CaV1 and CaV2 calcium channels mediate the release of distinct pools of synaptic vesicles
Figures
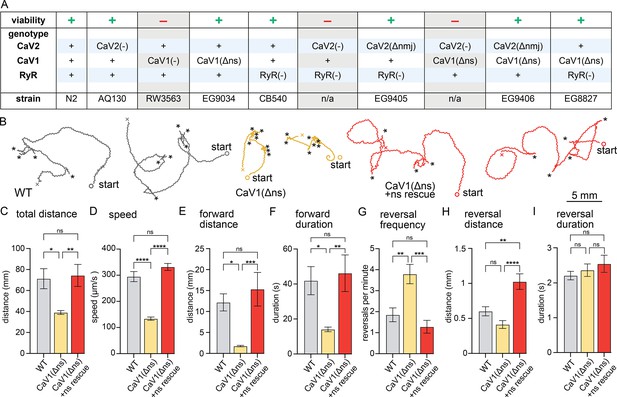
nervous system CaV1 is required for normal behavior and locomotion.
(A) Viability of calcium channel double mutants. (B) Worm Tracks. Healthy animals were tracked for 5 minutes with a frame rate of 8 frames per second. The path the animal created was plotted, starting point is indicated, asterisks represent reversal events. (C) Total average distance animals travelled per animal during the 5 minute interval by genotype. Wild-type 71.2±9.5 mm. CaV1(Δns) 39.0±1.8 mm. CaV1(Δns)+rescue 74.4±10.5 mm. (D) Average speed, including both forward and backward bouts but excluding pauses, for the duration of the assay. Wild-type 294.9 µm/s±19.4 µm/s. CaV1(Δns) 133.8 µm/s±6.3 µm/s. CaV1(Δns)+rescue 331.7 µm/s±13.3 µm/s. (E) Average distance of forward locomotion between reversal events that animals travelled by genotype. Wild-type 12.2 mm ±2.0 mm. CaV1(Δns) 17.7 mm ±1.7 mm. CaV1(Δns)+rescue 15.3 mm ±4.0 mm. (F) Average duration of forward run between reversal events. Wild-type 41.9±8 s. CaV1(Δns) 14.1±1.3 s. CaV1(Δns)+rescue 46.2±10.5 s. (G) Average number of reversal events per minute exhibited by animals by genotype. Wild-type 1.9±0.3 events. CaV1(Δns) 3.8±0.5 events. CaV1(Δns)+rescue 1.3±0.3 events. (H) Average distance travelled in reverse per animal by genotype. Wild-type 601.9±65.1 µm. CaV1(Δns) 413.2±56.8 µm. CaV1(Δns)+rescue 1026±111.1 µm. (I) Average duration of reversal run. Wild-type 2.2 +- 0.1 s. CaV1(Δns) 2.4+/-0.2 s. CaV1(Δns)+rescue 2.5 +- 0.2 s. Wild-type n=11, CaV1(Δns) n=16, CaV1(Δns)+rescue n=13. Error bars reported in SEM. Genotypes were blinded. One-way ANOVA with Tukey’s multiple comparisons was used to calculate p-value. *p<0.05, **p<0.005, ***p<0.001, ****p<0.0005. Data available as Figure 1—source data 1.
-
Figure 1—source data 1
Worm behaviorial data.
- https://cdn.elifesciences.org/articles/81407/elife-81407-fig1-data1-v2.zip
-
Figure 1—source data 2
Table of calcium channel mutant viability, genotype, and strain designation.
- https://cdn.elifesciences.org/articles/81407/elife-81407-fig1-data2-v2.zip
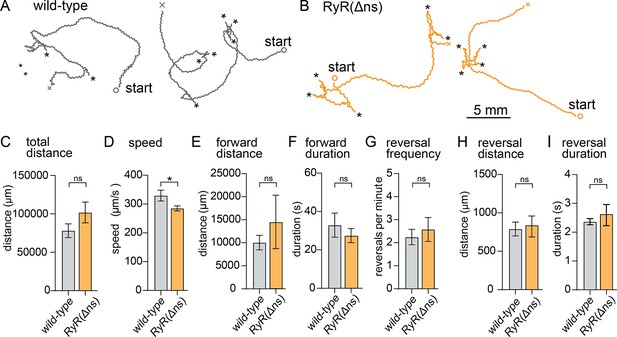
RyR neuronal isoform knockout moves slower than wildtype.
(A) Example track from WT. The path the animal created was plotted, starting point is indicated, asterisks represent reversal events. (B) Example track from RyR(Δns). The path the animal created was plotted, starting point is indicated, asterisks represent reversal events. (C) Total distance. Wild-type 78.2±8.9 mm RyR(Δns) 10.2±13.6 mm. (D) Average speed. Wild-type 329.3 µm/s±18.7 µm/s RyR(Δns) 285.1±8.8 µm/s. (E) Forward distance. Wild-type 10.0 mm ±1.6 mm RyR(Δns) 14.5±5.8 mm. (F) Forward run duration. Wild-type 32.8±6.3 s. RyR(Δns) 27.5±3.6 s. (RyR(Δns) n=11). (G) Reversals per minute. Wild-type 2.2±0.3 events. RyR(Δns) 2.6±0.5 events. (H) Distance travelled in reverse. Wild-type 789.0±90 µm. RyR(Δns) 838.8±136.7 µm. (RyR(Δns) n=11). (I) Duration of reversal run. Wild-type 2.4 +- 0.1 s. RyR(Δns) 2.6±0.4 s. Wild-type n=15 RyR(Δns) n=17. Error bars reported in SEM. Genotypes were blinded. Brown-Forsyth ad Welch ANOVA with Dunnett’s T3 multiple comparisons was used to calculate p-value. *p<0.05, **p<0.005, ***p<0.001, ****p<0.0005.
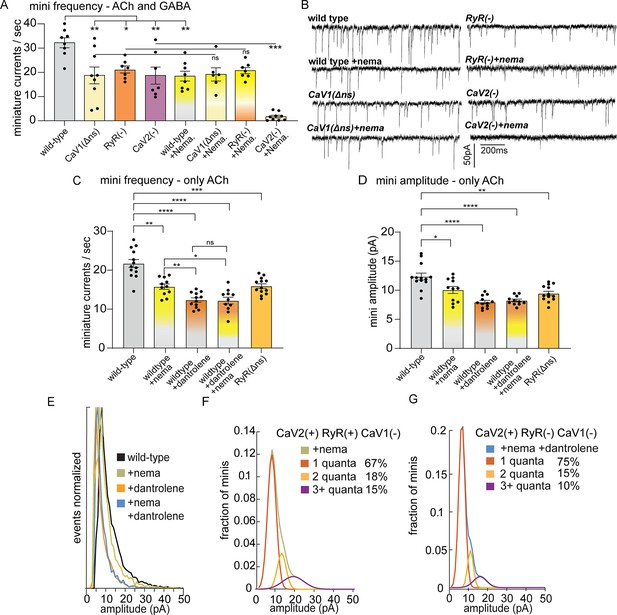
CaV2 and CaV1-RyR fuse different vesicle pools.
