc progeny in brc-1 mutants (Figure 1E, Figure 1—figure supplement 3). Specific mutation signatures are separated by horizontal dashed grey lines. The wild type unc-5 locus sequence at the site of Mos1 excision and the DSB product generated by Mos1 excision are displayed on the top of panel (B). Blue letters indicate a duplicated TA at the site of Mos1 insertion in the unc-5(ox171) locus, while yellow letters indicate the 3 nt 3’ overhangs left following Mos1 excision (Robert et al., 2008). In the panels displaying mutations identified, purple letters with bars indicate complementary bases flanking the deletion site. Red letters struck through with red lines indicate bases in the damaged locus that were deleted to produce the final product. Green letters indicate sites of nucleotide substitution mutations.
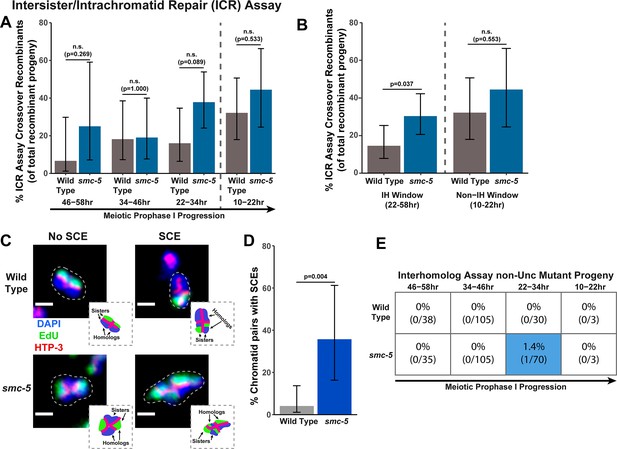
SMC-5 represses intersister crossovers.
(A) Bar plot displaying the percent of crossover recombinant progeny identified in wild type and smc-5 ICR assays out of all recombinant progeny scored within individual 12 hr timepoint periods. Frequencies of recombinants identified overall in ICR assays is displayed in Figure 2—figure supplement 1. Wild type ICR assay data is shared in Figures 1A, 2A, B, 4A, Figure 1—figure supplement 2, Figure 2—figure supplement 1, and Figure 4—figure supplement 1. The total number of progeny scored in each dataset are (10–22 hr/22–34 hr/34–46 hr/46–58 hr timepoints) wild type: 28/25/22/15, smc-5(ok2421) 18/37/21/8. Specific progeny counts separated by experimental replicate are presented in Figure 2—source data 1. (B) Bar plot displaying the percent of crossover recombinant progeny identified in wild type and smc-5 ICR assays out of all recombinant progeny scored within the interhomolog window (22–58 hr post heat shock) and non-interhomolog window (10–22 hr post heat shock). Data is shared with panel (A). (C) Images of wild type and smc-5(ok2421) mutant bivalent chromosomes displaying an absence or presence of SCEs. Scale bars represent 1 μm. Dashed bordered insets contain cartoon depictions of the SCE and non-SCE bivalents that are outlined with dashed lines in the images to aid in visualizing exchange events. (D) Frequency of SCEs identified among wild type (n=49) or smc-5(ok2421) mutant (n=14) chromatid pairs scored. (E) Table displaying the percent of sequenced non-Unc progeny in wild type and smc-5 IH assays that showed signatures of mutagenic repair. Numbers in parentheses indicate the number of mutant worms out of the total number of sequenced progeny. Colored boxes indicate timepoints in which mutant progeny were identified. The overall frequency of interhomolog assay non-Unc progeny is displayed in Figure 2—figure supplement 2. Note that only mutations that allow for translation of functional UNC-5 protein can be detected (see Methods). In all panels, error bars represent 95% Binomial confidence intervals, dashed vertical lines delineate between timepoints within the interhomolog window (22–58 hr post heat shock) and non-interhomolog window (10–22 hr post heat shock), and p values were calculated using Fisher’s Exact Test.
-
Figure 2—source data 1
The source data for Figure 2A is provided.
The total number of ICR assay progeny with GFP+ or non-GFP+ phenotypes are listed. Wild type data is shared with Figure 1, Figure 1—figure supplement 2, Figure 2, Figure 2—figure supplement 1, Figure 4, and Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/80687/elife-80687-fig2-data1-v2.xlsx
-
Figure 2—source data 2
The source data for Figure 2D is provided.
The number of scorable chromatid pairs with SCE or no SCE events are listed for each image assessed in generating this dataset. Wild type data is shared with Figure 1C.
- https://cdn.elifesciences.org/articles/80687/elife-80687-fig2-data2-v2.xlsx
-
Figure 2—source data 3
The source data for Figure 2E is provided.
