Fitness advantage of Bacteroides thetaiotaomicron capsular polysaccharide in the mouse gut depends on the resident microbiota
Figures
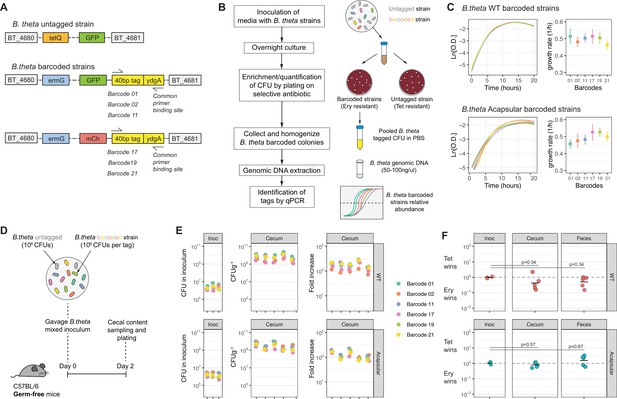
Barcoded B. theta strains have similar fitness for growing in vitro and in vivo.
(A) Schematic representation of insertions in B. theta genome. The barcoded strains carried the barcode, erythromycin resistance cassette, and fluorescent protein in the genome. The untagged B. theta carried a tetracycline resistance cassette together with a fluorescent protein inserted at the same position in the genome. (B) Workflow for barcoded strain enrichment and quantification. (C) Growth curves of B. theta WT and acapsular barcoded strains in BHIS media (n = 3, replicative cultures per strain) and growth rates (1/hr, mean and 95% confidence interval) per barcode. (D) Experimental design of in vivo competitions to confirm equal fitness of the barcoded strains. Each strain (B. theta untagged and six B. theta barcoded) were mixed in an equal ratio (inoculum: 106 CFU per strain). (E) Barcode distribution during colonization among six B. theta barcoded strains either WT or acapsular. Plots show distribution of barcodes in the inoculum, in cecal content of individual mice after 48 hr of colonization and fold increase of each barcode per mouse compared to the inoculum (n = 5 mice colonized). (F) Competitive index of tetracycline-resistant untagged strain (Tet) over the erythromycin-resistant barcoded strain (Ery) in vivo after 48 hr of colonization of B. theta WT or acapsular in cecal content and in feces (n = 2 replicative cultures in inoculum; n = 5 mice colonized). Points represent individual mice, and the horizontal line is the mean. p-Values were obtained by one-way ANOVA followed by Tukey’s honest significance test. Data are included in Figure 1—source data 1.
-
Figure 1—source data 1
Barcoded B. theta strains have similar fitness for growing in vitro and in vivo.
- https://cdn.elifesciences.org/articles/81212/elife-81212-fig1-data1-v2.xlsx
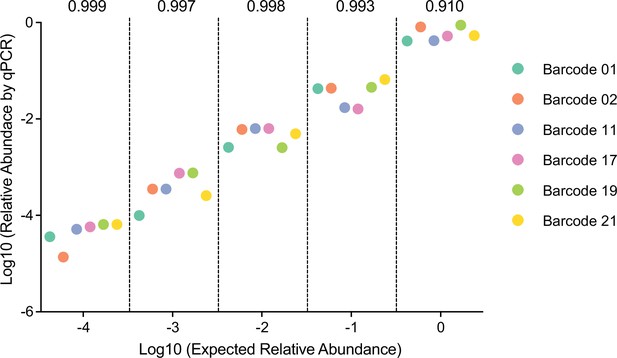
Barcoded B. theta strains can be accurately detected in vitro.
Barcoded WT B. theta were mixed serially diluted 100–10–4-fold. Randomly one dilution of each strain was selected for each dilution point and mixed (i.e., 100 barcode wild-type isogenic tagged strains [WITS] 1, 10–1 barcode WITS 17, 10–2 barcode WITS 11, 10–3 barcode WITS 21, and 10–4 barcode WITS 2). We repeated this process until each strain is present in a mix at least once on each dilution point. Approximately 106 CFU of these mixtures were inoculated into 5 ml BHIS. Relative abundance after overnight growth in BHIS (y-axis) was calculated by qPCR and CFU determination. Pielou’s evenness (numbers above the graph) was estimated with a maximum possible value of Hmax = ln(6) for all data points (six barcoded strains) at each dilution. Data are included in Figure 1—source data 1.
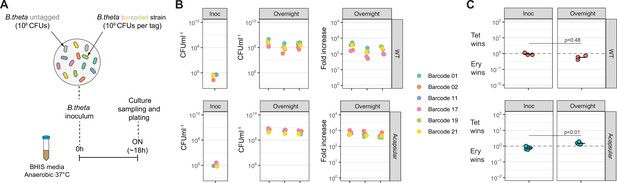
Barcoded B. theta strains have similar fitness for growing in vitro.
(A) Experimental design of in vitro competitions. Each strain (B. theta untagged, tetracycline resistant, and six B. theta barcoded, erythromycin resistant) were mixed in an equal ratio (inoculum: 106 CFU per strain). (B) Tags distribution after in vitro competition among six B. theta barcoded strains either WT or acapsular to demonstrate neutrality in vitro. Plots show distribution of barcodes in the inoculum, in individual overnight cultures, and fold increase of each barcode per culture compared to the inoculum (n = 3 replicates overnight cultures). (C) Competitive index of tetracycline-resistant untagged strain (Tet) over the erythromycin-resistant barcoded strain (Ery) in vitro after overnight cultures (n = 3 for replicative cultures in inoculum and overnight cultures). Points represent individual overnight cultures, and the horizontal line is the mean. p-Values were obtained by Welch t-test. Data are included in Figure 1—source data 1.
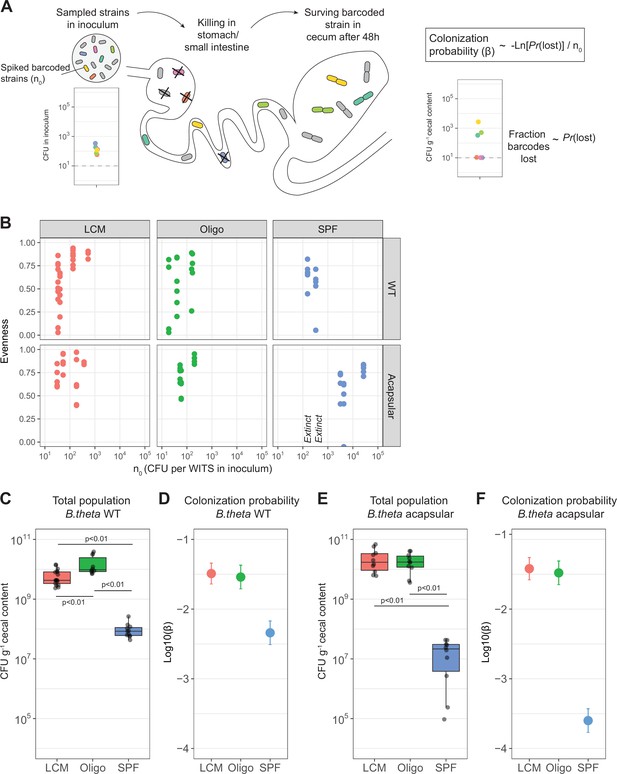
Colonization probability of B. theta strains in low-complexity microbiota (LCM), Oligo Mouse Microbiota (Oligo-MM12), and specific pathogen free (SPF) mice.
