Structure and flexibility of the yeast NuA4 histone acetyltransferase complex
Figures
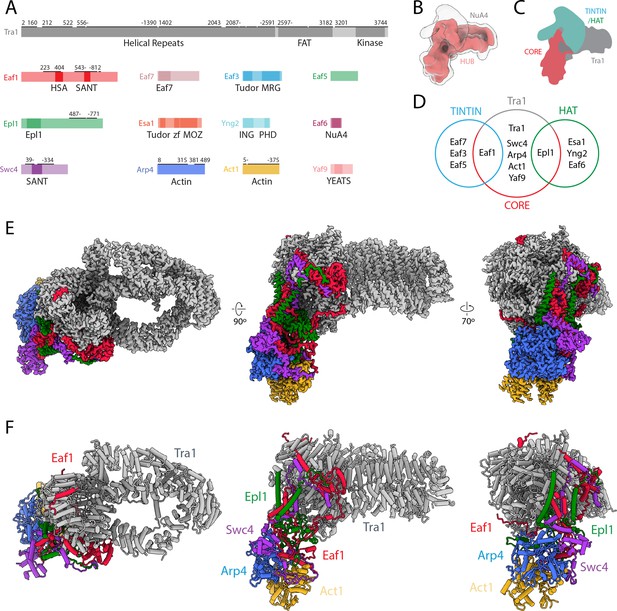
Structure of NuA4.
(A) Domain map of NuA4 subunits. Modeled regions are marked with a black bar; numbers indicate starting and ending residues. (B) Cryo-electron microscopy (cryo-EM) map in red showing the best-defined parts of NuA4. A transparent lower-threshold cryo-EM map is overlaid to show the flexible density likely corresponding to the histone acetyltransferase (HAT)/Trimer Independent of NuA4 involved in Transcription Interactions with Nucleosomes (TINTIN) modules. (C) Cartoon representation of NuA4 modules. (D) Venn diagram showing NuA4 subunit organization across different complex modules. Subunits in the core attach to Tra1 and act to tether the TINTIN and HAT modules to the complex. (E) Cryo-EM map of the NuA4 hub with individual subunits colored. (F) Model of the NuA4 hub with individual subunits colored and labeled.
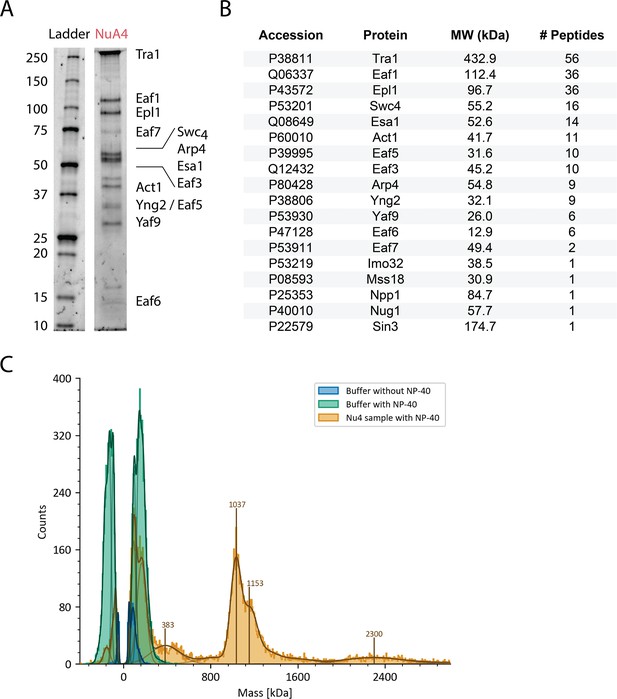
Purification of NuA4.
(A) Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) (Bio-Rad 4–20%) of purified S. cerevisiae NuA4, stained with Flamingo (Bio-Rad). (B) Mass spectrometry analysis showing the presence of all NuA4 subunits in purified sample. (C) Mass photometry analysis showing the molecular weights of particle species in NuA4 purification.
-
Figure 1—figure supplement 1—source data 1
Uncropped – sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) (Bio-Rad 4–20%) of purified S. cerevisiae NuA4, stained with Flamingo (Bio-Rad).
For panel A.
- https://cdn.elifesciences.org/articles/81400/elife-81400-fig1-figsupp1-data1-v2.zip
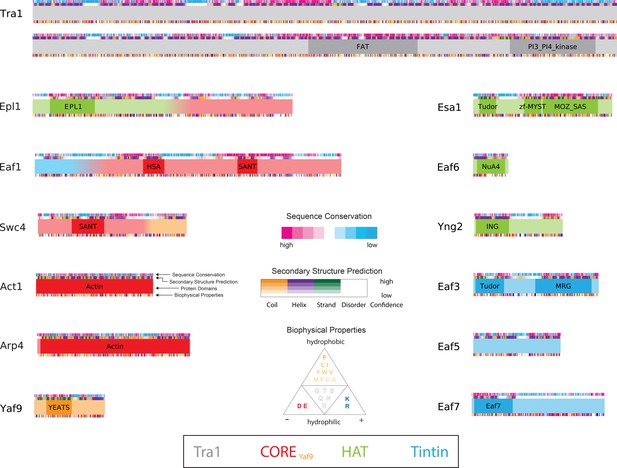
Protein domains of NuA4.