(A) Spontaneous miniature currents mediated by CaV1 and RyR are inhibited by nemadipine. Wild-type: 32.2±2.1 minis/s n=8, wild type with nemadipine (10 µM): 18.4±2.1 mini/s n=8. CaV1(Δns): 18.7±3.1 minis/s n=9, with nemadipine 19.1±2.7 mini/s n=6. RyR(-): 20.9±1.5 minis/s n=7, with nemadipine 20.7±1.3 mini/s n=7. CaV2(-): 18.7±3.5 minis/s n=7, with nemadipine 1.7±0.6 mini/s n=9. One-way ANOVA with Dunnett’s multiple comparisons test and one-way ANOVA with Tukey’s multiple comparisons tests were used to calculate significance. GABA and ACh release generated inward currents in this preparation. (B) Sample traces of spontaneous release in 0.5 mM extracellular calcium. GABA and ACh release generated inward currents in this preparation. (C) Spontaneous miniature currents from acetylcholine release are reduced with pharmacological block of CaV1 and RyR. Wild-type: 21.7+/-1 mini/s n=13. Wild-type +nemadipine (10 µm): 15.8+/-0.7 mini/s n=11. Wild-type +dantrolene: 12.36+/-0.6 mini/s n=12. Wild-type +nemadipine and dantrolene: 12.2+/-0.9 mini/s n=11. RyR(ns-): 15.9+/-0.6 mini/s n=13. Brown-Forsythe and Welch ANOVA with T3 Dunnett’s multiple comparisons test were used to calculate significance. (D) Dantrolene reduces miniature current amplitude from acetylcholine release. Wild-type: 12.4+/-0.6 pA n=13. Wild-type +nemadipine (10 µm): 10.1+/-0.6 pA n=11. Wild-type +dantrolene: 8.0+/-0.3 pA n=12. Wild-type +nemadipine and dantrolene: 8.2+/-0.24 pA n=11. RyR(ns-): 9.5+/-0.4 pA n=13. Brown-Forsythe and Welch ANOVA with T3 Dunnett’s multiple comparisons test were used to calculate significance. (E) Frequency distribution of mini amplitudes from acetylcholine release in wild-type and with pharmacological block of CaV1 or RyR, normalized to mode. (F) Quantal analysis of post-synaptic amplitudes from wild-type animals treated with nemadipine. Mini amplitudes were transformed into 1 pA bins. The wild-type +nema distribution of amplitudes was fit with a three-term Gaussian convolution to isolate 1, 2 and 3 quantal events, each centered around the mode: 7±1 pa, 14±2 pa and 21±3 pa (khaki). 1-quanta (rust) accounted for 67% of fusions. 2-quanta (butterscotch) 18%. 3-quanta (violet) 15%. (G) Quantal analysis of post-synaptic amplitudes from wild-type animals treated with dantrolene and nemadipine. Mini amplitudes were transformed into 1 pA bins. The wild-type +nema distribution of amplitudes was fit with a three-term Gaussian convolution to isolate 1, 2, and 3 quantal events, each centered around the mode: 6±1 pa, 12±2 pa and 18±3 pa (blue). (blue). 1-quanta (rust) accounted for 75% of fusions. 2-quanta (butterscotch) 15%. 3-quanta (violet) 10%. For all recordings, Vm = –60 mV, 0.5 mM calcium. Error bars reported in SEM. *p<0.05, **p<0.005, ***p<0.001, ****p<0.0005. Data available as Figure 2—source data 1.
-
Figure 2—source data 1
Electrophysiology data from calcium channel mutants with pharmacological block of calcium channels.
- https://cdn.elifesciences.org/articles/81407/elife-81407-fig2-data1-v2.zip
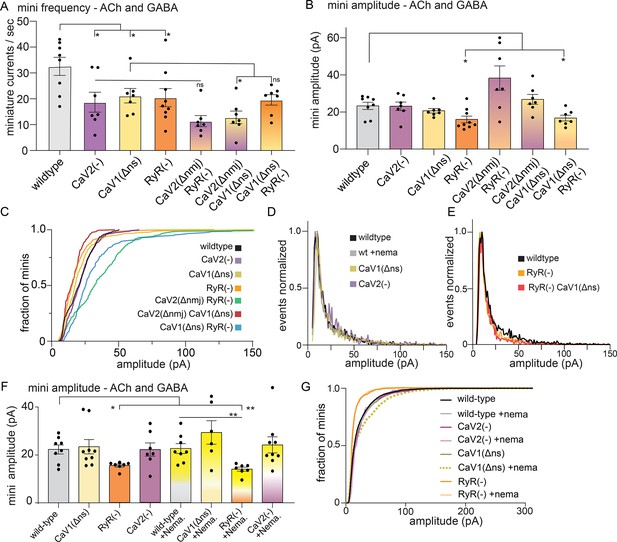
The ryanodine receptor acts in parallel to CaV2.
(A) CaV2 and CaV1-RyR contribute additively to tonic release. GABA and ACh release generated inward currents in these preparations. Muscles were voltage-clamped, and tonic miniature synaptic currents recorded in 0.5 mM extracellular calcium: wild type: 33±4 minis/s, n=8; CaV2(-): 19±4 minis/s n=7, CaV1(Δns): 21±3 minis/s n=7, RyR(-): 20±4 minis/s n=9. CaV2(Δnmj) RyR(-): 11±2 minis/s n=7, CaV2(Δnmj) CaV1(Δns): 13±2 minis/s n=7, CaV1(Δns) RyR(-): 20±2 minis/s n=7. Welch’s t-test was used to calculate significance (B) RyR is required for large-amplitude spontaneous events. At 0.5 mM calcium, wild-type 23±2 pA n=8, and CaV2(-): 23±2 pA n=7, CaV1(Δns): 21±1 pA n=7, RyR(-): 16±2 pA n=9. CaV2(Δnmj) RyR(-): 38±3 pA n=7, CaV2(Δnmj) CaV1(Δns): 26±3 pA n=7, and CaV1(Δns) RyR(-): 17±2 pA n=7. Welch’s t-test was used to calculate significance. (C) Cumulative distribution plot of mutant amplitudes. (D) Frequency distribution plot of CaV mutant miniature current amplitudes normalized to the mode. (E) Frequency distribution plot of ryanodine mutants with reduced amplitudes, normalized to mode. (F) The ryanodine receptor mediates large amplitude tonic miniature currents. Wild type 22.3±1.9 pA n=8, wild type with nemadipine 22.6±2.1 pA n=8. CaV2(-): 22.1±3.5 pA n=7, CaV2(-) with nemadipine 24.1±3.4 pA n=9. CaV1(Δns): 23.4±3.1 pA n=9, CaV1(Δns) with nemadipine 29.4±.9 pA n=6. RyR(-): 15.5±0.6 pA n=7, RyR(-) with nemadipine 14.0±0.9 pA n=7. One-way ANOVA with Dunnett’s multiple comparisons test and Welch’s t-test were used to calculate significance. GABA and ACh release generated inward currents in this preparation. (G) Cumulative distribution plot of miniature current amplitudes. GABA and ACh release generated inward currents in this preparation. Error bars reported in SEM. *p<0.05, **p<0.005, ***p<0.001, ****p<0.0005 Data available as Figure 2—figure supplement 1—source data 1.