The total number of IH assay nonrecombinant, recombinant, mutant, or undetermined non-Unc progeny are listed. Wild type data is shared with Figure 1, Figure 1—figure supplement 3, and Figure 2—figure supplement 2.
- https://cdn.elifesciences.org/articles/80687/elife-80687-fig2-data3-v2.xlsx
Intersister/intrachromatid repair (ICR) assay GFP+ progeny are elevated in smc-5 mutants.
Stacked bar plots displaying the percent of all progeny scored in wild type and smc-5(ok2421) (A) or smc-6(ok3294) (B) ICR assays that were determined to be GFP+ noncrossover or crossover recombinants. Error bars represent the 95% Binomial confidence intervals for the frequencies of GFP+ progeny. Wild type ICR assay data is shared in Figures 1A, 2A, B, 4A, Figure 1—figure supplement 2, Figure 2—figure supplement 1, and Figure 4—figure supplement 1. smc-5 ICR assay data is shared in Figure 2 and Figure 2—figure supplement 1. The total number of progeny scored in each dataset are (10–22 hr/22–34 hr/34–46 hr/46–58 hr timepoints) wild type: 3921/3359/3664/2383, smc-5(ok2421) 1350/1771/1450/449, smc-6(ok3294) (10–22 hr/22–46 hr) 1855/2129. Specific progeny counts separated by experimental replicate are presented in Figure 2—source data 1. p values were calculated by Fisher’s Exact test comparing the sum totals of GFP+ progeny between groups. Vertical dashed lines demarcate the interhomolog window (≥22 hr post heat shock) and non-interhomolog window (10–22 hr post heat shock) timepoints.
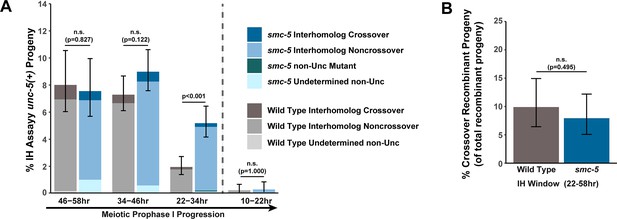
Interhomolog repair is largely unperturbed in smc-5 mutants.
(A) Stacked bar plots displaying the percent of all progeny scored in wild type and smc-5 IH assays that were determined to be noncrossover recombinants, crossover recombinants, non-Unc mutants, or undetermined non-Unc. (B) Percent of all recombinant progeny identified within the interhomolog window of wild type and smc-5 IH assays that were crossover recombinants. Error bars represent the 95% Binomial confidence intervals for the frequencies of non-Unc progeny. Wild type data is shared between Figures 1E and 2E, Figure 1—figure supplement 3, and Figure 2—figure supplement 2. The total number of progeny scored in each dataset are (10–22 hr/22–34 hr/34–46 hr/46–58 hr timepoints) wild type: 1308/1695/1592/562, smc-5 1031/1466/1369/596. Specific progeny counts separated by experimental replicate are presented in Figure 2—source data 3. p values were calculated by Fisher’s Exact Test comparing the overall frequency of non-Unc progeny (A) or proportion of crossover recombinants of all confirmed recombinant progeny (B, see Materials and methods). Vertical dashed lines demarcate the interhomolog window (22–58 hr post heat shock) and non-interhomolog window (10–22 hr post heat shock) timepoints.
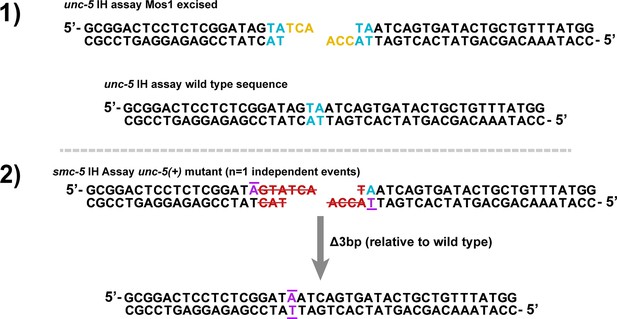
Illustrations of mutants identified in smc-5 IH assays.
Illustrations of unc-5 lesions identified in IH assay non-Unc progeny in smc-5 mutants (Figure 2, Figure 2 - figure supplement 2). Specific mutation signatures are numbered and are separated by horizontal dashed grey lines. The wild type unc-5 locus sequence at the site of Mos1 excision and the DSB product generated by Mos1 excision are displayed on the top (Numbered 1). Blue letters indicate a duplicated TA at the site of Mos1 insertion in the unc-5(ox171) locus, while yellow letters indicate the 3 nt 3’ overhangs left following Mos1 excision (Robert et al., 2008). Purple letters with bars indicate complementary bases flanking the deletion site. Red letters struck through with red lines indicate bases in the damaged locus which were deleted to produce the final product.