Domain maps of NuA4 subunits. For each bar, from top to bottom, are Consurf, PSIPRED/DISOPRED, PFAM, and amino acid biophysical properties (legend in the center bottom of the figure). Conservation scores ranging from high to low (pink to white to blue) were generated using the Consurf server (Ashkenazy et al., 2010; Ashkenazy et al., 2016). Predicted secondary structure based on PSIPRED and DISOPRED3 results (purple for helix, green for strand, orange for coil, and white for disordered) (Buchan et al., 2013; Buchan and Jones, 2019; Jones and Cozzetto, 2015). PFAM-predicted domains are labeled in bold (El-Gebali et al., 2019). Biophysical properties are colored based on amino acids hydrophilicity and charge: white-gold (hydrophilic–hydrophobic), red (negatively charged) and blue (positively charged).
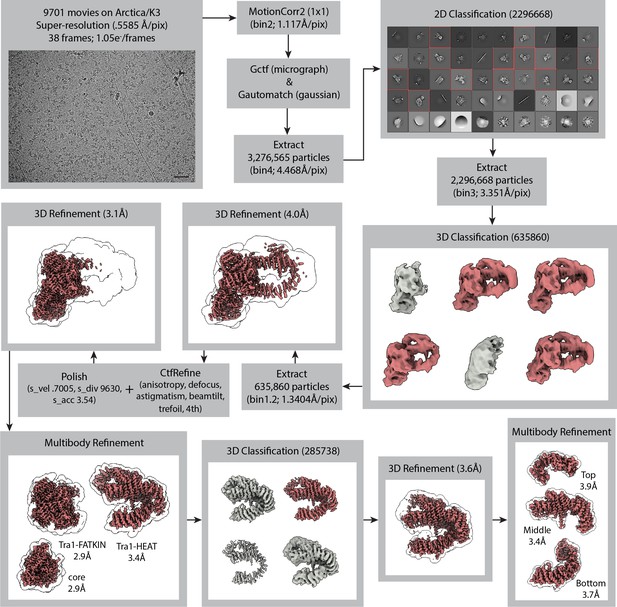
Cryo-electron microscopy (cryo-EM) data collection and processing for NuA4.
Cryo-EM data collection and processing for NuA4. Particles from class averages outlined in red and 3D classes colored red were selected for further processing. Particles that went into gray classes were eliminated from further processing. The final global refinement yielded a map with an overall resolution of 3.07 Å. To improve map quality in the flexible region of Tra1, multibody refinement was performed for three sections, followed by 3D classification of the partially signal-subtracted particles. The lobes of NuA4, termed core, Tra1-FATKIN, and Tra1-HEAT were refined to resolutions 2.9, 2.9, and 3.4 Å, respectively. The regions of Tra1-HEAT, termed top, middle, and bottom were refined to resolutions 3.9, 3.4, and 3.7 Å, respectively. The scale bar in bottom right of the micrograph image is 500 Å.
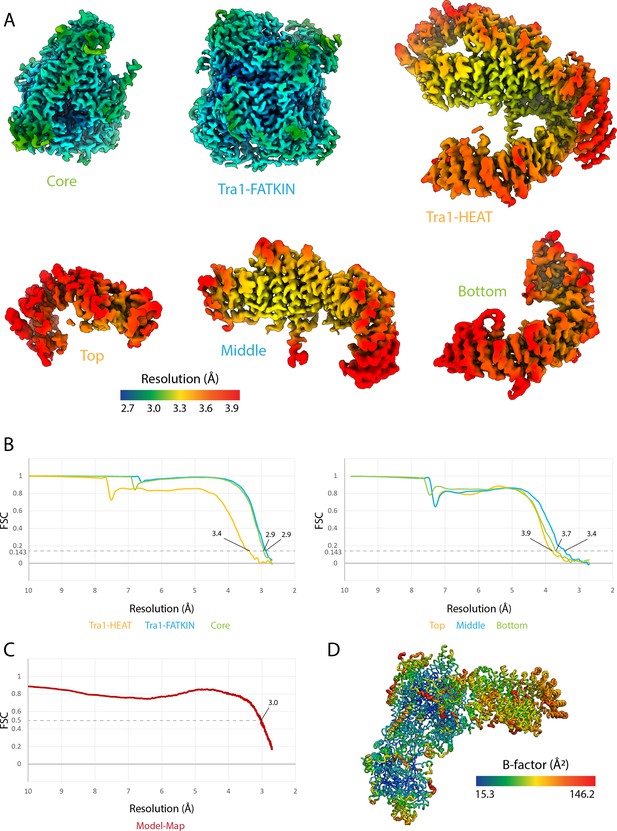
NuA4 structure model validation.