-
Figure 2—figure supplement 1—source data 1
Electrophysiology data from calcium channel double mutants.
- https://cdn.elifesciences.org/articles/81407/elife-81407-fig2-figsupp1-data1-v2.zip
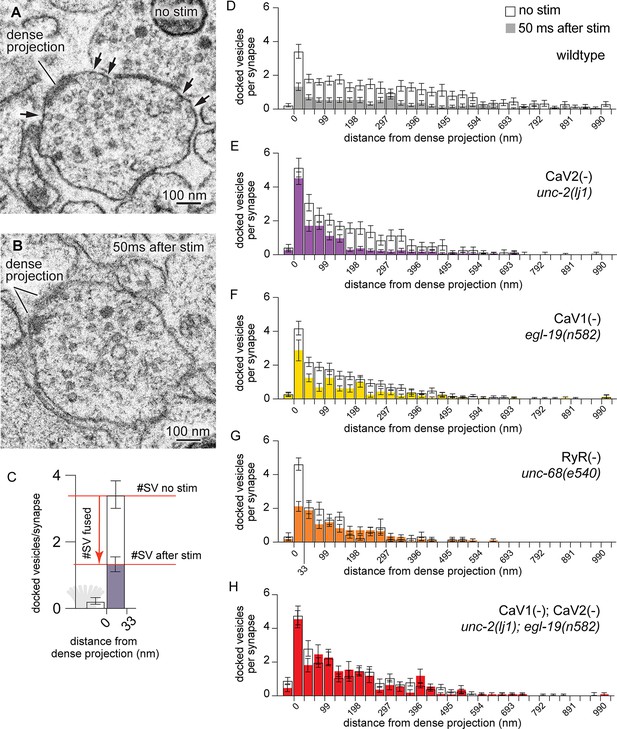
CaV2 and CaV1-RyR act at distinct vesicle release sites.
(A) Docked vesicles (black arrows) are present at synapses in electron micrographs of unstimulated animals. (B) Docked vesicles are reduced 50ms after channelrhodopsin stimulation. (C) Vesicle fusion. The number of synaptic vesicles that fuse can be calculated as the number of docked vesicles lost by stimulation. Dense projection indicated in gray in the 0 nm bin. (D–H) Average number of docked vesicles per synapse at a given distance from the dense projection with, or without, light stimulation of channelrhodopsin. (D) Wild-type animals exhibit fewer docked vesicles at all locations after stimulation. Wild type (no stimulation), n=26 synapses. Wild type (stimulated) n=24 synapses. (E) In the CaV2 null mutant unc-2(lj1), vesicles fuse greater than 33 nm from the dense projection; vesicle fusion is reduced directly adjacent to the dense projection. No stimulation n=14, stimulated n=27 synapses. (F) The CaV1 hypomorphic mutant egl-19(n582) exhibits reduced fusions at all distances. No stimulation n=29 synapses, stimulated n=16 synapses. (G) The RyR mutant unc-68(e540) exhibits fusions adjacent to the dense projection, but lacks fusions of lateral vesicles. No stimulation n=11 synapses, stimulated n=17 synapses. (H) The CaV1 CaV2 double mutant, egl-19(n582) unc-2(lj1), lacks fusion of all docked vesicles after stimulation. No stimulation n=24 synapses, stimulated n=17 synapses. Micrographs were segmented blind to treatment and genotype. Bin size was fixed at 33 nm to be consistent with our section thickness in case 3D reconstruction of synapses is required. Error bars SEM, N=2 animals for each condition. Data available as Figure 3—source data 1.
-
Figure 3—source data 1
Quantification of the number of docked vesicles in calcium channel mutants.
- https://cdn.elifesciences.org/articles/81407/elife-81407-fig3-data1-v2.zip
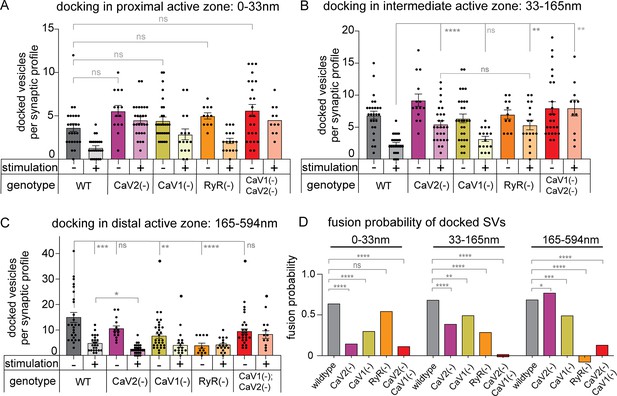
Statistical analysis of synaptic vesicle fusion in domains of the active zone.