(A) Schematic representation of experimental estimation of colonization probability. The untagged strain was tetracycline-resistant, and all barcoded strains were erythromycin-resistant. (B) Pielou’s evenness vs. n0. Pielou’s evenness was estimated with a maximum possible value of Hmax = ln(6) for all data points (six barcoded strains). Each dot represents the evenness calculated per mouse. Values of n0 for which all barcodes were lost, and therefore no evenness could be estimated, were marked with ‘Extinct.’ Total inoculum size was maintained at 107 CFU. The exact inocula compositions are shown in Figures 3 and 4. (C, E) Total B. theta population in the cecum at 48 hr post-colonization for (C) WT and (E) acapsular strains. Points represent individual mice and boxplot quartiles provide summary statistics. p-Values were obtained by one-way ANOVA followed by Tukey’s honest significance test. (D, F) Probability of colonization (β) in (D) WT and (F) acapsular in the cecum after 48 hr of colonization using the loss method. Circles represent the best estimate and vertical line the higher and lower bound of the 95% confidence interval. Estimation based on six barcodes times the number of mice (LCM = 17, OligoMM12 = 10, SPF = 11). See ‘Methods’ for parameter estimations. Data are included in Figure 2—source data 1.
-
Figure 2—source data 1
Colonization probability of B. theta strains in low-complexity microbiota (LCM), Oligo Mouse Microbiota (Oligo-MM12), and specific pathogen free (SPF) mice.
- https://cdn.elifesciences.org/articles/81212/elife-81212-fig2-data1-v2.xlsx
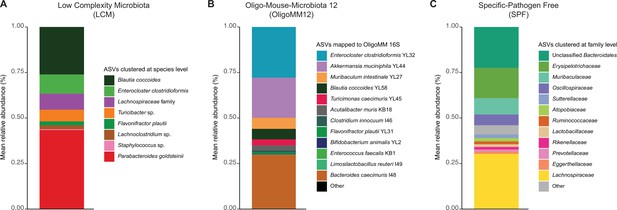
Fecal microbiota composition of different resident microbiotas.
(A) Low-complexity microbiota (LCM). Amplicon sequence variants from 16S sequencing were matched at the species level. (B) Oligo Mouse Microbiota (OligoMM12). Amplicon sequence variants from 16S sequencing were matched to the reported 12 species. (C) Specific pathogen free (SPF). Amplicon sequence variants from 16S sequencing were matched at the family level. Raw sequencing data is available in the ENA repository under project ID PRJEB57876 (SPF and LCM) and PRJEB53981 (OligoMM12). Data are included in Figure 2—source data 1.
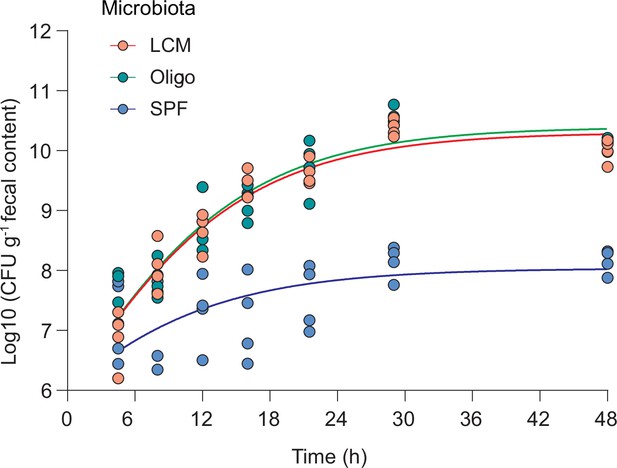
In vivo growth curve of B. theta WT.
Mice with three different resident microbiotas (low-complexity microbiota [LCM], Oligo Mouse Microbiota [OligoMM12], specific pathogen free [SPF]; n = 4 per group) were inoculated with 107 CFU of B. theta WT strain (erythromycin-resistant barcode 1), and fecal samples were collected and plated at the depicted time points. Data are included in Figure 2—source data 1.

Distribution of B. theta WT barcoded strains in inoculum and after 48 hr of colonization.
Abundance of each barcoded strain (n0) spiked in the total inoculum and in the cecum of low-complexity microbiota (LCM), Oligo Mouse Microbiota (OligoMM12), and specific pathogen free (SPF) at 48 hr post colonization. In all experiments, we used 107 CFUs of the untagged strain in the inoculum. Data are included in Figure 2—source data 1.

Distribution of B. theta acapsular barcoded strains in inoculum and after 48 hr of colonization.
Abundance of each barcoded strain (n0) spiked in the total inoculum and in the cecum of low-complexity microbiota (LCM), Oligo Mouse Microbiota (OligoMM12), and specific pathogen free (SPF) at 48 hr post colonization. In all experiments, we used 107 CFUs of the untagged strain in the inoculum. Data are included in Figure 2—source data 1.
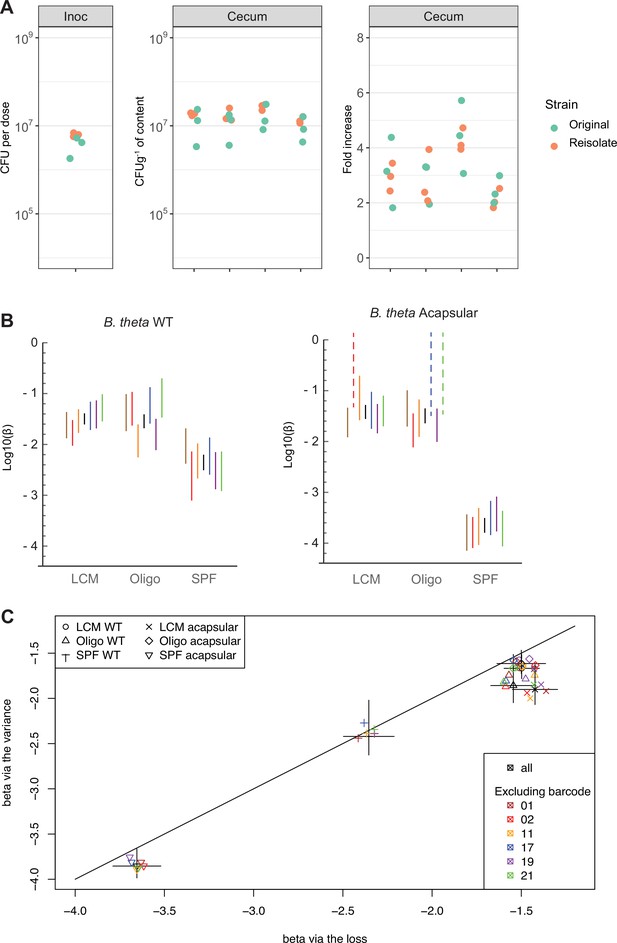
Barcoded strain persistence is not link to fitness advantage.