(A) Multibody maps colored by local resolution (Zivanov et al., 2018). Maps in the top row are from multibody refinement of NuA4. Maps in the bottom row are from multibody refinement of the Tra1-HEAT region. (B) Half-map FSC curves of the NuA4 multibody regions (left) and Tra1-HEAT multibody regions (right) (Rosenthal and Henderson, 2003). (C) Model-map FSC curve (map generated from a composite of multibody maps: core, Tra1-FATKIN, Tra1-HEAT). (D) Model of the core of RSC colored according to refined B-factors.
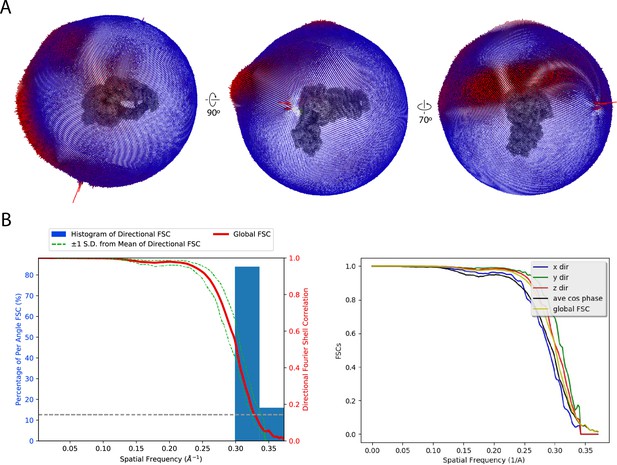
Euler angle distribution and map anisotropy.
(A) Euler angle distribution plots shown in three views. (B) 3DFSC plot generated using 3DFSC server for NuA4 consensus refinement (Tan et al., 2017).
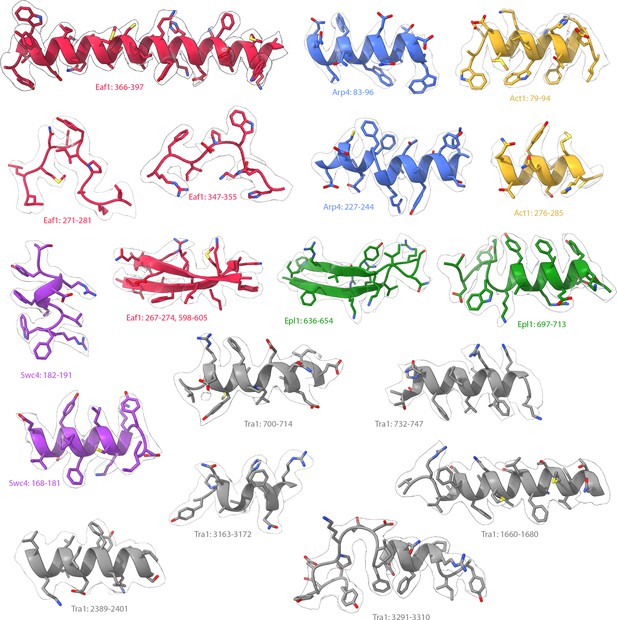
Model fit for NuA4.
Transparent regions of the consensus map of NuA4, segmented, with built atomic models shown.
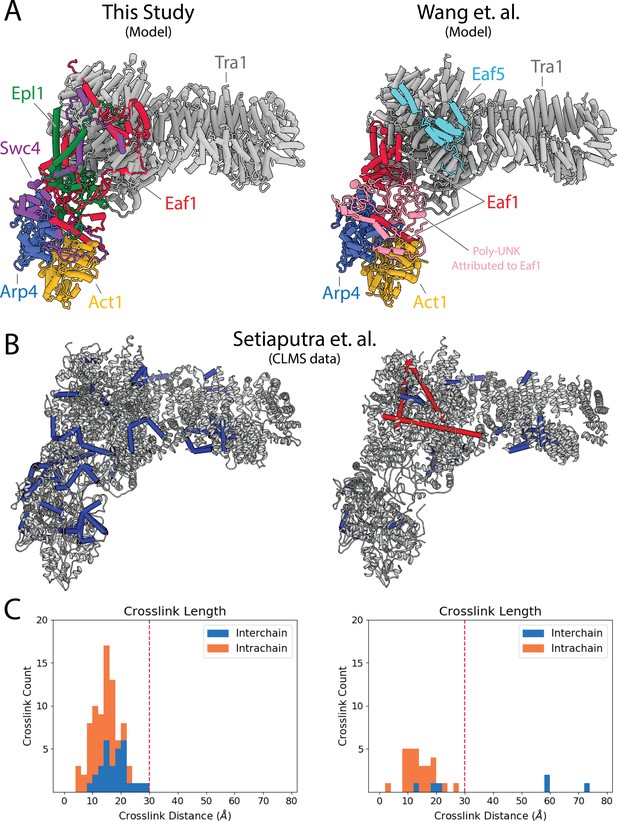
Comparison with previous NuA4 model and CX-MS validation.