(A) Docked vesicles adjacent to the dense projection in calcium channel mutants. Unstimulated: wild type: 3.7+/-0.5 vesicles. CaV2(-): 5.6+/-0.6 vesicles. CaV1(-): 4.5+/-0.4. vesicles. RyR(-): 5.0+/-0.4. CaV2(-) CaV1(-): 5.6+/-0.7. Stimulated: wild type: 1.3+/-0.2. CaV2(-): 4.8+/-0.4. CaV1(-): 3.1+/-0.7. RyR(-): 2.3+/-0.4. CaV2(-) CaV1(-): 5.0+/-0.6. (B) Docked vesicles in the intermediate active zone in calcium channel mutants. Unstimulated: wild type: 3.1+/-0.6 vesicles. CaV2(-): 9.2+/-1.0 vesicles. CaV1(-): 6.4+/-0.7 vesicles. RyR(-): 7.0+/-0.7. CaV2(-) CaV1(-): 5.2+/-0.7 vesicles. Stimulated: wild type: 1.5+/-0.3 vesicles. CaV2(-): 2.8+/-0.5 vesicles. CaV1(-): 3.2+/-0.4. RyR(-): 4.9+/-0.6. CaV2(-) CaV1(-): 8.0+/-1.2 vesicles. (C) Docked vesicles in the lateral active zone in calcium channel mutants. Unstimulated: wild-type: 16.7+/-1.9 vesicles. CaV2(-): 12.2+/-1.2 vesicles. CaV1(-): 7.6+/-0.9 vesicles. RyR(-): 3.8+/-1.0 vesicles. CaV2(-) CaV1(-): 9.4+/-1.0 vesicles. Stimulated: wild-type: 3.1+/-0.6 vesicles. CaV2(-):3.4+/-0.4 vesicles. CaV1(-): 3.9+/-0.8 vesicles. RyR(-): 4.1+/-0.6 vesicles. CaV2(-) CaV1(-): 8.2+/-1.5 vesicles. (D) Fusion probability of vesicles docked across the active zone in calcium channel mutants. To calculate the fusion probability at a zone, the mean number of docked vesicles after fusion was divided by the mean number of docked vesicles before fusion, and that result was subtracted from 1. Comparisons made with Chi-squared test. Wild type (no stimulation), n=26 synapses. Wild type (stimulated) n=24 synapses. CaV1(-) (no stimulation) n=29 synapses. CaV1(-) (stimulated) n=16 synapses. CaV2(-) (no stimulation) n=14, CaV2(-) (stimulated) n=27 synapses. RyR(-) (no stimulation) n=11 synapses. RyR(-) (stimulated) n=17 synapses. CaV2(-) CaV1(-) (control) n=24 synapses. CaV2(-) CaV1(-) (stimulated) n=17 synapses All multiple comparisons made with Brown Forsythe and Welch ANOVA with Dunnett’s T3. All single comparisons made with Welch’s T-test. Comparisons of fusion rates were performed using Chi-squared test. Error bars reported in SEM. *p<0.05, **p<0.005, ***p<0.001, ****p<0.0005.
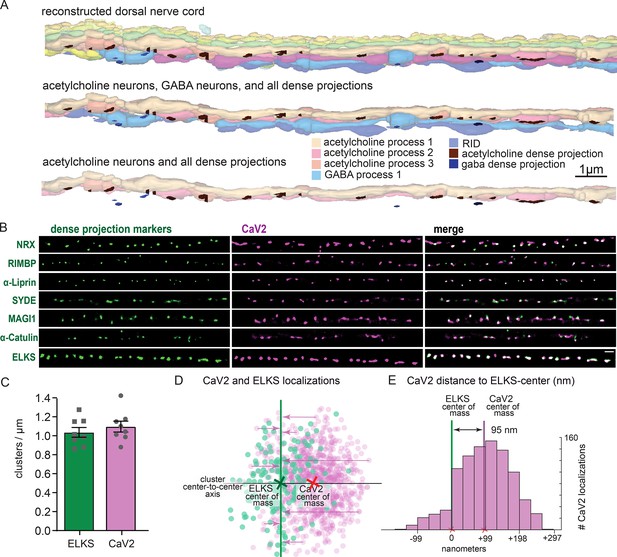
Dorsal nerve cord reconstruction and candidate dense projection markers.
(A) 20 micron reconstruction of the wild-type C. elegans dorsal nerve cord. Dense projections are highlighted to compare to superresolution images below. Scale bar 1 µm, section thickness 100 nm. (B) CaV2 colocalizes with cytomatrix active zone proteins. Super-resolution images of Skylan-S-tagged cytomatrix protein homologs in C. elegans NRX-1, RIMB-1, SYD-2, SYD-1, MAGI-1, CTN-1, ELKS-1 compared to CaV2-HALO in the same animal. (C) ELKS and CaV2 clusters form approximately 1 / µm along the dorsal nerve cord from super-resolution image analysis. Clusters were quantified for over dorsal nerve cords with an average length of 17.8 µm, N=8 animals (D) Localization plot tool (Proberuler) example diagram of a single ELKS (green) and CaV2 (purple) synapse. Cluster centers are marked by solid lines. (E) Histogram of CaV2 localization distance to ELKS center of mass axis from example ELKS and CaV2 synapse.
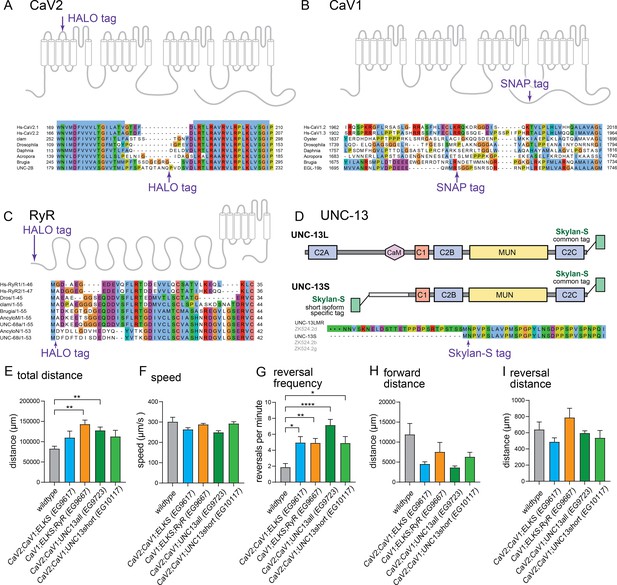
Tagging sites for calcium channels and UNC-13.