BRC-1 is required for long noncrossover gene conversion.
(A–B) Plots of conversion tracts sequenced from recombinant ICR assay loci. Vertical grey lines indicate the positions of polymorphisms in the ICR assay with bp measurements given 3’ relative to the site of Mos1 excision (Toraason et al., 2021b; Toraason et al., 2021a;). Each horizontal line represents a single recombinant sequenced, ordered from smallest tract to largest tract within the interhomolog and non-interhomolog windows. Filled in points represent fully converted polymorphisms, while points with white interiors represent heteroduplex DNA sequences identified in conversion tracts. Tracts containing heteroduplex are marked with asterisks. High opacity horizontal lines within plots represent the minimum conversion tract length, or the distance from the most proximal to the most distal converted polymorphisms. Low opacity horizontal lines indicate the maximum conversion tract, extending from the most distal converted polymorphism to its most proximal unconverted polymorphism. Tracts from noncrossover recombinants are displayed in A, while tracts from crossover recombinants are displayed in (B). (C–D) Frequency of short noncrossover tracts (C, minimum tract length 1 bp converted at only the 12 bp polymorphism) or short crossover tracts (D, minimum tract length 198 bp) as a proportion of all tracts identified from progeny laid within the interhomolog and non-interhomolog windows. Error bars represent the 95% binomial confidence intervals of these proportions and p values were calculated using Fisher’s Exact Test. Diagrams above bar plots depict the sizes of tracts considered ‘long’ or ‘short’ in each respective group. In all panels, dashed grey lines delineate between the interhomolog window (22–58 hr post heat shock) and non-interhomolog window (10–22 hr post heat shock) timepoints.
-
Figure 3—source data 1
The source data for Figure 3 is provided.
The polymorphism conversions scored in individual sequenced ICR assay conversion tracts are listed.
- https://cdn.elifesciences.org/articles/80687/elife-80687-fig3-data1-v2.xlsx
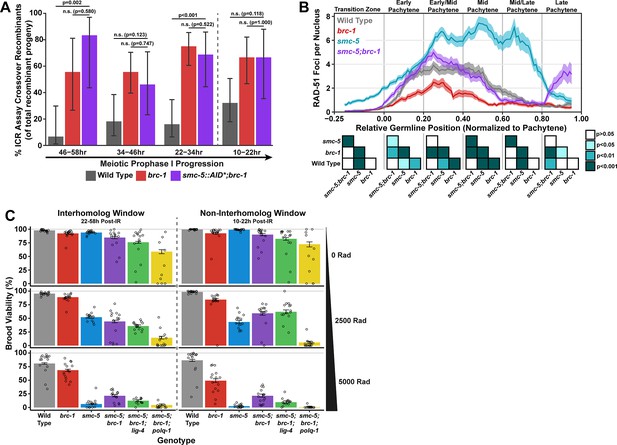
Interactions of SMC-5/6 and BRC-1 in meiotic DSB repair.
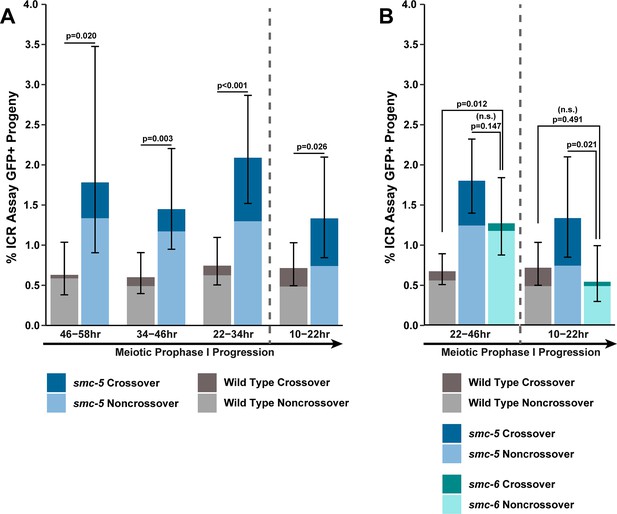
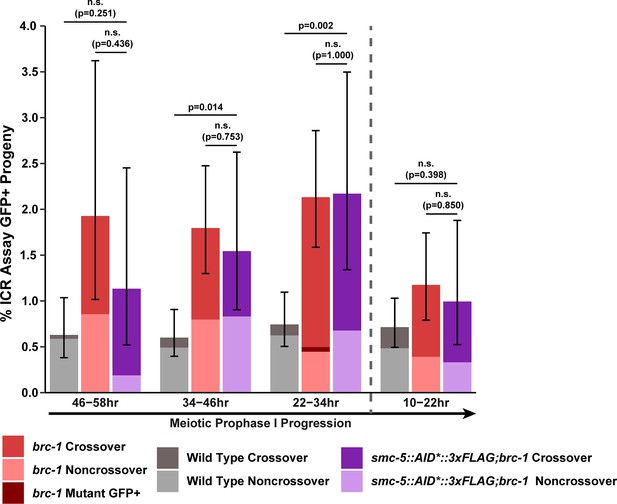
(A) Bar plot displaying the percent of crossover recombinant progeny identified in wild type, brc-1, and smc-5::AID*::3xFLAG;brc-1 ICR assays out of all recombinant progeny scored within individual 12 hr timepoint periods. Frequencies of recombinants identified overall in ICR assays is displayed in Figure 4—figure supplement 1. Wild type ICR assay data is shared in Figures 1A, 2A, B, 4A, Figure 1—figure supplement 2, Figure 2—figure supplement 1, and Figure 4—figure supplement 1. brc-1 ICR assay data is shared in Figure 1A, Figure 1—figure supplement 2, and Figure 4—figure supplement 1. The total number of recombinant progeny scored in each dataset are (10–22 hr/22–34 hr/34–46 hr/46–58 hr timepoints) wild type: 28/25/22/15, brc-1(xoe4) 24/43/36/9, smc-5::AID*::3xFLAG;brc-1 9/16/13/6. Specific progeny counts separated by experimental replicate are presented in Figure 4—source data 3. (B) RAD-51 foci per nucleus displayed as a