(A) Comparison between the NuA4 model in this study (left) and that of Wang et al. (right) (Wang et al., 2018). In contrast to the previous model, we identify Swc4 and Epl1 within the rigid hub, while do not assign any density to Eaf5. (B) Cross-linking mass spectrometry (CX-MS) data of NuA4 (Setiaputra et al., 2018) mapped on the two models. Crosslinks with a distance >30 Å between alpha carbons are colored red, those with a distance <30 Å are colored blue (Kosinski et al., 2015). (C) Graph displaying the distribution of crosslink distances between the two models (Goddard et al., 2018). Interchain crosslinks are shown in blue, intrachain crosslinks in orange. A vertical line indicates a cutoff at 30 Å, the distance considered reasonable for DSS crosslinks.
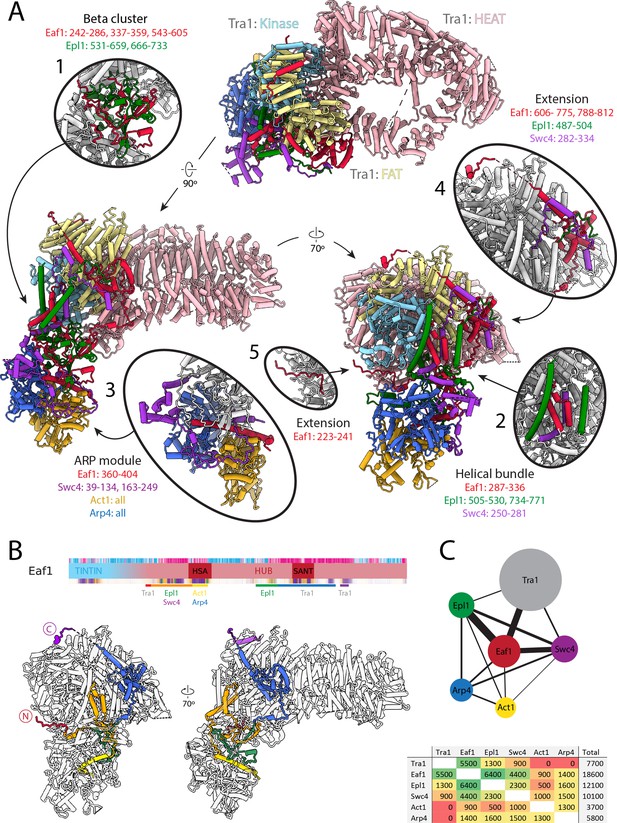
Architecture of the NuA4 central hub.
(A) Structure of NuA4 with subunits of the core and Tra1 domains in different colors. Within Tra1, the pseudo-kinase domain is colored light blue, the FAT domain is colored pale yellow, and the HEAT domain is colored pink. The FAT and pseudo-kinase domains form the bulk of the interactions with the NuA4 core, highlighted in subpanels (1, 2, 4, and 5). Organization of the NuA4 core can be seen in subpanels (1, 2, and 3). Arp module containing Arp4 and Act1 assemble onto the HSA helix of Eaf1 (seen in subpanel 3). (B) Top: Eaf1 domain map, as introduced in Figure 1—figure supplement 2. Bars underneath are colored to indicate its protein interactions. Bottom: depiction of Eaf1 interaction with NuA4 subunits. Sections of Eaf1 are colored in a rainbow from N to C terminus with different colors representing regions with different protein–protein interactions. (C) Top: schematic representation of contacts between NuA4 subunits. The width of each line is proportional to the contact area between subunits. Bottom: table showing the contact area (Å2) between NuA4 subunits, colored from red (minimal contact) to green (maximal contact).