(A–C), Tagging strategies and sites used for CRISPR/Cas9 tagging of the endogenous loci for CaV2, CaV1, and RyR. Regions with low conservation were targeted for insertion of tags into the genomic locus of each gene, unc-2, egl-19, and unc-68, respectively. (D) Tagging strategy at the endogenous locus of unc-13 CRISPR / Cas9. The C-terminal tag labels all isoforms of UNC-13. The N-terminal tag labels the UNC-13S isoform. In addition, it will label a rare transcript UNC-13-LMR (~2% of transcripts), that includes the C2A domain, sequences upstream of UNC-13S, and all sequences included in UNC-13S (https://wormbase.org/ version ws284). Behavioral analysis of imaging strains. (E) Total distance travelled: wild-type 82.7±6.1 mm. EG9617 110.0±16.1 mm. EG9667 143.1±10.1 mm. EG9723 127.7±8.3 mm. EG10117 112.7±15.3 mm. (F) Speed: wild-type 301.6±22.1 µm/s. EG9617 263.5±8.8 µm/s. EG9667 288.0±5.4 µm/s. EG9723 249.0±8.3 µm/s. EG10117 292.4±9.2 µm/s. (G) Reversals per minute: wild-type 1.9±0.5 reversals/min. EG9617 5.0±0.8 reversals/min. EG9667 4.9±0.6 reversals/min. EG9723 7.1±0.7 reversals/min. EG10117 4.9±0.8 reversals/min. (H) Average length of forward run: wild-type 11.9±2.8 mm. EG9617 4.5±0.6 mm. EG9667 7.5±2.4 mm. EG9723 3.6±0.4 mm. EG10117 6.3±1.1 mm.(I) Average length of backward run: wild-type 0.64±0.09 mm. EG9617 0.49±0.05 mm. EG9667 0.78±0.1. EG9723 0.6±0.03 mm. EG10117 0.54±0.09 mm. wild-type n=7. EG9617 n=15. EG9667 n=13. EG9723 n=15. EG10117 n=14. Error bars reported in SEM. Genotypes were blinded. Brown-Forsyth ad Welch ANOVA with Dunnett’s T3 multiple comparisons was used to calculate p-value. *p<0.05, **p<0.005, ***p<0.001, ****p<0.0005.
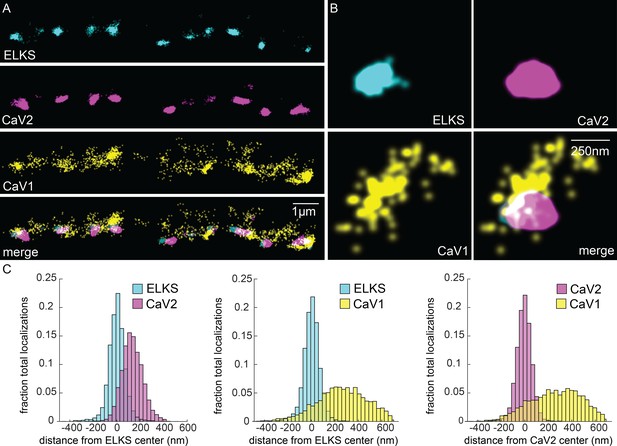
CaV1 is excluded from the dense projection, and dispersed in the active zone.
(A, B) Localization microscopy plots of the dorsal nerve cord. ELKS is tagged with Skylan-S. The CaV2-HALO ligand is HTL-JF646, and the CaV1-SNAP tag ligand is STL-JF549pa. (A) CaV2 (magenta) colocalizes with dense projections labeled with ELKS (cyan). CaV1-SNAP (yellow) is largely excluded from the dense projection; and scattered in the synaptic varicosity. Scale bar = 1 µm. (B) Distributions of CaV2 and CaV1 in a synapse. Dense projections labeled with ELKS (cyan) colocalize with CaV2 (magenta), but not CaV1 (yellow). Scale bar = 250 nm. (C) Quantitation of protein localizations from multiple synapses. The center of mass of localizations was calculated from 2D plots. An axis between the centers was fixed and all localizations collapsed onto the axis. Localizations were combined into 33 nm bins, to match the electron microscopy analysis, and plotted as the fraction of total localizations. Data were collected and combined from n=26 synapses, N=5 animals. Data available as Figure 5—source data 1.
-
Figure 5—source data 1
Particle files for ELKS/CaV1/CaV2 SMLM data and distance measurements.
- https://cdn.elifesciences.org/articles/81407/elife-81407-fig5-data1-v2.zip
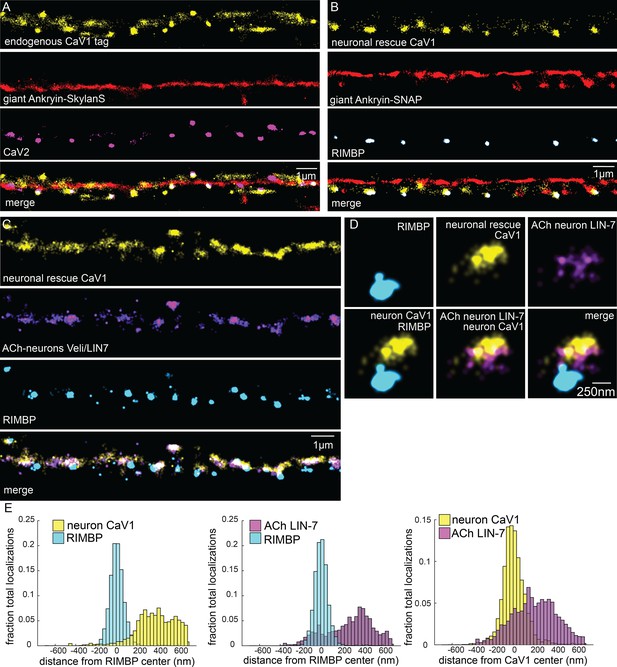
Tagged CaV1 expressed in neurons forms clusters at presynaptic boutons.
Comparison of endogenously-tagged CaV1 and neuronally expressed rescue of CaV1. (A) Endogenous CaV1 tag: Localization microscopy images of dorsal nerve cord with CaV1-SNAP stained with STL-JF549cp (yellow), CaV2-HALO stained with HTL-JF646 (purple), and Giant Ankyrin-Skylan-S (red). scale bar = 1 µm. (B) Exogenous CaV1 in the CaV1(Δns) strain. In the CaV1(Δns) background, CaV1 was rescued in neurons using a single copy transgene insertion of Psnt-1 promoter driving CaV1-HALO in neurons. Localization microscopy images of dorsal nerve cord. Dorsal cord of animals labelled with neuronal CaV1-HALO stained with HTL-JF646 (purple), Giant Ankyrin-SNAP stained with TMR-Star (red) and RIMBP-Skylan-S (cyan). scale bar = 1 µm. (C) Neuronal CaV1 colocalizes with LIN7 expressed in acetylcholine neurons. In the CaV1(Δns) background, CaV1 was rescued in neurons using a single copy transgene insertion of Psnt-1 promoter driving CaV1-HALO in neurons and stained with HTL-JF646. LIN7 was tagged with SNAP and stained with STL-JF549cp, and was expressed in acetylcholine neurons using the unc-129 promotor as an extrachromosomal array. scale bar = 1 µm. Localization microscopy images of dorsal nerve cord. Dense projections are marked by RIMBP-Skylan-S. (D) Single synapse analysis of LIN7, CaV1, RIMBP. Dense projections are marked by RIMBP-Skylan-S. In the CaV1(Δns) background, CaV1 was rescued in neurons using a single copy transgene insertion of the Psnt-1 promoter driving CaV1-HALO, and stained with HTL-JF646. LIN7 was expressed in acetylcholine neurons using the Punc-129 promotor as an extrachromosomal array, and tagged with SNAP and stained with STL-JF549cp and. Scale bar = 250 nm. (E) Cumulative distribution plot of Psnt-1:CaV1-HALO to RIMBP-Skylan-S center, and LIN7 tagged with SNAP to RIMBP-Skylan-S center measured from synaptic regions. n=24 synapses, N=5 animals. Data available as Figure 5—figure supplement 1—source data 1.
