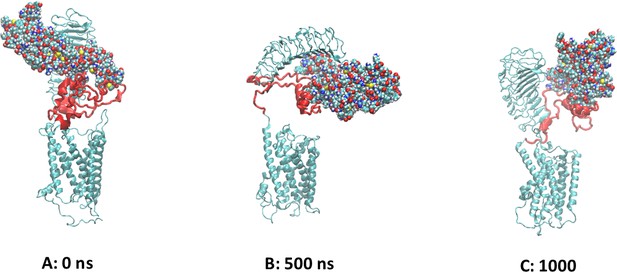
Computational model of the full-length TSH receptor
Figures
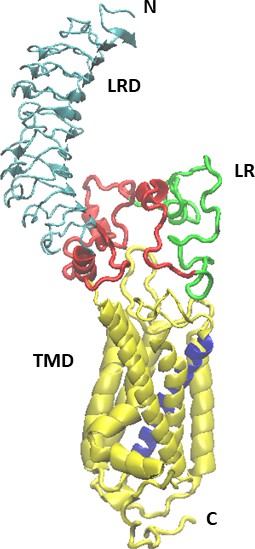
The initial model of the full-length TSH receptor (TSHR) (leucine-rich domain [LRD]: blue, linker region [LR:] red, and transmembrane domain [TMD]: yellow) derived from combination of the LRD and LR of the Alphafold2 program and the TMD of our earlier ‘TRIO’ model (Mezei et al., 2021).
Helix 3 of the TMD is shown in purple.
Initial model of the full-length TSHR receptor.
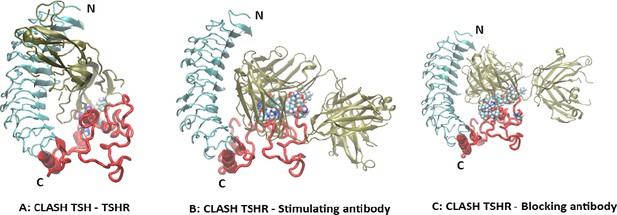
(A) The extracellular part of the full-length model from Figure 1 is shown in combination with the TSH ligand. The leucine-rich domain (LRD) region is shown in gray, the linker region (LR) backbone is shown in red, the ligand is green, and several LR residues clashing with the TSH are shown as spheres colored by atom types (partly obscured). For clarity, the transmembrane domain (TMD) has been removed in this and subsequent illustrations. (B) Similarly, the LR model is shown clashing with a stimulating TSH receptor (TSHR) monoclonal antibody (MS-1) based on the crystal structure (PDB id 3g04) with even more clashes than with TSH. (C) Here, the LR is clashing with a blocking TSHR monoclonal antibody (K1-70) based on the crystal structure (PDB id 2xwt) which once again shows many clashes.
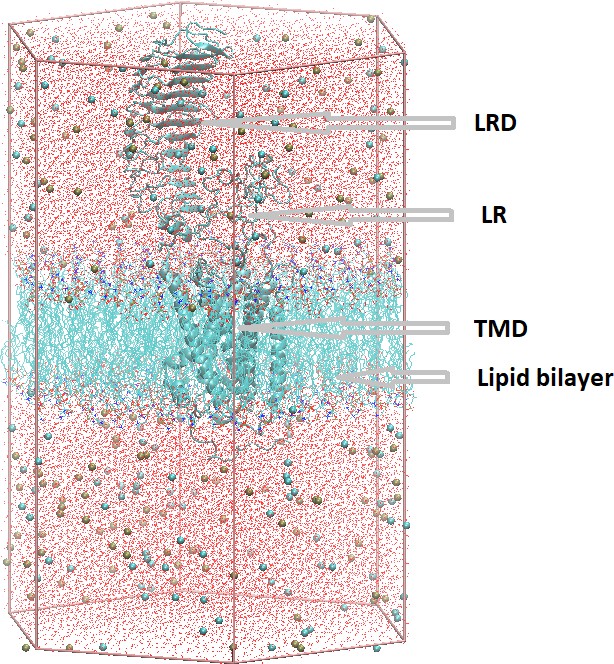
The full simulation cell prepared by Charmm-gui.
The TSH receptor (TSHR) is shown in gray cartoon representation, lipids are shown as lines without hydrogens, ions as tan or cyan spheres representing K+ or Cl− ions, respectively, and the water oxygens as red dots. The hexagonal prism edges defined the initial simulation cell.