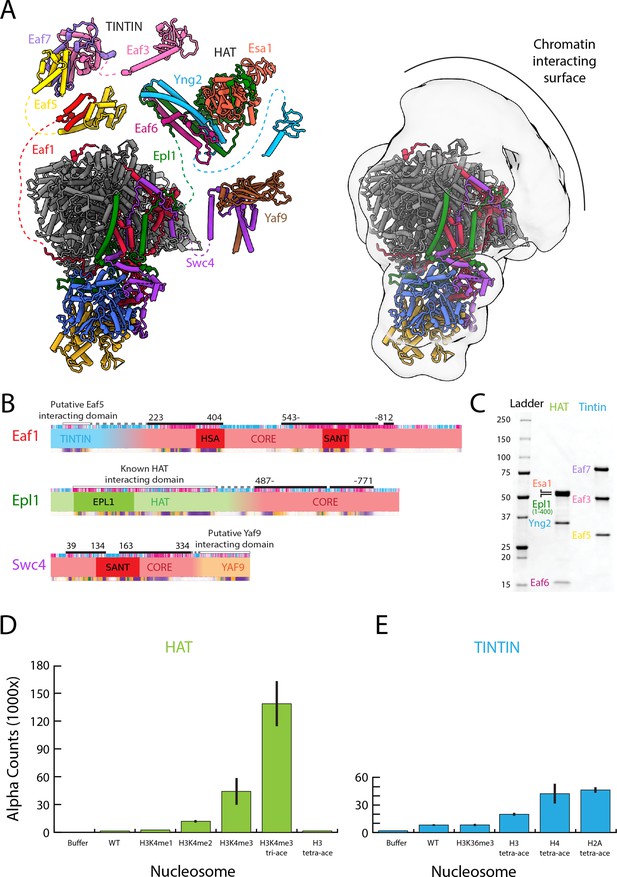
The NuA4 central hub tethers the nucleosome-interacting modules.
(A) Model for NuA4 complex organization (NuA4 central hub model from the present cryo-electron microscopy (cryo-EM) structure, with the rest of the models from AlphaFold2 prediction) (Jumper et al., 2021; Mirdita et al., 2022). Spatial constraints are imposed on the position of flexible modules by the length of the linker to the connecting amino acids resolved in the structure. Additional low-resolution density adjacent to the NuA4 hub suggests the approximate location of the flexible modules. (B) Domain map showcasing the subunits that link the Trimer Independent of NuA4 involved in Transcription Interactions with Nucleosomes (TINTIN), histone acetyltransferase (HAT), and YAF9 modules to the HUB. (C) Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) (Bio-Rad 4–20%) of purified NuA4 TINTIN and HAT modules, stained with InstantBlue (Expedeon). (D) dCypher assay results of nucleosome discovery screen for the purified HAT module. Error bars calculate from duplicate experiments, (E) dCypher assay result of nucleosome discovery screen for the purified TINTIN module. Error bars calculate from duplicate experiments.
-
Figure 3—source data 1
Uncropped – sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) (Bio-Rad 4–20%) of purified NuA4 Trimer Independent of NuA4 involved in Transcription Interactions with Nucleosomes (TINTIN) and histone acetyltransferase (HAT) modules, stained with InstantBlue (Expedeon).
For panel C.
- https://cdn.elifesciences.org/articles/81400/elife-81400-fig3-data1-v2.zip
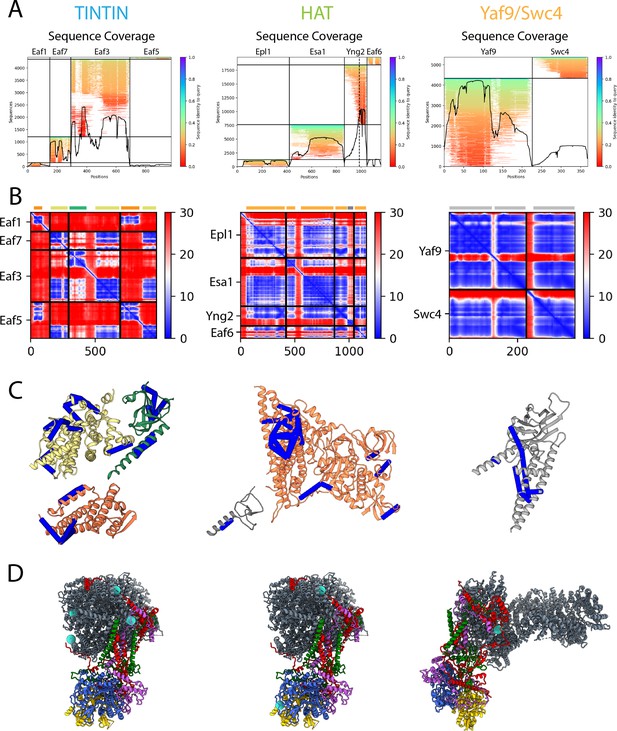
AlphaFold2 prediction and validation of the flexible NuA4 modules.
AlphaFold2 prediction for the module structures of Trimer Independent of NuA4 involved in Transcription Interactions with Nucleosomes (TINTIN) (left), histone acetyltransferase (HAT) (middle), and Yaf8/Swc4 (right) (Jumper et al., 2021; Mirdita et al., 2022). (A) MSA coverage (MSA search and alignment using Jackhammer). (B) Predicted alignment error (blue high, red low). Colored bars on top of the graph indicate separate regions of interaction within the modules. (C) AlphaFold2 models (ordered regions) with crosslinks mapped (blue <30 Å, red >30 Å) (Setiaputra et al., 2018; Kosinski et al., 2015). Models colored based on the AlphaFold2 predicted interacting regions (as seen in panel B). (D) NuA4 crosslinking position with the given module indicated by cyan spheres (Setiaputra et al., 2018).