(A) In vivo competition among B. theta WT barcoded strains (erythromycin-resistant) reisolated from specific pathogen free (SPF) mice vs. original stock barcoded strains. Plots show distribution of B. theta original and isolated strains in the inoculum (106 CFUs per strain), in cecal content of individual mice after 48 hr of colonization and fold increase of each barcode compared to the inoculum (n = 4 mice recolonized). (B, C) To exclude excessive influence of individual barcoded strains on estimates of colonization probability, we compared results using all data, individual barcodes, and all-data-minus-one barcode. (B) Inference of colonization probability was carried out using data from each barcode individually, using the ‘barcode loss’ method. Lines represent the 95% confidence interval of the colonization probability. Black lines represent the estimates using all data. Each color represents the estimates using the data from only one barcoded strain across all mice. Note than in the case of acapsular colonization of low-complexity microbiota (LCM) and Oligo Mouse Microbiota (OligoMM12) mice, some barcoded strains showed no loss in any animals, so an estimate is here shown assuming loss of one barcode at the lowest n0 number. (C) Colonization probability, from the loss and the variance methods estimated using all data (black) or all data except for one barcode (colors, as indicated in the legend). Lines on each axis represent the 95% confidence interval of the colonization probability estimated by each method. Data are included in Figure 2—source data 1.
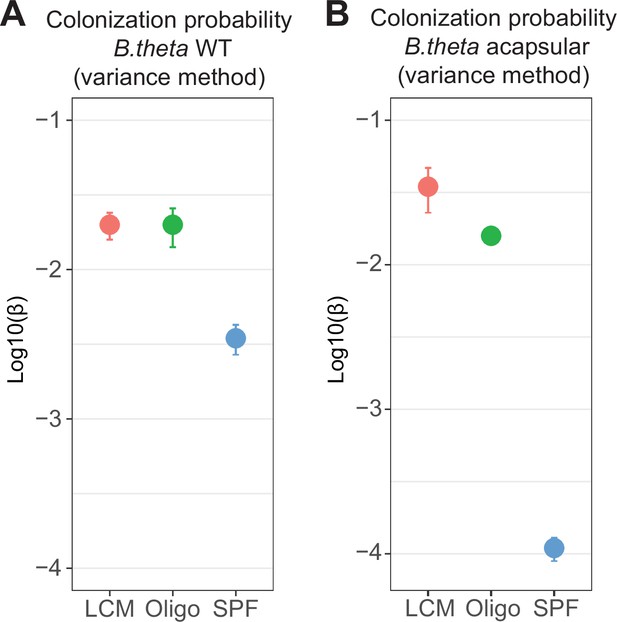
Probability of colonization (β) using variance of barcoded strains before (inoculum) and after colonization (cecum).
Estimates of β in (A) WT and (B) acapsular in the cecum after 48 hr of colonization. Circles represent the best estimate and vertical line the higher and lower bound of the 95% confidence interval. Estimation based on six barcodes × m mice (low-complexity microbiota [LCM] = 17, Oligo Mouse Microbiota [OligoMM12] = 10, specific pathogen free [SPF] = 11) as in Figure 2D and F. Data are included in Figure 2—source data 1.
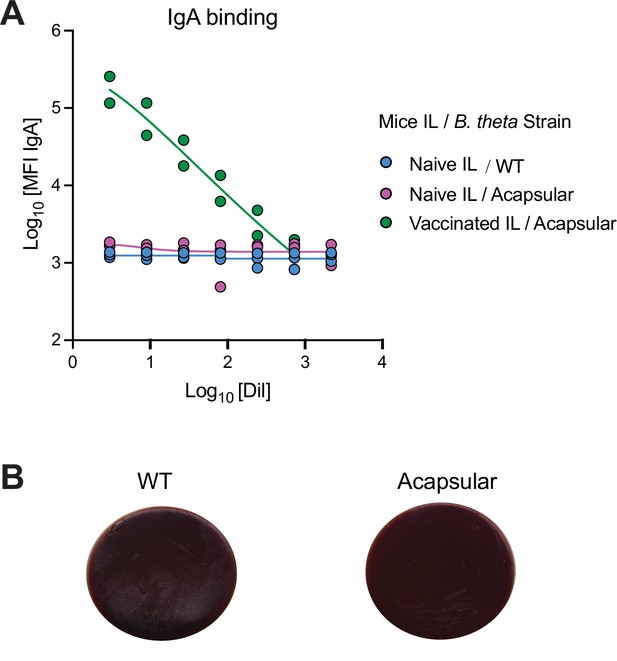
Acapsular B. theta strain is not targeted by IgA or phages in naïve specific pathogen free (SPF) mice.
(A) Bacterial flow cytometry binding curves (median fluorescence intensity) of IgA from intestinal lavage (IL) from naïve SPF mice against B. theta WT and acapsular strains (blue and magenta). As a positive control for high-avidity IgA antibodies binding to B. theta, we included an intestinal lavage from two germ-free (GF) mice that had been vaccinated orally once weekly for 4 weeks with 1011 particles of inactivated B. theta acapsular strain (green). Curves are four-parameter logistic regression fits. (B) Log phase WT and acapsular B. theta strains (300 μl) were added into 3 ml of fresh BHIS media and spiked with 100 μl of sterile-filtered homogenized cecum content from SPF mice. After 15 min of incubation, serial dilutions were plated in BHIS agar plates and incubated overnight. Plates showed no plaques neither in the WT or acapsular strains. Data are included in Figure 2—source data 1.
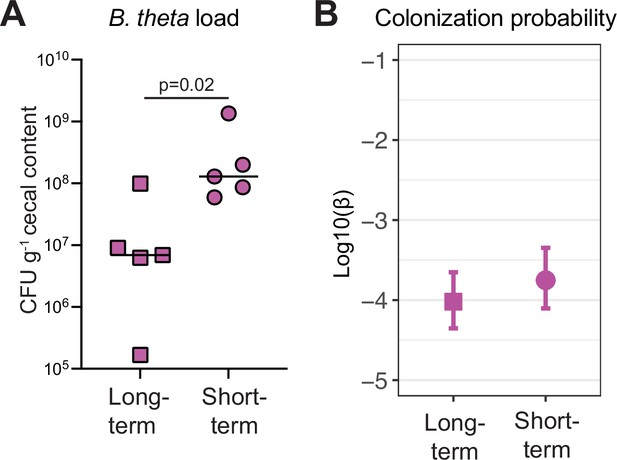
Time of colonization with specific pathogen free (SPF) microbiota in germ-free (GF) mice do not affect colonization probability of B. theta acapsular.
GF mice were colonized with an SPF microbiota by co-housing with SPF mice for 14 days (long-term) or 2 days (short-term). Mice were then colonized with 107 CFU untagged acapsular B. theta, tetracycline resistant, and an n0 approximately 1.5 × 103 CFU per barcoded erythromycin-resistant acapsular (B. theta). (A) Total load of acapsular B. theta in cecum of SPF ex-GF mice at 48 hr post B. theta inoculation. Points represent individual mice and boxplot quartiles provide summary statistics. p-Values were obtained by Welch t-test. (B) Colonization probability of B. theta acapsular in SPF ex-GF mice (mean and 95% confidence interval). Data are included in Figure 2—source data 1.