sliding window (window width 0.1 position units, step size 0.01 position units) across the length of the germline with germline position normalized to the length of pachytene (see Methods). Lines represent the mean RAD-51 foci per nucleus while the shaded area represents SEM within a given window. RAD-51 foci were compared between genotypes within six bins across the germline divided by germline distance: transition zone (−0.25–0), early pachytene (0–0.2), early/mid pachytene (0.2–0.4), mid pachytene (0.4–0.6), mid/late pachytene (0.6–0.8), and late pachytene (0.8–0.1). Heat maps below each bin display the p values for pairwise Mann-Whitney U tests with Holm-Bonferroni correction for multiple comparisons within each bin. For each genotype, n≥7 germlines derived from at least three experimental replicates were analyzed and combined. The number of nuclei scored in each bin (TZ, EP, E/MP, MP, M/LP, LP) are: wild type 258/137/135/121/114/95, smc-5 402/395/348/285/217/189, brc-1 231/170/161/186/159/121, smc-5;brc-1 202/182/167/157/122/82. The per-nucleus RAD-51 counts from this analysis are available in Figure 4—source data 2. (C) Brood viabilities of irradiated hermaphrodites. Bar plots represent the population brood viability and error bars indicate the 95% Binomial confidence interval of this value. The brood viabilities of individual hermaphrodites scored are indicated by data points. For all genotypes and radiation treatments presented in this panel, three experimental replicates were performed with n=5 hermaphrodites scored per condition in each replicate for a total of n=15 hermaphrodite broods scored for each genotype and condition combination. Individual hermaphrodite progeny counts are presented in Figure 4—source data 3. Statistical analysis of the data presented in this panel is depicted in Figure 4—figure supplement 3. Localization of BRC-1::GFP and SMC-5::AID*::3xFLAG in respective smc-5 and brc-1 mutants and in the presence of ionizing radiation is presented in Figure 4—figure supplements 4 and 5. A proposed molecular model for the interactions between BRC-1 and SMC-5/6 is presented in Figure 4—figure supplement 6.
-
Figure 4—source data 1
The source data for Figure 4A is provided.
The total number of ICR assay progeny with GFP+ or non-GFP+ phenotypes are listed. Wild type data is shared with Figure 1—figure supplement 2, Figure 2, Figure 2—figure supplement 1, Figure 4, and Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/80687/elife-80687-fig4-data1-v2.xlsx
-
Figure 4—source data 2
The source data for Figure 4B is provided.
The number of RAD-51 foci per nucleus scored are listed.
- https://cdn.elifesciences.org/articles/80687/elife-80687-fig4-data2-v2.xlsx
-
Figure 4—source data 3
The source data for Figure 4C is provided.
The number of hatched (Live), unhatched (Dead), or unfertilized F1 progeny scored in the brood viability experiment data used to generate Figure 4C and Figure 4—figure supplement 3. Wild type and smc-5(ok2421) data is shared with Figure 4—figure supplement 2A.
- https://cdn.elifesciences.org/articles/80687/elife-80687-fig4-data3-v2.xlsx
Intersister/intrachromatid repair (ICR) assay defects in brc-1 mutants are not exacerbated by SMC-5 depletion.