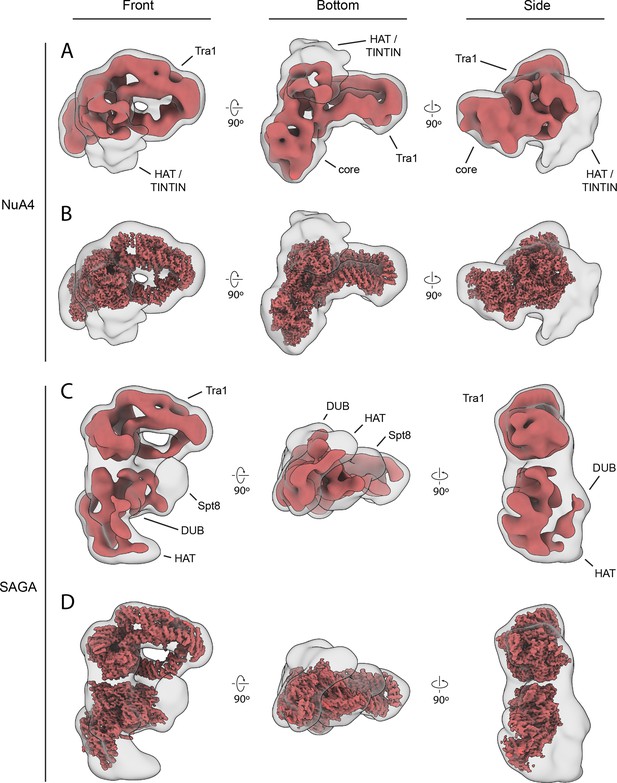
NuA4 and SAGA both have flexible nucleosome engaging domains.
Negative stain reconstructions of NuA4 (A) and SAGA (C) displayed with two thresholds: high (solid red) and low (transparent white). The low threshold maps show extra density for the flexible chromatin-interacting modules of the two complexes. SAGA domain assignments are based on previous structural studies (Setiaputra et al., 2015; Papai et al., 2020; Wang et al., 2020). Cryo-electron microscopy (cryo-EM) reconstructions (solid red) of NuA4 (B) and SAGA (D) fit into low threshold negative stain reconstructions (transparent white). SAGA cryo-EM reconstructions from EMD-10412 (Wang et al., 2020).
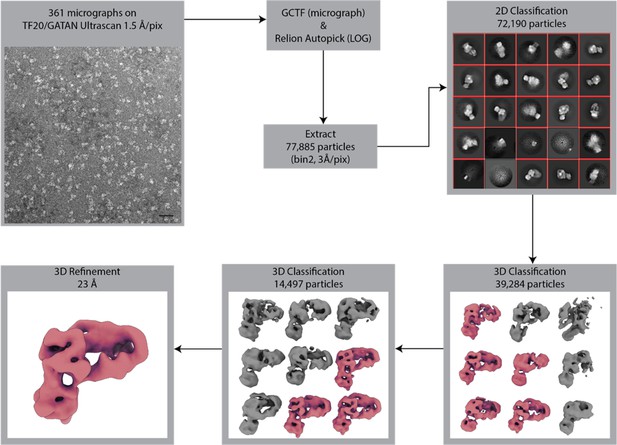
NuA4-negative stain data processing.
Negative stain EM data collection and processing for NuA4. Collected images were subjected to contrast transfer function (CTF) estimation using GCTF and particles were picked and analyzed by 2D and 3D classification using Relion (Zhang, 2016; Zivanov et al., 2018). The scale bar in bottom right of the micrograph image is 500 Å.
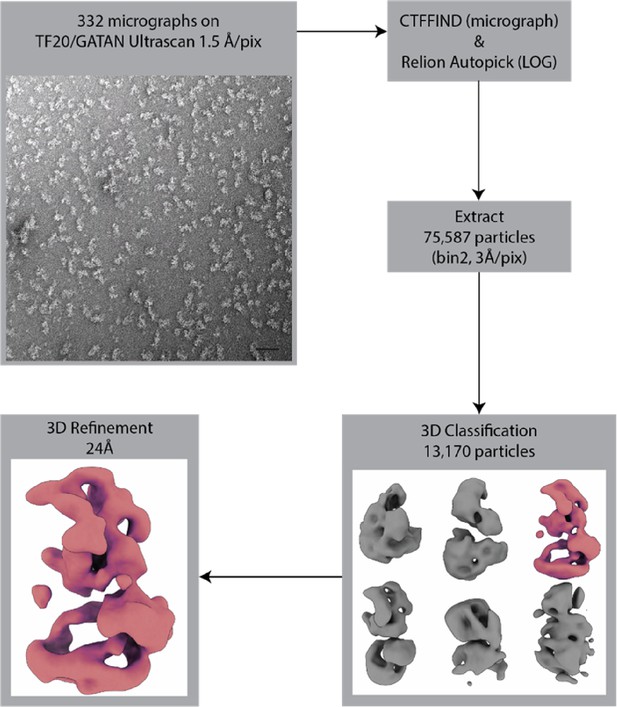
SAGA-negative stain data processing.