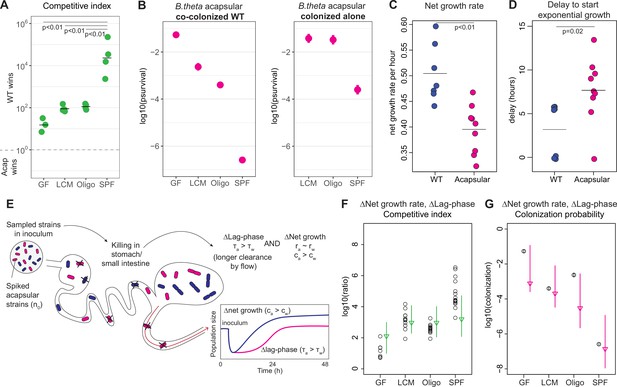
Competitive colonization with acapsular and WT strains.
(A) Competitive index (ratio between WT over acapsular) in the cecum after 48 hr of colonization starting at a 1:1 ratio (inoculum: approximately 106 CFU of each: erythromycin-resistant WT and tetracycline-resistant acapsular; germ-free [GF] = 3, low-complexity microbiota [LCM] = 4, Oligo Mouse Microbiota [OligoMM12] = 4, specific pathogen free [SPF] = 4). Points represent individual mice, and the horizontal line is the mean. p-Values were obtained by one-way ANOVA followed by Tukey’s honest significance test. (B) (Left) Probability of colonization by the acapsular strain during co-colonization with the WT strain. Circles represent the best estimate and vertical line the higher and lower bound of the 95% confidence interval. Barcoded erythromycin-resistant acapsular strains were spiked into a WT untagged strain inoculum (inoculum: 107 of untagged tetracycline-resistant B. theta WT and n0 CFU of erythromycin-resistant barcoded B. theta acapsular adjusted to each microbiota background: n0GF ~ 10, n0LCM ~ 103, n0Oligo ~ 103, n0SPF ~ 106; number of mice per group: GF = 7, LCM = 13, OligoMM12=12, SPF = 16). (Right) Probability of colonization by the acapsular strain when colonizing alone. Graph generated using the same data as Figure 2F. (C, D) Estimation of (C) net growth rate and (D) and delay to start exponential growth (see Figure 3—figure supplement 2 and Appendix 1 for fitting function; n = 7 for WT and n = 9 for acapsular). Points represent estimations of individual mice, and the horizontal line is the mean. p-Values were obtained by Welch t-test. (E) Schematic representation of competitive advantage of the WT over the acapsular B. theta having a similar initial probability of colonization of the cecum: difference in lag phase (mean time to growth commencement in acapsular (τa) and WT (τw)) and difference in net growth rate (growth rate in acapsular (ra) and WT (rw); clearance rate in acapsular (ca) and WT (cw)). Clearance can be due to both flow/loss in the fecal stream and death. (F, G) Estimation of the (F) competitive index and (G) colonization probability of the acapsular strain assuming a mean 4.5 hr difference in lag phase and the estimated difference in net growth rate between the WT and acapsular strains. Circles represent experimental data from (B) and Figure 1C. Triangles represent the best estimate and vertical line the higher and lower bound of the 95% confidence interval. See 'Methods' for parameter estimations. Data are included in Figure 3—source data 1.
-
Figure 3—source data 1
Competitive colonization with acapsular and WT strains.
- https://cdn.elifesciences.org/articles/81212/elife-81212-fig3-data1-v2.xlsx
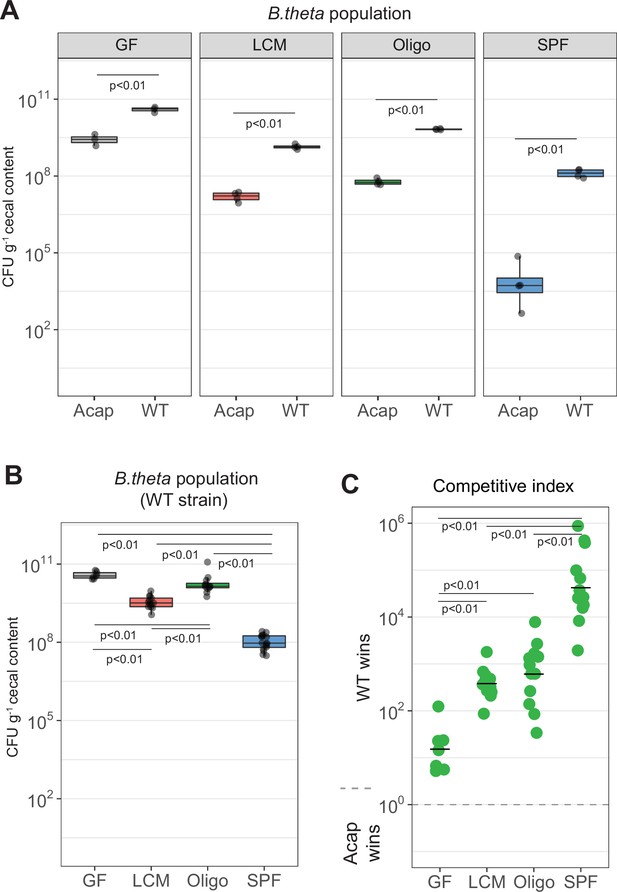
B. theta acapsular is outcompeted by WT during co-colonization.
(A) Competition between acapsular and WT strains during colonization starting at a 1:1 ratio. Population density of the WT (erythromycin resistant) and acapsular (tetracycline resistant) strains in cecum after 48 hr of colonization (inoculum and groups as described in Figure 3A). Points represent individual mice and boxplot quartiles provide summary statistics. p-Values were obtained by one-way ANOVA followed by Tukey’s honest significance test. (B, C) Estimation of probability of colonization of the cecum by the acapsular strain using a WT (tetracycline resistant) strain inoculum spiked with erythromycin-resistant barcoded acapsular strains. (B) Population density of the WT strain. Points represent individual mice and boxplot quartiles provide summary statistics. p-Values were obtained by one-way ANOVA followed by Tukey’s honest significance test. (C) Competitive index after correction with the initial WT/acapsular ratio in the inoculum (inoculum as described in Figure 3B). Points represent individual mice, and the horizontal line is the mean. p-Values were obtained by one-way ANOVA followed by Tukey’s honest significance test. Data are included in Figure 3—source data 1.