Stacked bar plots displaying the percent of all progeny scored in wild type, brc-1(xoe4), and smc-5(syb3065);brc-1(xoe4) ICR assays that were determined to be GFP+ noncrossover recombinants, crossover recombinants, or mutants. Error bars represent the 95% Binomial confidence intervals for the frequencies of GFP+ progeny. smc-5(syb3065);brc-1 assays were performed in the presence of 10 mM auxin (see methods). Wild type ICR assay data is shared in Figures 1A, 2A, B, 4A, Figure 1—figure supplement 2, Figure 2—figure supplement 1, and Figure 4—figure supplement 1. brc-1 ICR assay data is shared in Figures 1A and 4A, Figure 1—figure supplement 1, and Figure 4—figure supplement 1. smc-5;brc-1 ICR assay data is shared in Figure 4A and Figure 4—figure supplement 1. The total number of progeny scored in each dataset are (10–22 hr/22–34 hr/34–46 hr/46–58 hr timepoints) wild type 3921/3359/3664/2383, brc-1(xoe4) 2042/2017/2005/467, smc-5(syb3065);brc-1(xoe4) 905/737/842/529. Specific progeny counts separated by experimental replicate are presented in Figure 2—source data 1. p values were calculated by Fisher’s Exact Test comparing the sum totals of GFP+ progeny between groups. Vertical dashed lines demarcate the interhomolog window (≥22 hr post heat shock) and non-interhomolog window (10–22 hr post heat shock) timepoints.
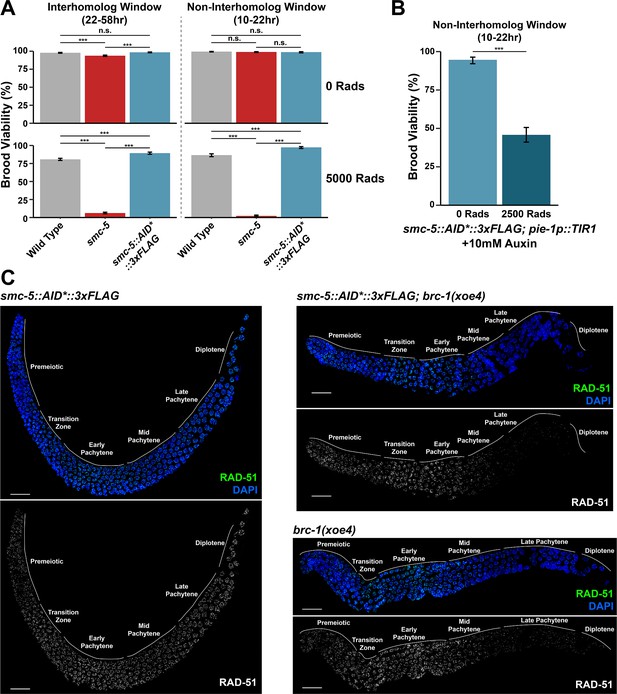
Loss of BRC-1, but not endogenous tagging of SMC-5::AID*::3xFLAG, inhibits RAD-51 localization to irradiation-induced DSBs.
(A) Brood viabilities of wild type, smc-5(ok2421), or smc-5::AID*::3xFLAG (PHX3065) worms exposed to ionizing radiation. Wild type and smc-5 brood viability data is shared with Figure 4C. (B) Brood viability of DLW220 worms following SMC-5 depletion on 10 mM auxin and exposure to ionizing radiation. In (A) and (B), columns represent the population brood viability, error bars represent 95% Binomial confidence intervals, and p values were calculated by Fisher’s Exact Test (n.s.=not significant p>0.05, *** p<0.001). (C) Deconvolved images of whole extruded germlines stained for RAD-51 and DAPI. All germlines were exposed to 5000 Rads of ionizing radiation and were dissected within 1 hr of the radiation treatment. Loss of brc-1 impedes RAD-51 localization in mid/late pachytene (Janisiw et al., 2018; Li et al., 2018), and this phenotype is not recapitulated nor enhanced by the smc-5(syb3065) allele. Grey lines and labels demarcate the mitotic and meiotic stages of the germline. Scale bars represent 20 μm.
-
Figure 4—figure supplement 2—source data 1
The source data for Figure 4—figure supplement 2B is provided.
The number of hatched (Live), unhatched (Dead), or unfertilized F1 progeny scored in the brood viability experiment data used to generate Figure 4—figure supplement 2B. Wild type and smc-5(ok2421) data is shared with Figure 4—figure supplement 2A–B.
- https://cdn.elifesciences.org/articles/80687/elife-80687-fig4-figsupp2-data1-v2.xlsx
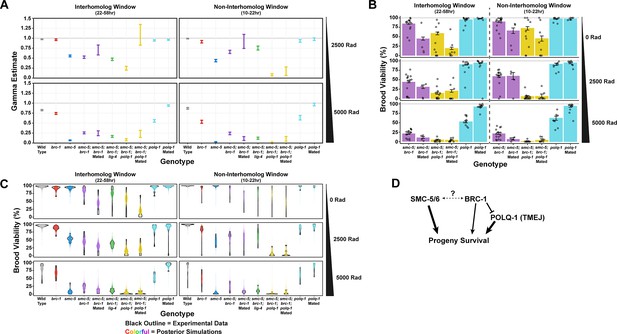
Bayesian analysis of brood viability following ionizing radiation treatment.