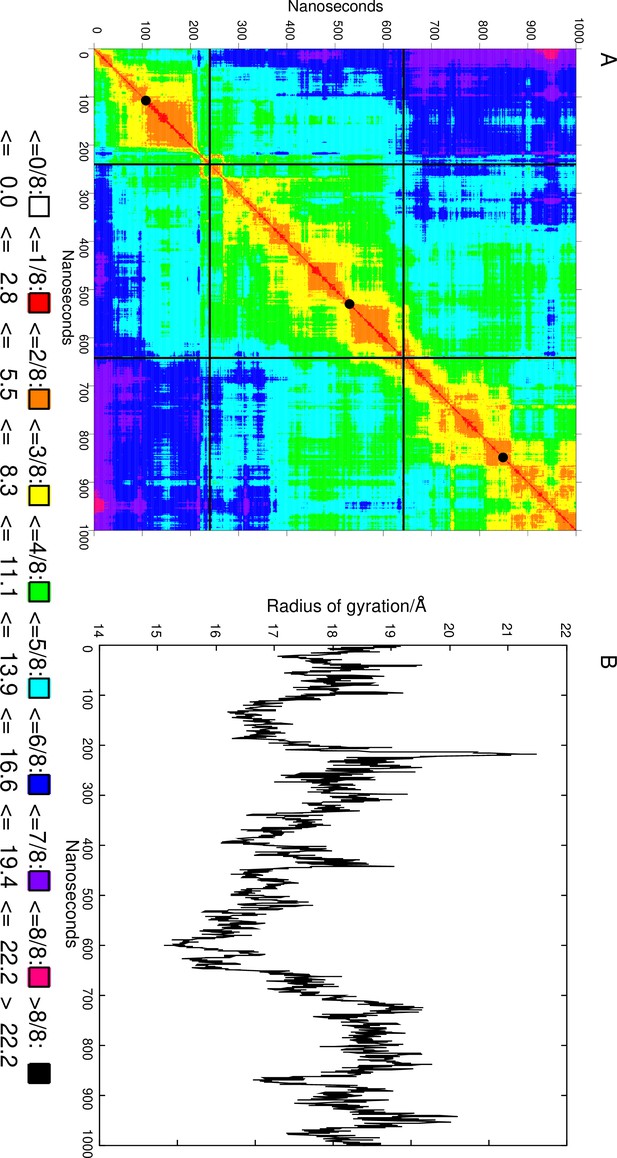
2D RMSD map of the linker region (LR) during the 1000 ns simulation.
(A) RMSD is in Å. Black lines delineate the three clusters, and the black discs on the diagonal indicate the most representative structure. (B) The radius of gyration (in Å) of the LR during the simulation.
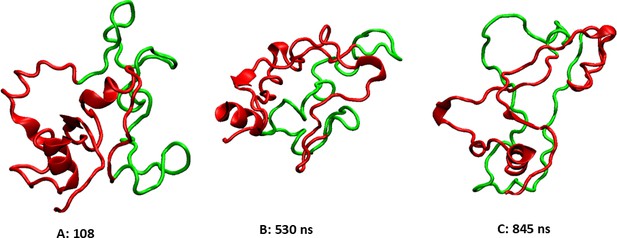
These three clusters are representative of the highly flexible structures of the linker region (LR) backbone at different times during the 1000 ns simulation.
The 50 amino acid (AA) cleaved segment is shown in green, the rest of the LR is in red.
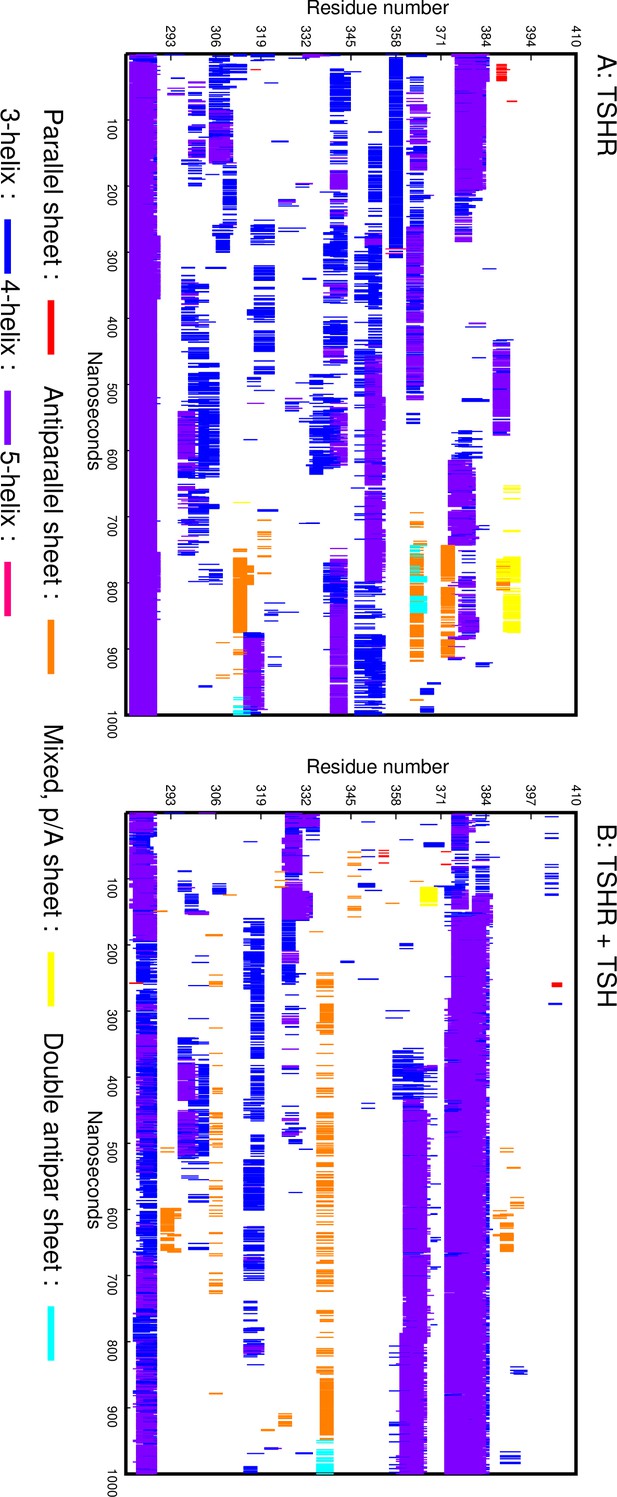
DSSP plots.