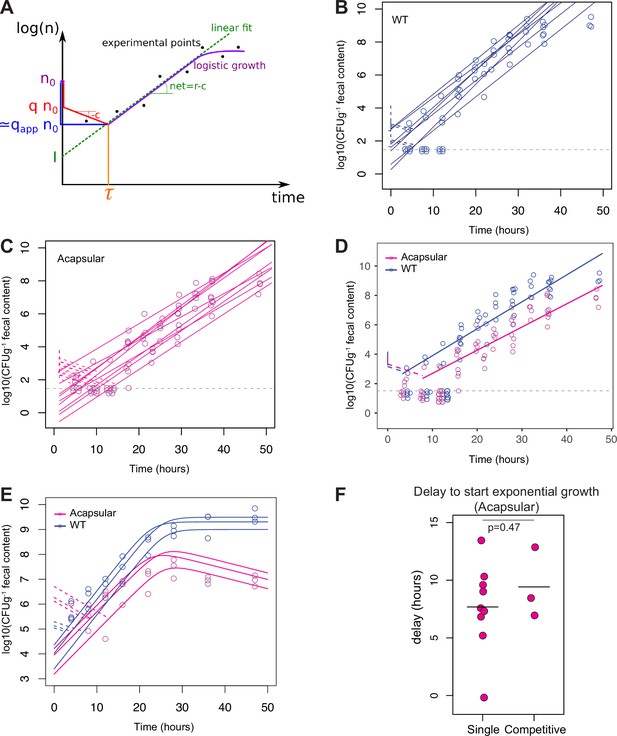
Net growth of B. theta during colonization of Oligo Mouse Microbiota (OligoMM12) mice.
(A) Schematic of the analysis of the growth curves with delay (r = absolute growth rate, c = clearance rate, n0 = inoculum size, q = real colonization probability, qapp = observed colonization probability, τ = estimated lag phase, I = concentration value when t = 0). (B, C) Fecal bacterial load of (B) B. theta WT (erythromycin-resistant barcode 1) or (C) acapsular (erythromycin-resistant barcode 1) during single-strain colonization in OligoMM12 mice (n = 7 for WT and n = 9 for acapsular; inoculum n0 ~ 104 CFU of each strain). Estimation of net growth rate (solid line) and delay to start exponential growth (dotted line). Gray dotted line represents limit of detection at Log10(CFUg–1) = 1.5. Values below detection limit have been jittered across the x and y axes and values over the detection limit have been only jittered in the x axis for better visualization. (D) Overlay of fecal bacterial load of B. theta WT (blue) or acapsular (magenta) and net growth rate (solid line) and lag-phase time base (dotted line) based on average estimates from Figure 3D and E. Gray dotted line represents limit of detection at Log10(CFUg–1) = 1.5. Values below detection limit have been jittered across the x and y axes and values over the detection limit have been only jittered in the x axis for better visualization. (E) Fecal bacterial load of B. theta WT (blue, tetracycline-resistant) or acapsular (magenta, erythromycin resistant) during competitive colonization in OligoMM12 mice (n = 3 per group, inoculum n0 ~ 5 × 106 CFU of each strain). Delay to start exponential growth (dotted line) based on fixed net growth rate parameters obtained in (B) and (C). (F) Comparison of delay to start exponential growth of acapsular strain during single and competitive colonization. Estimations obtained from panel (C) (single colonization) and panel (E) (completive colonization with WT strain). Data are included in Figure 3—source data 1.
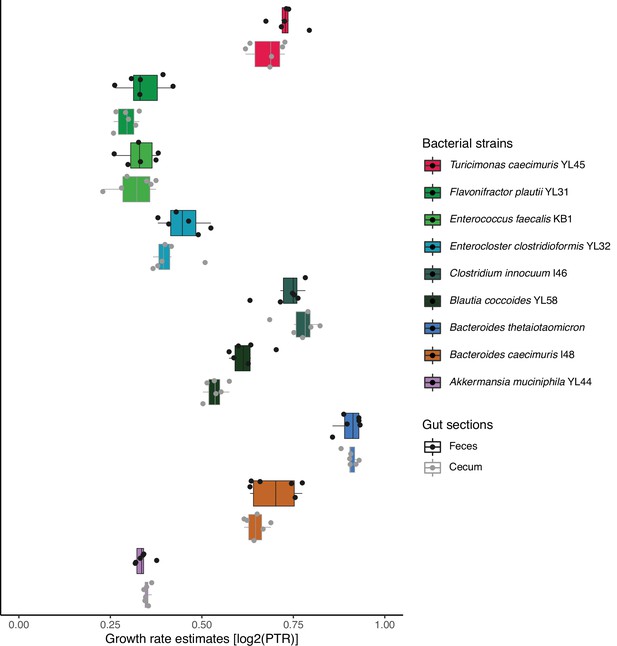
Growth of B. theta and Oligo Mouse Microbiota (OligoMM12) in vivo.
Estimation of growth rate of B. theta WT and other OligoMM12 strains in the cecum mice 8 hr after colonization using peak-to-trough analysis of metagenome sequencing data (n = 6). Points represent individual mice and boxplot quartiles provide summary statistics. Raw sequencing data is available in the ENA repository under project ID PRJEB57876.
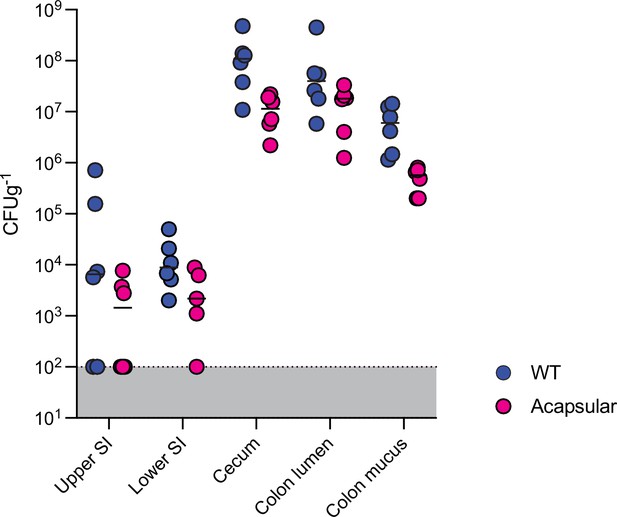
B. theta load across gut sections of Oligo Mouse Microbiota (OligoMM12) mice.
Single colonization with WT (erythromycin-resistant barcode 1) or acapsular (erythromycin-resistant barcode 1) strains (inoculum ~107 CFU of each strain). Load quantified 8 hr after inoculation (n = 6 per group). Points represent individual mice, and the horizontal line is the mean. Data are included in Figure 3—source data 1.
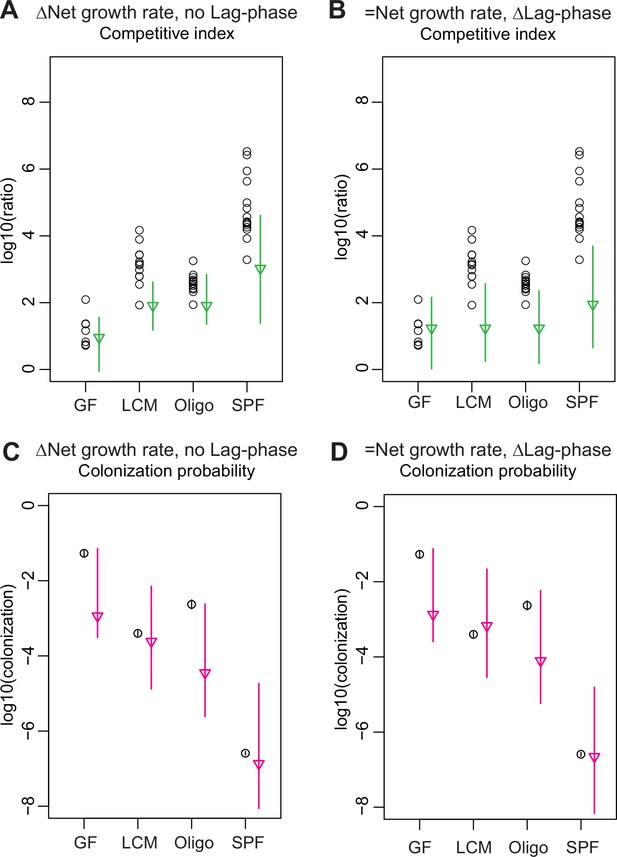
Models for competitive index and colonization probability with fixed parameters.