-
Figure 5—figure supplement 1—source data 1
Particle files for RIMBP/CaV1/LIN-7 SMLM data and distance measurements.
- https://cdn.elifesciences.org/articles/81407/elife-81407-fig5-figsupp1-data1-v2.zip
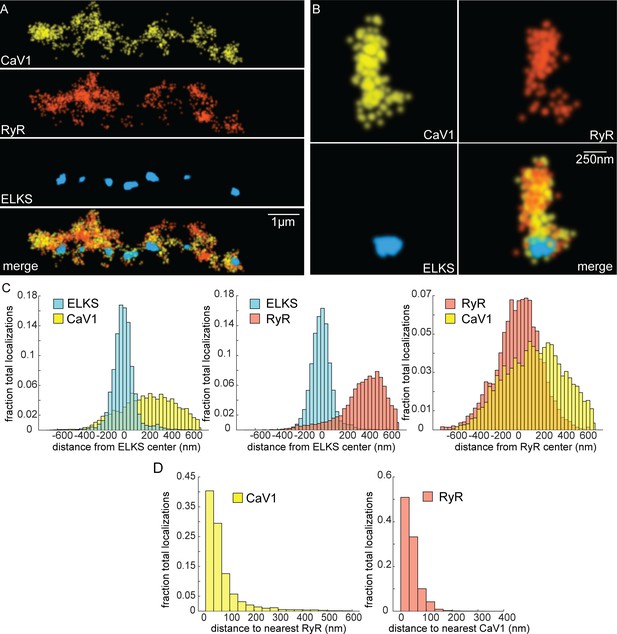
CaV1 and RyR are adjacent.
(A) CaV1 and RyR are adjacent along the dorsal nerve cord, lateral to the dense projection. Animals and HTL-JF646. Scale bar = 1 µm. CaV1-SNAP is labelled with STL-JF549pa, RyR-HALO is labelled with HTL-JF646, and dense projections are labeled by ELKS-Skylan-S. (B) RyR and CaV1 colocalize within synapses. Labelling as in ‘A’. Scale bar = 250 nm. (C) Distances from CaV1-SNAP localizations to center of ELKS-Skylan-S cluster versus ELKS localizations to ELKS center. Distances from RyR-HALO localizations to center of ELKS-Skylan-S cluster versus ELKS localizations to ELKS center. Distances from CaV1-SNAP localizations to the center of the RyR-HALO cluster versus RyR-HALO localizations to the RyR center. N=5 animals, n=25 synapses. (D) RyR and CaV1 are adjacent. Left, nearest neighbor analysis was performed on CaV1-SNAP localizations to find the nearest RyR-HALO localization. Right, nearest neighbor distances from RyR-HALO to CaV1-SNAP were calculated. n=5 animals, 25 synapses. Data available as Figure 6—source data 1.
-
Figure 6—source data 1
Particle files for ELKS/CaV1/RyR SMLM data and distance measurements.
- https://cdn.elifesciences.org/articles/81407/elife-81407-fig6-data1-v2.zip
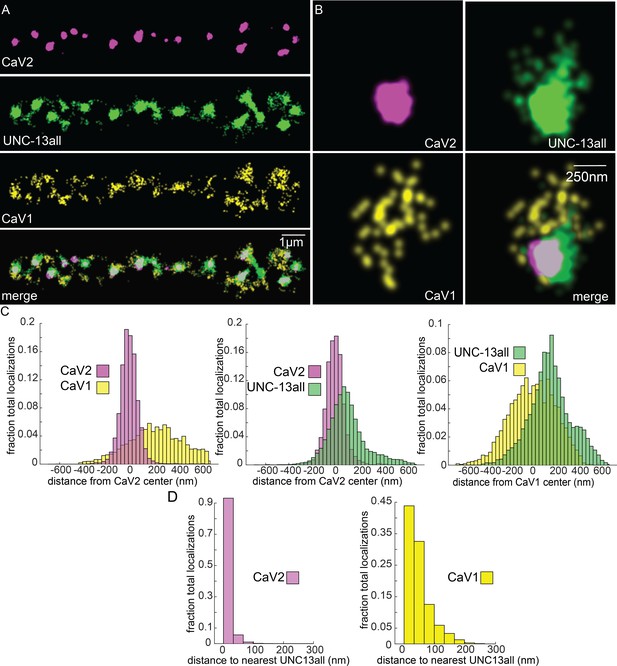
UNC-13 isoforms colocalize with CaV2 and CaV1 calcium channels.
Localization microscopy identifies CaV1 and CaV2 associated with ‘UNC13all’, which labels a C-terminal site common to all UNC-13 isoforms. (A) UNC-13all colocalizes with CaV1 and CaV2 along the dorsal nerve cord. Proteins are labelled with CaV2-HALO stained with HTL-JF646, CaV1-SNAP stained with STL-JF549, and UNC13all-Skylan-S. (B) UNC-13all colocalizes with CaV1 and CaV2 within synapses. Staining as in ‘A’. (C) Left, distances from CaV1-SNAP localizations to the center of the CaV2-HALO cluster, and CaV2-HALO localizations to the center of the CaV2-HALO cluster. Middle, distances from UNC13all-Skylan-S localizations to the center of the CaV2-HALO cluster. Right, distances from UNC13all-Skylan-S localizations to the center of the CaV1-SNAP cluster, n=5 animals, 25 synapses. (D) Left, nearest-neighbor distances between UNC13all and CaV1 and CaV2 localizations. Right, nearest neighbor analysis between UNC13all-Skylan-S and CaV2-HALO or CaV1-SNAP measured from synaptic regions, n=5 animals, 25 synapses. Data available as Figure 7—source data 1.
-
Figure 7—source data 1
Particle files for UNC-13all/CaV1/CaV2 SMLM data and distance measurements.