Negative stain EM data processing for SAGA. Collected images were subjected to contrast transfer function (CTF) estimation using GCTF and particles were picked and analyzed by 3D classification using Relion (Zhang, 2016; Zivanov et al., 2018). The scale bar in bottom right of the micrograph image is 500 Å.
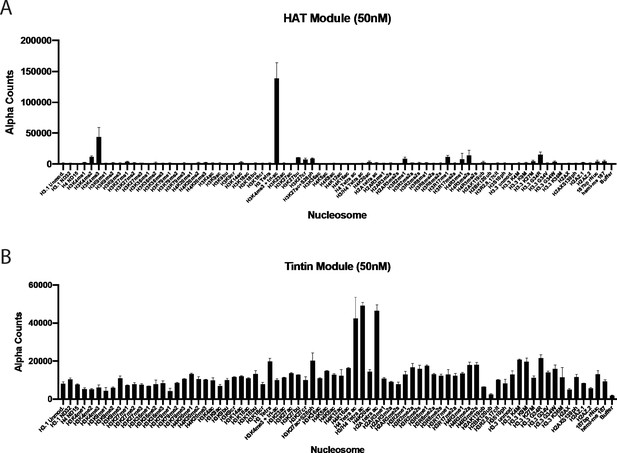
Complete results of dCypher nucleosome discovery screen.
(A) dCypher nucleosome discovery screen-binding profile for purified histone acetyltransferase (HAT) module. Abbreviations for histone modifications are as follows: me, methyl; ac, acetyl; bu, butyryl; cr, crotonyl; ph, phosphoryl; ub, ubiquitin; cit, citrulline. Error bars calculate from duplicate experiments, (B) dCypher nucleosome discovery screen-binding profile for purified Trimer Independent of NuA4 involved in Transcription Interactions with Nucleosomes (TINTIN) module. Same abbreviations as panel A. Error bars calculate from duplicate experiments.
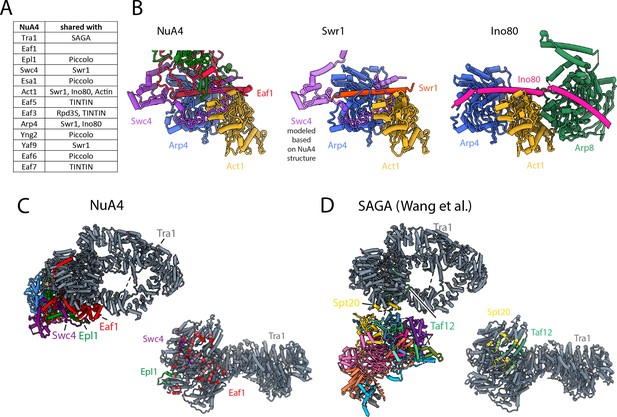
NuA4 subunits shared with other Saccharomyces cerevisiae complexes.
(A) Table showing protein subunits of NuA4 and the complexes in yeast that share these subunits. (B) Comparison of Arp4–Act1 interactions with HSA helix in NuA4 (Eaf1 subunit), SWR1 (Swr1 subunit), and INO80 (Ino80 subunit) (Cao et al., 2016; Knoll et al., 2018). (C) Left/top: model of NuA4 depicting attachment of the core to Tra1. Right/bottom: the Tra1 subunit of NuA4 colored according to which protein chain contacts it. (D) Left/top: model of SAGA depicting attachment of the core to Tra1. Right: the Tra1 subunit in SAGA colored according to which protein chain contacts it (Papai et al., 2020; Wang et al., 2020).