(A–D) Estimation of the competitive index and colonization probability of the acapsular strain (germ-free [GF] = 7, low-complexity microbiota [LCM] = 13, Oligo Mouse Microbiota [OligoMM12] = 12, specific pathogen free [SPF] = 16): (A, C) assuming no lag phase and estimated difference in net growth rate between the WT and acapsular strains and (B, D) assuming a mean 4.5 hr difference in lag phase and identical net growth rate between the WT and acapsular strains. Circles represent experimental data from Figure 3B and Figure 1C. Triangles represent the best estimate and vertical line the higher and lower bound of the 95% confidence interval. See 'Methods' for parameter estimations. Data are included in Figure 3—source data 1.
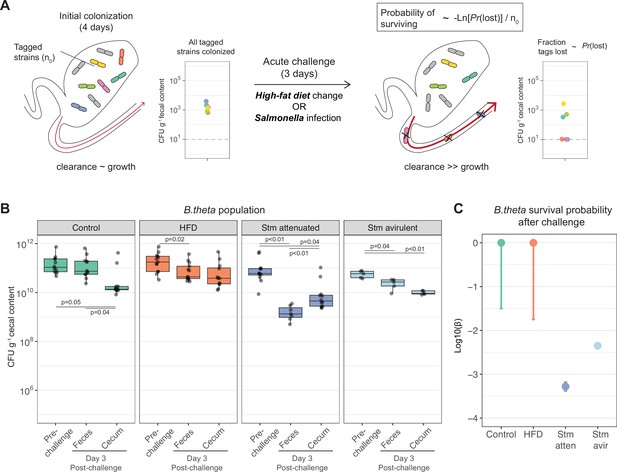
Acute challenges imposed a population bottleneck in the resident B. theta population.
(A) Schematic representation of experimental estimation of colonization survival probability after acute challenges. (B, C) Germ-free (GF) mice were colonized with an inoculum of ~109 CFU untagged tetracycline-resistant B. theta WT spiked with barcoded erythromycin-resistant WT strains. The number of spiked CFU of each individual barcode per experiment (n0) is described in Figure 4—figure supplement 1. (B) Population of B. theta in monocolonized ex-GF mice kept under standard chow (control) and during acute challenge with high-fat diet (HFD), infection with attenuated Salmonella (Stm) (ΔssaV) or avirulent (ΔssaVΔinvG) Salmonella. Points represent individual mice and boxplot quartiles provide summary statistics. p-Values were obtained by one-way ANOVA followed by Tukey’s honest significance test. (C) Probability of surviving in the cecum after 3 days of the acute challenge. Estimation based on six barcodes timer the total number of mice (Control = 12, HFD = 13, Stm attenuated = 14, Stm avirulent = 5). Circle represent the best estimate and vertical line the higher and lower bound of the 95% confidence interval. Data are included in Figure 3—source data 1.
-
Figure 4—source data 1
Acute challenges imposed a population bottleneck in the resident B. theta population.
- https://cdn.elifesciences.org/articles/81212/elife-81212-fig4-data1-v2.xlsx
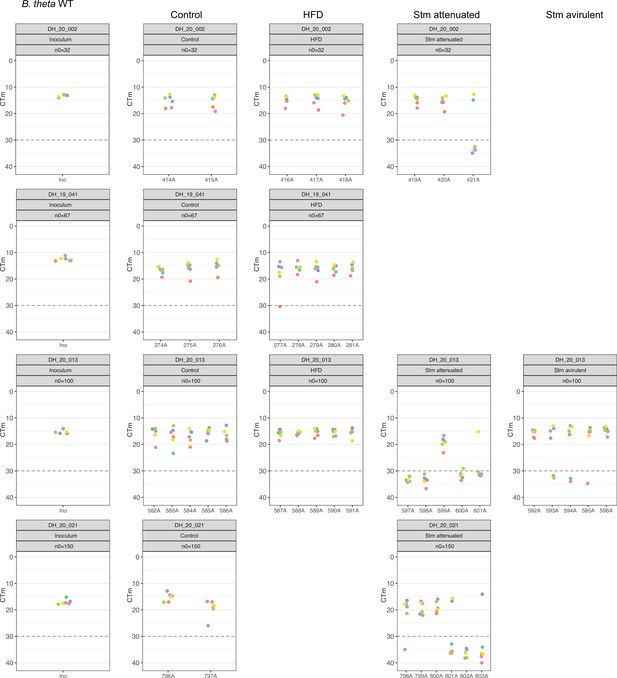
Distribution of B. theta WT barcoded strains in inoculum and after challenge.
Relative abundance of each barcoded strain spiked in the total inoculum, and in the cecum, of germ-free (GF) mice after 3 days of the challenge measure by qPCR. Gray dotted line marked the detection limit, set by qPCR amplification of GF mice cecum content without barcoded B. theta strains. On each experiment, n0 represents the approximate number of each barcoded strain spiked in the inoculum. In all experiments, we used 109 CFUs of the untagged strain in the inoculum to achieve the required starting abundance of each barcoded strain. Data are included in Figure 4—source data 1.
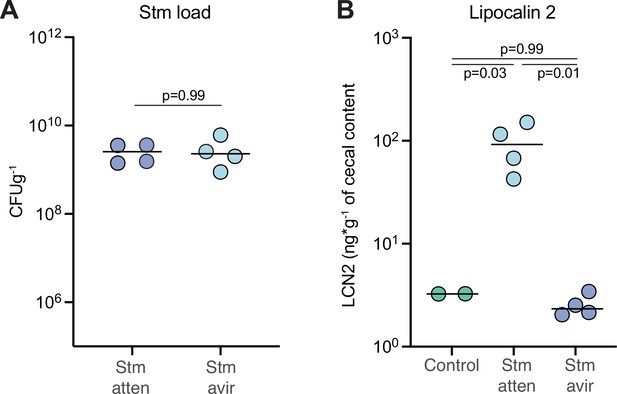
Inflammation induced by Stm attenuated challenge.