- https://cdn.elifesciences.org/articles/81407/elife-81407-fig7-data1-v2.zip
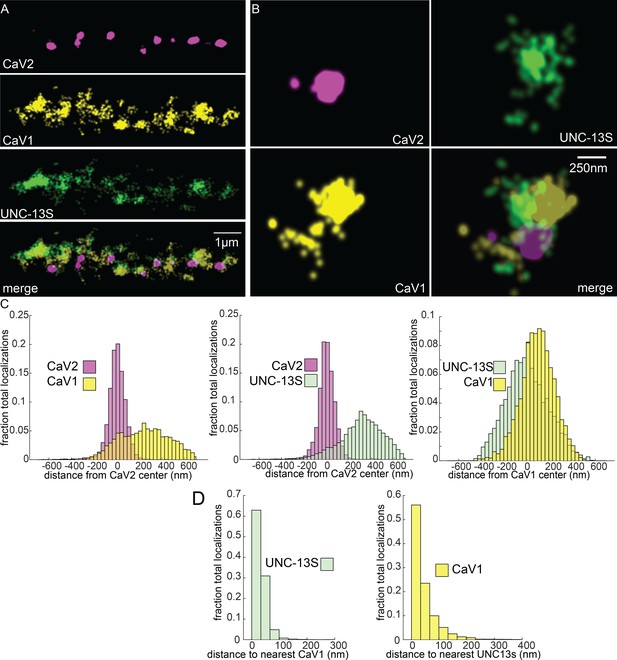
UNC-13S is associated with CaV1 calcium channels.
Localization microscopy identifies CaV1 associated with UNC-13S which labels a n-terminal site common to a short isoform. (A) UNC-13S localizes with CaV1 along the dorsal cord, but not with CaV2. The endogenous protein tags CaV2-HALO was stained with HTL-JF646, CaV1-SNAP with STL-JF549, and imaged with Skylan-S-UNC-13S using single-molecule localization microscopy. (B) UNC-13S localizes with CaV1 within synapses. Strain was labelled as in ‘A’. (C) Left, distances from CaV1-SNAP localizations to the center of the CaV2-HALO cluster compared to CaV2-HALO localizations to their own center. Middle, distances from Skylan-S-UNC-13S localizations to the center of the CaV2-HALO cluster. Right, distances from Skylan-S-UNC-13S localizations to the center of the CaV1-SNAP cluster. N=5 animals, 25 synapses (D) Nearest-neighbor distances of CaV1-SNAP to Skylan-S-UNC-13S localizations. Nearest neighbor analysis of Skylan-S-UNC-13S to CaV1-SNAP measured from synaptic regions. N=5 animals, 25 synapses. Data available as Figure 8—source data 1.
-
Figure 8—source data 1
Particle files for UNC-13S/CaV1/CaV2 SMLM data and distance measurements.
- https://cdn.elifesciences.org/articles/81407/elife-81407-fig8-data1-v2.zip
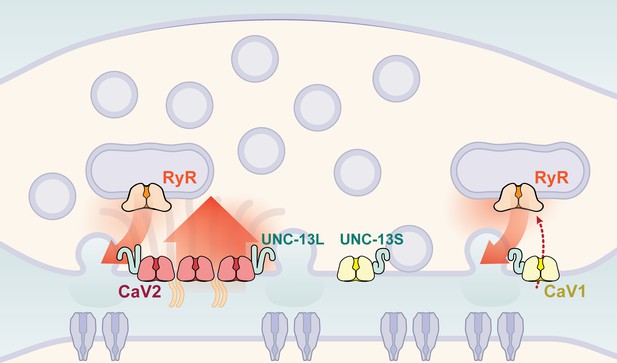
Two independent release sites for synaptic vesicles.
Voltage-gated calcium channels localize to two distinct zones at the neuromuscular synapse of C. elegans. The CaV2 channel localizes to the dense projection along with ELKS, RIMBP, Neurexin, Liprin-alpha, SYDE, MAGI1, alpha-Catulin and the SNARE priming protein UNC-13L. CaV2 is required to fuse synaptic vesicles are docked directly adjacent to the dense projection. The second channel CaV1 is at a lateral site centered ~300 nm from the dense projection but can span hundreds of nanometers. CaV1 requires coupling to RyR to synaptic vesicles at the lateral site. These near and far pools utilize specific release machinery. Most UNC-13all localizes to the dense projection. However, some UNC-13 localizes with CaV1 at the lateral site. Isoform specific tagging shows UNC-13S localized with lateral site.
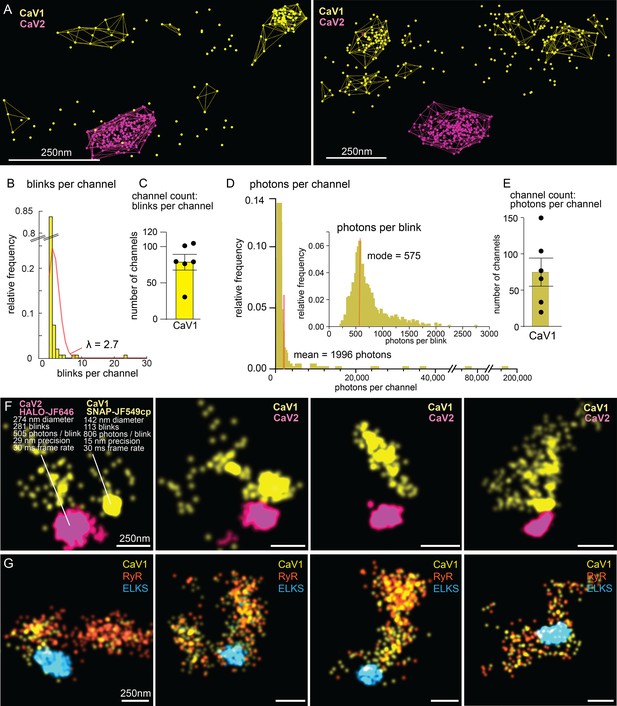
Counting calcium channels and example synapses.
(A) Superresolution images of C. elegans expressing CaV1::SNAP, CaV2::HALO. Animals were stained with HTL-JF646, STL-JF549cp. Clusters of calcium channels are indicated with colored wireframes. Scale bar = 250 nm. (B) Poisson distribution fitted to frequency distribution plot of number of blinks per channel. (C) CaV1 channel count based on number of total number of blink over blinks per channel. (D) Frequency distribution of number of photons per channel cluster. Inlay: Frequency distribution of number of photons per blink. (E) CaV1 channel count based on number of total photons over the mode of photons per channel. (F) Four example superresolution images of C. elegans expressing CaV1::SNAP, CaV2::HALO. Animals were stained with HTL-JF646, STL-JF549cp. A sample cluster of CaV1 was 142 nm in diameter, contained 113 blinks that on average emitted 806 photons. These blinks were localized with 15 nm precision at a 30ms frame rate. A sample CaV2 cluster was 274 nm in diameter, contained 281 blinks with an average emissions of 505 photons per blink. Blinks were localized with 29 nm precision at a 30ms frame rate. (G) Four example superresolution images of C. elegans expressing CaV1::SNAP, RyR::HALO. Animals were stained with HTL-JF646, STL-JF549pa. n=6 synapses N=3 animals. Error bars reported in SEM. CaV1 data available as Figure 10—source data 1. CaV2 data available as Figure 10—source data 2.