(A) Gamma metric estimates of radiation sensitivity inferred from a Beta-Binomial model fit to the irradiation data presented in Figure 4C and (B). For details on this calculation, see Methods. Bars indicate the bounds of the 95% Credible Interval for each Gamma estimate. (B) Brood viability results following irradiation at doses of 0, 2500, or 5000 Rads for nematodes mated to wild type unirradiated males or using endogenous sperm following irradiation. Bars represent the population brood viability, while points represent the brood viabilities of individual hermaphrodites scored. Error bars indicate 95% Binomial confidence intervals of the population brood viability. Parameter estimate summary statistics are presented in Figure 4—figure supplement 3—source data 1. The broods of three separate replicates of n=5 hermaphrodites were scored for each unirradiated condition (total n=15 hermaphrodite broods per condition), the broods of n=2 separate replicates of n=5 hermaphrodites were scored for polq-1 mated and smc-5;brc-1;polq-1 mated conditions (total n=10 hermaphrodite broods per condition), and n=1 replicate of n=5 hermaphrodite broods were scored for the smc-5;brc-1 mated condition. (C) Violin plots of the brood viabilities from individual hermaphrodites scored (black outlines, the same data is displayed as points in Figure 4B and in panel B) and 1500 simulated broods generated using the posterior parameter estimates of the Beta-Binomial model fit to the data (see Methods). (D) Genetic interaction diagram inferred from estimates presented in panel A. SMC-5/6 and BRC-1 both contribute to progeny viability following meiotic exposure to exogenous DSBs. However, BRC-1 also inhibits error prone repair, which can compensate for the DSB defects of smc-5 mutants when brc-1 is also ablated.
-
Figure 4—figure supplement 3—source data 1
The source data for Figure 4—figure supplement 3A is provided.
Summary statistics of Beta-Binomial model parameter estimates (see Materials and methods) output by RStan (see Figure 4—figure supplement 3—source code 1) are provided.
- https://cdn.elifesciences.org/articles/80687/elife-80687-fig4-figsupp3-data1-v2.xlsx
-
Figure 4—figure supplement 3—source code 1
The source code for Figure 4-figure supplement 3A and 3C is provided.
R code to perform Beta-Binomial model fitting and posterior predictive simulations is provided.
- https://cdn.elifesciences.org/articles/80687/elife-80687-fig4-figsupp3-code1-v2.zip
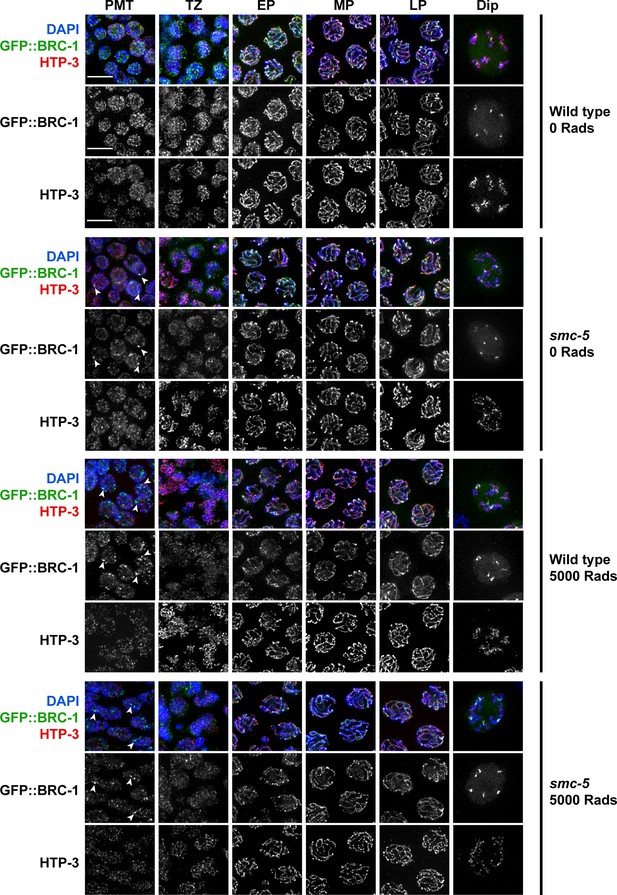
SMC-5/6 is not required for GFP::BRC-1 localization.