B. theta WT monocolonized mice, shown in Figure 4, were challenged with Stm avirulent (SPI-1/SPI-2 KO) or Stm attenuated (SPI2-KO). (A) Stm load in feces at day 3 of after infection. Points represent estimations of individual mice, and the horizontal line is the mean. p-Vvalues were obtained by Welch t-test. (B) Lipocalin-2 levels in cecum at day 3 post Stm infection. Points represent estimations of individual mice, and the horizontal line is the mean. p-Values were obtained by one-way ANOVA followed by Tukey’s honest significance test. Data are included in Figure 4—source data 1.
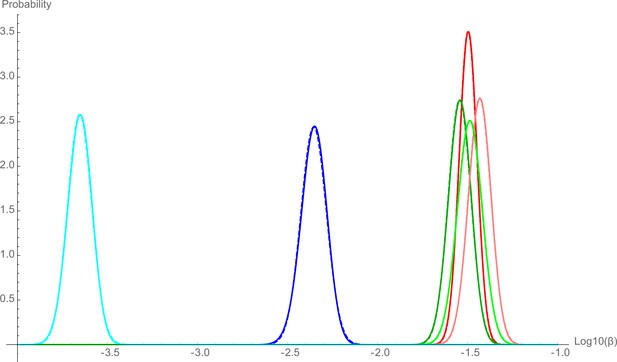
Distribution of the probability estimation of .
The colors represent experiments: low-complexity microbiota (LCM) WT (red), Oligo WT (green), specific pathogen free (SPF) WT (blue), LCM acapsular (pink), Oligo acapsular (light green), and SPF acapsular (light blue). Solid lines represent the numerical result for the probability distribution of using renormalized equation (6), the dashed lines represent the normal distribution with the same mean and same variance than the numerical distribution.
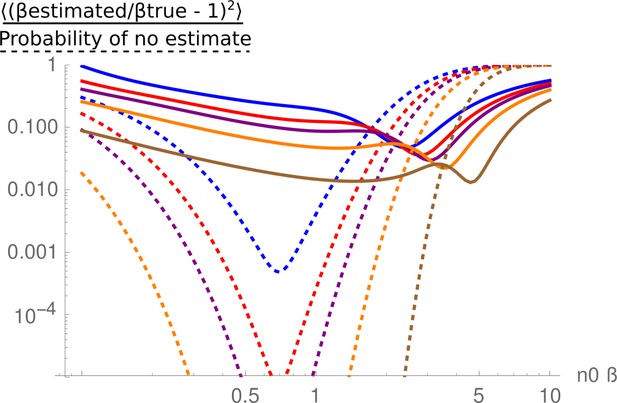
Mean relative square error in the estimate of , as a function of .
The solid-colored lines are the expression (17) for 12 (blue), 18 (red), 24 (purple), 40 (orange), and 120 (brown). The dotted lines are the probability that no estimate is given, either because all the tags are lost, or no tag is lost. They are the sum of equations (15) and (16).
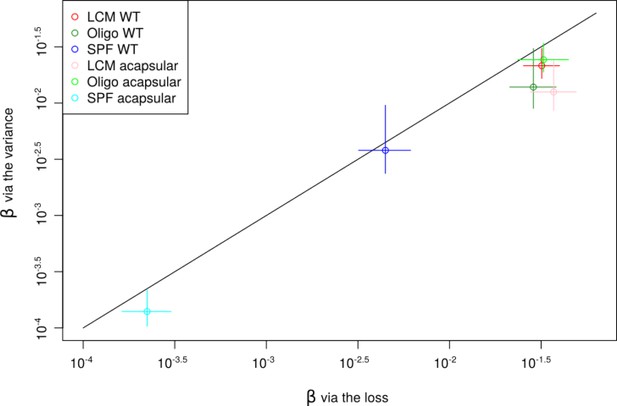
Comparison between the lineage survival probability estimated via the loss method and via the variance method.
The colors represent experiments: low-complexity microbiota (LCM) WT (red), Oligo WT (green), specific pathogen free (SPF) WT (blue), LCM acapsular (pink), Oligo acapsular (light green), and SPF acapsular (light blue). Circles represent best estimation and bars the error. The error bars are given via the direct study of the estimation probability for the loss method via the standard error between mice for the variance method.
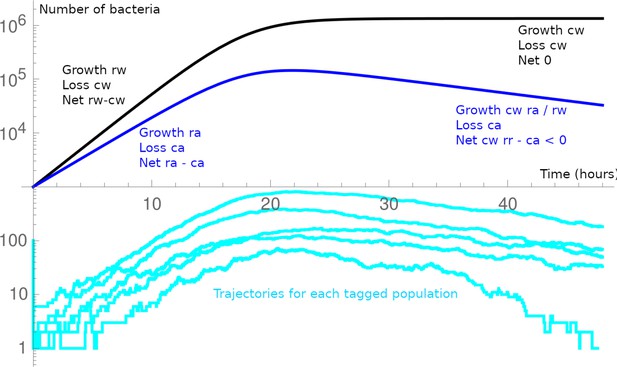
Schematic of the general dynamics in the minimal model.
The WT (black) and acapsular (blue) population are in competition. After a first bottleneck (at ), as the acapsular has a larger loss rate, its net growth rate is smaller. As the bacteria only interact through food, they follow their own dynamics until carrying capacity is reached, then WT is with a null net growth rate, and the acapsular, with its higher loss rate, has a negative net growth rate (we usually assume the absolute growth rate are equal, i.e., ). A barcoded population may be lost at the first bottleneck; or in the initial dynamics at low numbers; or towards the end of the experiment when the acapsular population decreases.
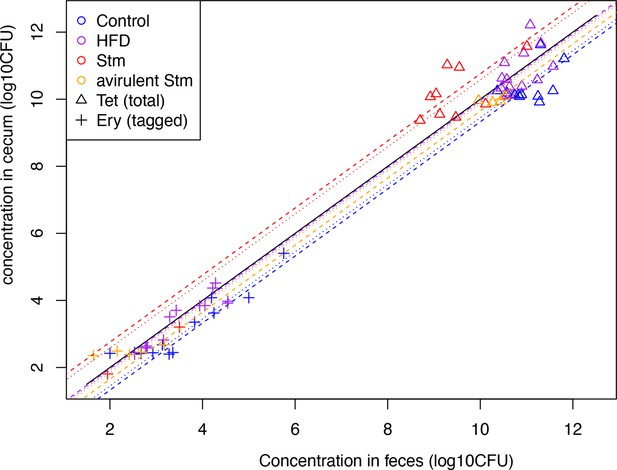
Concentration of B. theta in the cecum and the feces.