-
Figure 10—source data 1
Quantification of CaV1 channels by spatial constraint and photon flux.
- https://cdn.elifesciences.org/articles/81407/elife-81407-fig10-data1-v2.zip
-
Figure 10—source data 2
Particle files of CaV2 localization data for channel count estimations.
- https://cdn.elifesciences.org/articles/81407/elife-81407-fig10-data2-v2.zip
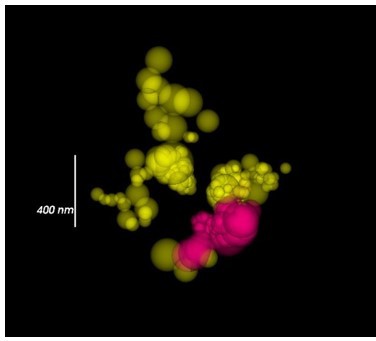
Calcium channel localizations visualized by sphere sized by radial precision.
It should be noted that large dot size corresponds to less-certain localizations rather than the size of the calcium channel cluster. The most accurate localizations are small dots. CaV1 yellow. CaV2 magenta.
Tables
Summary of strain nomenclature and alleles.
| Common name | Usage | CaV2 | CaV1 | RyR | CaV2 rescue | CaV1 rescue |
|---|---|---|---|---|---|---|
| Wildtype N2 Bristol | ePhys EM behavior | wt | wt | wt | n/a | n/a |
| CaV1(rescue) | behavior SMLM | wt | st556 | wt | n/a | oxTi1047[Pset-18::egl-19b] oxTi1049[Psnt-1::HALO::egl-19b] |
| CaV1(Δns) | behavior ePhys | wt | st556 | wt | n/a | oxTi1047[Pset-18::egl-19b] |
| CaV1(Δns) RyR(-) | ePhys | wt | st556 | e540 | n/a | oxEx2017[Pset-18::eGFP::egl-19b] |
| CaV2(Δnmj) CaV1(Δns) | ePhys | lj1 | st556 | wt | oxEx2096[Punc-17h::SNAP::unc-2] | oxTi1047[Pset-18::egl-19b] |
| CaV2(Δnmj) RyR(-) | ePhys | lj1 | wt | e540 | oxEx2096[Punc-17h::SNAP::unc-2] | n/a |
| CaV2(-) | ePhys EM | lj1 | wt | wt | no | n/a |
| RyR(-) | ePhys EM | wt | wt | e540 | n/a | n/a |
| CaV1(-) CaV2(-) | EM | lj1 | n582 | wt | no | no |
| CaV1(-) | EM | wt | n582 | wt | n/a | no |
Super-resolution alleles generated for this study.
| Allele | Gene | Common name | sgRNA | Repair template | Tag | Chr | Terminus |
|---|---|---|---|---|---|---|---|
| ox672 | unc-2 | CaV2 | pSAM429 (ACAGACCGCCAACCAACCGG) | pSAM593 | HALO | X | internal |
| ox704 | rimb-1 | RIMBP | TGGGTAAATCGATAAATCG | pSAM514 | SKY-S | III | c |
| ox719 | nrx-1 | Neurexin | TTTTCTTTGCCACCCCATTC | pSAM534 | SKY-S | V | c |
| ox721 | unc-68 | RyR | pSAM488 (gattagttagttccaagaaA) | pSAM593h | HALO | V | n |
| ox727 | ctn-1d | α-Catulin | CATCCAATGTAATCGGC | pSAM598 | SKY-S | III | c |
| ox728 | egl-19 | CaV1 | CTTCTCATCCATTGCTC | pSAM604 | SNAP | IV | internal |
| ox729 | syd-1 | SYDE | GCACTGCGATTCCGAGACAT | pSAM545 | SKY-S | II | c |
| ox730 | syd-2 | α-Liprin | TTGCTGTAGCTCATatttct | pSAM549 | SKY-S | X | n |
| ox747 | elks-1 | ELKS/CAST | gagcagtacaatATGGCACC | pSAM550 | SKY-S | IV | n |
| ox748 | unc-13all | UNC13all | gctttgaatccaacaaaaaa | pSAM613 | SKY-S | I | c |
| ox803 | magi-1 | MAGI | aagATGACCGACAAAACAGC | pSAM552 | SKY-S | IV | n |
| ox814 | unc-13b | UNC13s | GGAACTGCAAGACTTGGCAC | pSAM684 | SKY-S | I | n |
| ox802 | unc-44 | Giant Ankyrin | GCTGTTGGTCGTGCTCCCGA | pSAM546 | SKY-S | IV | c |
| ox708 | unc-44 | Giant Ankyrin | GCTGTTGGTCGTGCTCCCGA | pSAM557 | SNAP | IV | c |
Common names and nomenclature used in this study.
| Common name | Mammalian ortholog | C. elegans ortholog | Drosophila ortholog |
|---|---|---|---|
| CaV2 | CaV2.1 CaV2.2 CaV2.3 | UNC-2 | Cacophony |
| CaV1/L-type | CaV1.1 CaV1.2 CaV1.3 CaV1.4 | EGL-19 | DmCa1D |
| ELKS | ELKS/CAST | ELKS-1 | Bruchpilot |
| RyR | RYR1 RYR2 RYR3 | UNC-68 | dRyr |
| UNC13all | Munc13-1 ubMunc13-2 bMunc13-2 Munc13-3 | UNC-13L, UNC-13S | Unc13A Unc13B |
| UNC-13S | bMunc13-2 Munc13-3 | UNC-13S | UNC13B |
| RIMBP | RIMBP | RIMB-1 | Rbp |
| Veli/LIN7 | LIN7A | LIN-7 | Lin-7 |
| Giant Ankyrin | gAnkB | UNC-44L | AnkG |
| Neurexin/NRX | Neurexin 1 | NRX-1 | DNrx |
| α-Liprin | α-Liprin | SYD-2 | Liprin-α |
| SYDE | SYDE | SYD-1 | Syd-1 |
| α-Catulin | α-Catulin | CTN-1 | α-Cat |
| MAGI1 | MAGI1 | MAGI-1 | Magi |
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/81407/elife-81407-mdarchecklist1-v2.pdf
-
Source data 1
VCF file of CB540 unc-68(e540).
- https://cdn.elifesciences.org/articles/81407/elife-81407-data1-v2.zip