Deconvolved widefield images of germline nuclei stained for GFP (GFP::BRC-1), chromosome axis protein HTP-3, and DAPI (DNA) in a wild type or smc-5(ok2421) mutant background and treated with 0 or 5000 Rads of ionizing radiation. Scale bars represent 5 μm. Stages of meiotic nuclei were determined based on DAPI morphology and are listed on the top of the figure (PMT = premeiotic tip, TZ = transition zone, EP = early pachytene, MP = mid pachytene, LP = late pachytene, Dip = Diplotene). Arrowheads indicate GFP::BRC-1 foci.
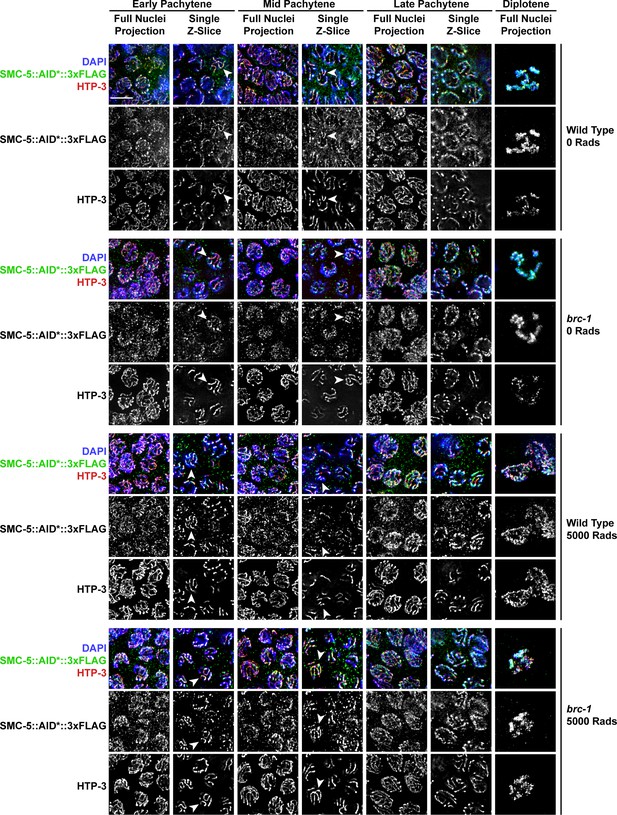
BRC-1 is not required for SMC-5::AID*::3xFLAG localization.
Deconvolved images of germline nuclei stained for AID* (SMC-5::AID*::3xFLAG), chromosome axis protein HTP-3, or DAPI (DNA) in a wild type or brc-1(xoe4) mutant background and treated with 0 or 5000 Rads of ionizing radiation. Scale bars represent 5 μm. Stages of meiotic nuclei are determined based on DAPI morphology and are listed at the top of the figure. For each image, a max intensity projection of whole nuclei and single z-slices are displayed to demonstrate the relative localization of SMC-5 and HTP-3. Arrowheads indicate examples of colocalization between HTP-3 and SMC-5::AID*3xFLAG.
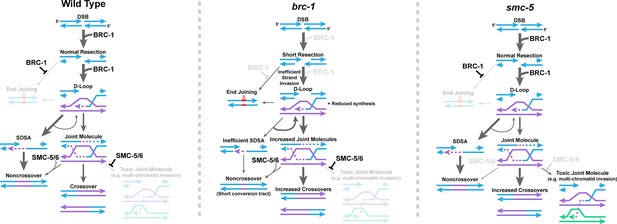
Proposed model of BRC-1 and SMC-5/6 function in C. elegans intersister DSB repair.