Concentration of untagged (triangle) and barcoded (cross) on day 3 after acute challenge (see color legend for each challenge). The dashed lines represent the average ratio considering only the untagged strain data, whereas the dotted lines represent the average of both untagged and barcoded strains (except the case of total loss of barcoded).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Bacteroides thetaiotaomicron) | B. theta WT | Porter et al., 2017 | Not applicable | tdk- Parent strain of B. theta VPI-5482 (ATCC 29148). Used to generate wild-type CPS mutants in this study. |
| Genetic reagent (B. thetaiotaomicron) | B. theta acapsular | Porter et al., 2017 | Not applicable | tdk- Δcps1-8 Acapsular B. theta with deletion of capsular polysaccharide locus. Used to generate acapsular mutants in this study. |
| Genetic reagent (B. thetaiotaomicron) | B. theta WT barcode 1 | This study | PRJEB57876 (ERP142888) | tdk-:: pNBU2-cat-ermG-GFP-WITS1 B. theta WT strain isogenic barcode 1; GFP tag; erythromycin resistant. |
| Genetic reagent (B. thetaiotaomicron) | B. theta WT barcode 2 | This study | PRJEB57876 (ERP142888) | tdk-:: pNBU2-cat-ermG-GFP-WITS2 B. theta WT strain isogenic barcode 2; GFP tag; erythromycin resistant. |
| Genetic reagent (B. thetaiotaomicron) | B. theta WT barcode 11 | This study | PRJEB57876 (ERP142888) | tdk-:: pNBU2-cat-ermG-GFP-WITS11 B. theta WT strain isogenic barcode 11; GFP tag; erythromycin resistant. |
| Genetic reagent (B. thetaiotaomicron) | B. theta WT barcode 17 | This study | PRJEB57876 (ERP142888) | tdk-:: pNBU2-cat-ermG-mCherry-WITS17 B. theta WT strain isogenic barcode 17; mCherry tag; erythromycin resistant. |
| Genetic reagent (B. thetaiotaomicron) | B. theta WT barcode 19 | This study | PRJEB57876 (ERP142888) | tdk-:: pNBU2-cat-ermG-mCherry-WITS19 B. theta WT strain isogenic barcode 19; mCherry tag; erythromycin resistant. |
| Genetic reagent (B. thetaiotaomicron) | B. theta WT barcode 21 | This study | PRJEB57876 (ERP142888) | tdk-:: pNBU2-cat-ermG-mCherry-WITS21 B. theta WT strain isogenic barcode 21; mCherry tag; erythromycin resistant. |
| Genetic reagent (B. thetaiotaomicron) | B. theta acapsular barcode 1 | This study | PRJEB57876 (ERP142888) | tdk- Δcps1-8:: pNBU2-cat-ermG-GFP-WITS1 B. theta acapsular strain with isogenic barcode 1; GFP tag; erythromycin resistant. |
| Genetic reagent (B. thetaiotaomicron) | B. theta acapsular barcode 2 | This study | PRJEB57876 (ERP142888) | tdk- Δcps1-8:: pNBU2-cat-ermG-GFP-WITS2 B. theta acapsular strain with isogenic barcode 1; GFP tag; erythromycin resistant. |
| Genetic reagent (B. thetaiotaomicron) | B. theta acapsular barcode 11 | This study | PRJEB57876 (ERP142888) | tdk- Δcps1-8:: pNBU2-cat-ermG-GFP-WITS11 B. theta acapsular strain with isogenic barcode 1; GFP tag; erythromycin resistant. |
| Genetic reagent (B. thetaiotaomicron) | B. theta acapsular barcode 17 | This study | PRJEB57876 (ERP142888) | tdk- Δcps1-8:: pNBU2-cat-ermG-mCherry-WITS17 B. theta acapsular strain with isogenic barcode 17; mCherry tag; erythromycin resistant. |
| Genetic reagent (B. thetaiotaomicron) | B. theta acapsular barcode 19 | This study | PRJEB57876 (ERP142888) | tdk- Δcps1-8:: pNBU2-cat-ermG-mCherry-WITS19 B. theta acapsular strain with isogenic barcode 19; mCherry tag; erythromycin resistant. |
| Genetic reagent (B. thetaiotaomicron) | B. theta acapsular barcode 21 | This study | PRJEB57876 (ERP142888) | tdk- Δcps1-8:: pNBU2-cat-ermG-mCherry-WITS21 B. theta acapsular strain with isogenic barcode 21; mCherry tag; erythromycin resistant. |
| Genetic reagent (B. thetaiotaomicron) | B. theta WT untagged | This study | PRJEB57876 (ERP142888) | tdk-:: pNBU2-bla-tetQb Untagged strain. B. theta WT strain with GFP insert; tetracycline resistant. |
| Genetic reagent (B. thetaiotaomicron) | B. theta acapsular untagged | This study | PRJEB57876 (ERP142888) | tdk- Δcps1-8:: pNBU2-bla-tetQb Untagged strain. B. theta acapsular strain with GFP insert; tetracycline resistant. |
| Genetic reagent (Salmonella enterica) | Stm attenuated (M3103) | Diard et al., 2017 | Not applicable | ΔssaV Salmonella enterica serovar Typhimurium (SL1344), attenuated (SPI-2 KO) |
| Genetic reagent (S. enterica) | Stm avirulent (M2702) | Diard et al., 2017 | Not applicable | ΔinvG ΔssaV Salmonella enterica serovar Typhimurium (SL1344), avirulent (SPI-1 KO and SPI-2 KO). |
| Sequence-based reagent | WITS01_F | Maier et al., 2014 | Forward primer barcoded strain 1 | acgacaccactccacaccta |
| Sequence-based reagent | WITS02_F | Maier et al., 2014 | Forward primer barcoded strain 2 | acccgcaataccaacaactc |
| Sequence-based reagent | WITS11_F | Maier et al., 2014 | Forward primer barcoded strain 11 | atcccacacactcgatctca |
| Sequence-based reagent | WITS17_F | Maier et al., 2014 | Forward primer barcoded strain 17 | tcaccagcccaccccctca |
| Sequence-based reagent | WITS19_F | Maier et al., 2014 | Forward primer barcoded strain 19 | gcactatccagccccataac |
| Sequence-based reagent | WITS21_F | Maier et al., 2014 | Forward primer barcoded strain 21 | acaaccaccgatcactctcc |
| Sequence-based reagent | ydgA_R | Maier et al., 2014 | Common reverse primer for all tagged strain | ggctgtccgcaatgggtc |
| Sequence-based reagent | BTt70-CHF | Jacobson, 2017 | pNBU2 vector genome integration test | TTCAAATTGCTCGGTAAAGCTC |
| Sequence-based reagent | BTt70-CHR | Jacobson, 2017 | pNBU2 vector genome integration test | AAAACCTTGATTTTACGGGAC |
| Sequence-based reagent | BTt71-CHF3 | Jacobson, 2017 | pNBU2 vector genome integration test | TTCGAGGAATGAAGCATCTCCGTA |
| Sequence-based reagent | BTt71-CHR3 | Jacobson, 2017 | pNBU2 vector genome integration test | ACCGTTCCGATTCAATTTCGT |
| Sequence-based reagent | IntN2BTt71-CHF3 | Jacobson, 2017 | pNBU2 vector genome integration test | TTTCCGGCTCTCCAATGCAA |
Additional files
-
Supplementary file 1
Genomic variants (SNPs, small insertions, and deletions) in B. theta strains whole-genome sequences.
Whole-genome sequencings of all barcoded and untagged strains (WT and acapsular) were mapped against Bacteroides thetaiotaomicron strain VPI 5482 genome (CP092641.1) to identify genetics variants. Data are included in Figure 4—source data 1.
- https://cdn.elifesciences.org/articles/81212/elife-81212-supp1-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/81212/elife-81212-mdarchecklist1-v2.pdf