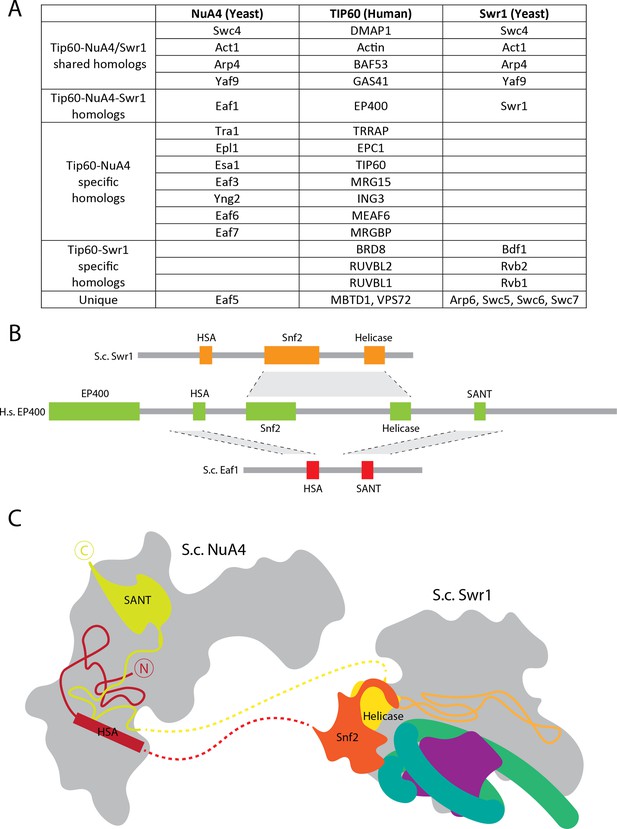
A model of H.s.TIP60.
(A) Table of homologous protein subunits in budding yeast (S.c.; Saccharomyces cerevisiae) NuA4, S.c. Swr1 and human (H.s.; Homo sapiens) TIP60. (B) Domain similarities between S.c. Swr1 and S.c. NuA4 with H.s. EP400. (C) Schematic for the proposed path of EP400 based on the structures of yeast NuA4 and Swr1 (Willhoft et al., 2018).
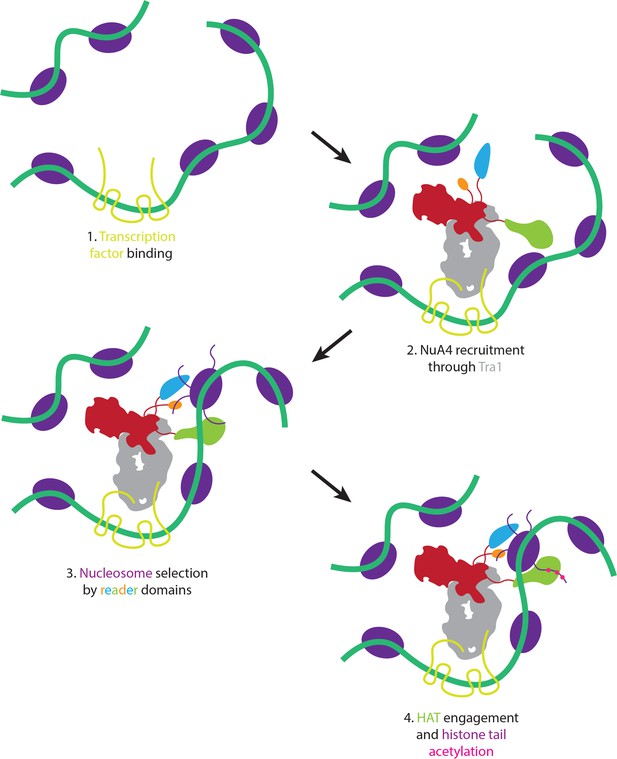
Proposed model of nucleosome selection and propagation of acetylation.
Model of NuA4 chromatin localization and histone acetylation. NuA4 is recruited to genomic loci through interaction of Tra1 with site-specific transcription factors. Once recruited, the flexible reader domains interrogate nearby nucleosomes. Nucleosomes bearing the proper chemical marks are preferentially acetylated.
Tables
Refinement table.
| Data collection, map and model refinement, validation | |
|---|---|
| Data collection | |
| Dataset | NuA4 |
| Microscope | Talos Arctica |
| Stage type | Autoloader |
| Voltage (kV) | 200 |
| Detector (mode) | Gatan K3 (super-resolution) |
| Pixel size (Å) | 1.117 (0.5585) |
| Electron dose (e−/Å2) | 38 |
| Reconstruction | |
| Software | RELION |
| Particles | 635,860 |
| Box size (pixels) | 320 |
| Map resolution (Å) | 3.1 |
| Map sharpening B-factor (Å2) | −78 |
| Coordinate refinement | |
| Software | PHENIX |
| Resolution cutoff (Å) | 3.0 |
| FSCmodel-vs-map = 0.5 (Å) | 3.0 |
| Model-data fit (CCmask) | 0.84 |
| Model | |
| Residues | |
| Protein | 5259 |
| B-Factors | |
| Protein | 65.28 |
| RMS deviations | |
| Bond lengths (Å) | 0.002 |
| Bond angles (°) | 0.507 |
| Validation | |
| Molprobity score | 1.72 (88th) |
| Molprobity clashscore | 6.88 (87th) |
| Rotamer outliers (%) | 0.02 |
| Cβ deviations (%) | 0 |
| Ramachandran plot | |
| Favored (%) | 95.03 |
| Allowed (%) | 4.97 |
| Outliers (%) | 0 |