Displayed is a proposed model for the functions of BRC-1 and SMC-5/6 in regulating intersister DSB repair in the C. elegans germline. Under wild type conditions, BRC-1 promotes efficient resection of the damaged chromatid (blue) and facilitates strand invasion and extension with the sister chromatid (purple). BRC-1 also inhibits TMEJ either through direct antagonism of this pathway or indirectly by promoting efficient recombination. Following strand extension, the majority of D-loop intermediates are dissolved and repaired through SDSA, which is efficient due to BRC-1 promoted resection of the second end of the DSB. A minority of D-loops will proceed to form joint molecules, which may potentially be preferentially resolved as noncrossovers via the action of SMC-5/6 or as crossovers in an SMC-5/6 independent manner. In addition, SMC-5/6 inhibits the formation of toxic joint molecule intermediates, such as multi-chromatid joint molecules. In a brc-1 mutant, DSBs are not resected to wild type levels and strand invasion is inefficient. Reduced resection limits the efficiency of second end capture in SDSA, reducing noncrossovers through this pathway. Further, limited strand extension reduces the extent of gene conversion in noncrossovers generated by successful SDSA or joint molecule dissolution. Failure in SDSA leads to increased DSB reinvasion of repair templates, contributing to the tandem duplications observed in mutants for BRCA1 (Chandramouly et al., 2013; Kamp et al., 2020). In addition, either due to absence of direct inhibition by BRC-1 or inefficiencies in recombination, end joining (particularly TMEJ) becomes activated to resolve DSBs. However, reduced resection does not inhibit joint molecule formation, leading to more of these intermediates which are preferentially resolved as crossovers. Finally, in an smc-5 mutant, early steps in DSB repair proceed normally. However, absence of SMC-5/6 results in unconstrained joint molecule formation, including toxic intermediates. Failure in SMC-5/6 action to promote noncrossover repair further increases the proportion of joint molecules which are resolved as crossovers.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (C. elegans) | brc-1 | https://wormbase.org/species/c_elegans/gene/WBGene00000264 | WormBase ID: WBGene00000264 | |
| Gene (C. elegans) | brd-1 | https://wormbase.org/species/c_elegans/gene/WBGene00000265 | WormBase ID: WBGene00000265 | |
| Gene (C. elegans) | smc-5 | https://wormbase.org/species/c_elegans/gene/WBGene00016151 | WormBase ID: WBGene00016151 | |
| Gene (C. elegans) | smc-6 | https://wormbase.org/species/c_elegans/gene/WBGene00010056 | WormBase ID: WBGene00010056 | |
| Gene (C. elegans) | polq-1 | https://wormbase.org/species/c_elegans/gene/WBGene00020964 | WormBase ID: WBGene00020964 | |
| Gene (C. elegans) | lig-4 | https://wormbase.org/species/c_elegans/gene/WBGene00002986 | WormBase ID: WBGene00002986 | |
| Gene (C. elegans) | unc-5 | https://wormbase.org/species/c_elegans/gene/WBGene00006745 | WormBase ID: WBGene00006745 | |
| Gene (C. elegans) | rad-51 | https://wormbase.org/species/c_elegans/gene/WBGene00004297 | WormBase ID: WBGene00004297 | |
| Strain, strain background (C. elegans) | For C. elegans alleles and strain information, see strain list in Methods (C. elegans strains and maintenance) | This paper | See strain list in Methods (C. elegans strains and maintenance) | |
| Genetic reagent (C. elegans) | Details on C. elegans CRISPR/Cas9 editing are detailed in the Methods (CRISPR/Cas9 genome editing) | This paper | CRISPR/Cas9 transgenics were generated by InVivo Biosystems | |
| Antibody | Anti-RAD-51 (chicken polyclonal) | Kurhanewicz et al., 2020 | IF (1:1000) | |
| Antibody | Anti-RAD-51 (rabbit polyclonal) | Yokoo et al., 2012 | IF (1:5000) | |
| Antibody | Anti-mini-AID M214-3 (mouse monoclonal) | MBL International | IF (1:500) | |
| Antibody | Anti-HTP-3 (rat polyclonal) | This study | IF (1:1000) | |
| Antibody | Anti-GFP (rabbit polyclonal) | Yokoo et al., 2012 | IF (1:500) | |
| Antibody | Alexa Fluor 488 anti-rabbit (goat polyclonal) | Thermo Fisher | Cat# A21428 | IF (1:200) |
| Antibody | Alexa Fluor 488 anti-mouse (goat polyclonal) | Thermo Fisher | Cat# A11001 | IF (1:200) |
| Antibody | Alexa Fluor 555 anti-rat (goat polyclonal) | Thermo Fisher | Cat# A48263 | IF (1:200) |
| Antibody | Alexa Fluor 647 anti-chicken (goat polyclonal) | Thermo Fisher | Cat# A21449 | IF (1:200) |
| Sequence-based reagent | PCR primers for amplifying ICR and IH assay recombinants are described in Methods (Sequencing and analysis of ICR assay conversion tracts, Interhomolog assay (IH assay)) | Toraason et al., 2021c, This Paper | PCR Primers | See Methods (Sequencing and analysis of ICR assay conversion tracts, Interhomolog assay (IH assay)) |
| Chemical compound, drug | Naphthaleneacetic acid (auxin) | PhytoTechnology Laboratories | Cat# N610 | 10 mM |
| Software, algorithm | Whole Gonad Analysis (R script) v1.0 | Toraason et al., 2021a; Toraason et al., 2021d | https://github.com/libudalab/Gonad-Analysis-Pipeline | |
| Software, algorithm | RStan | Stan Development Team | https://mc-stan.org/ | |
| Software, algorithm | Imaris 9 | Oxford Instruments | https://imaris.oxinst.com/products | |
| Software, algorithm | FIJI | Schindelin et al., 2012 | https://imagej.net/software/fiji/ | |
| Software, algorithm | FIJI plug in – Stitcher | Preibisch et al., 2009 | https://imagej.net/plugins/image-stitching |