(A) DSSP plot showing the secondary structure elements formed in the linker region (LR) during the simulation of the TSH receptor (TSHR) without ligand. The X axis is the simulation time, and the Y axis is the residue number. (B) DSSP plot showing the secondary structure elements formed in the LR during the simulation of the TSHR-TSH complex.
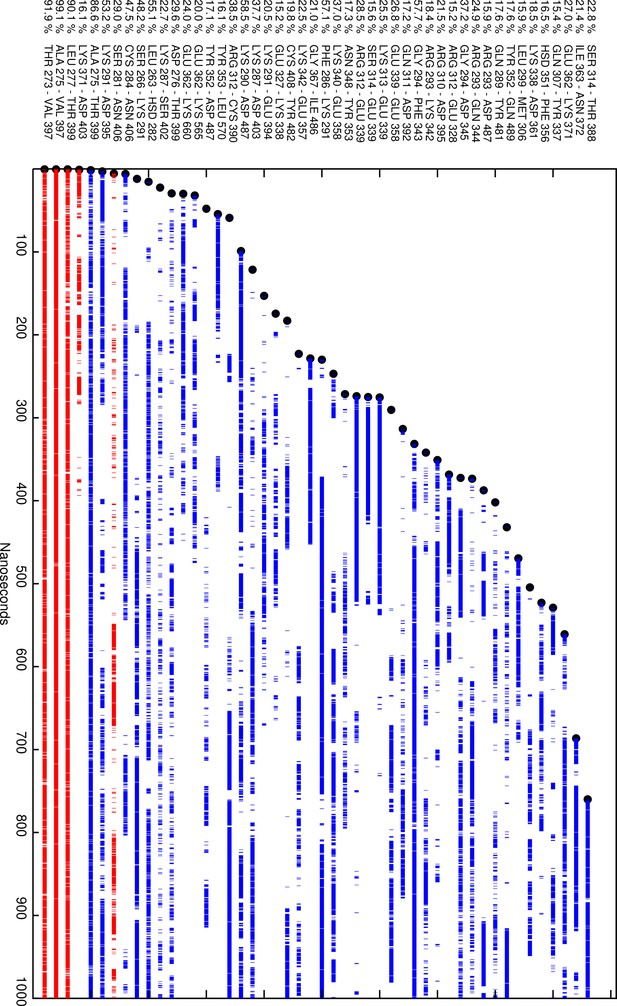
Plot of the residue pairs involving just the linker region (LR) that were hydrogen bonded at some parts of the simulation.
The lines are broken whenever the residue pair was not hydrogen bonded. Blue represents residue pairs within the LR, and red represents hydrogen bonds between residues in the LR and the leucine-rich domain (LRD). Note the unbroken lines between the LR and LRD while the LR itself is intrinsically unstable. Note: residue pairs have to be at least five residues apart (to exclude the many intra-helix hydrogen bonds) and be hydrogen-bonded at least 15% of simulation time to be represented.
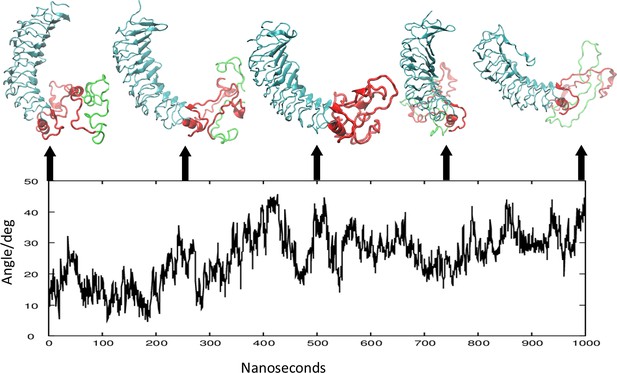
Cystein-cystein distances in the linker region (LR).
(A) Comparison of the relative orientation of the leucine-rich domain (LRD) with the transmembrane domain (TMD) at 250 ns intervals. The structures are aligned by the TMD that is not shown. The linker region (LR) is shown in red with the 50 AA unique insert (316-366) that may be cleaved during post-translational processing is shown in green. (B) The instability of the LR is further illustrated by changes in the angle (in degrees) between the first principal axis of the LRD and the Z axis, over 1000 ns.
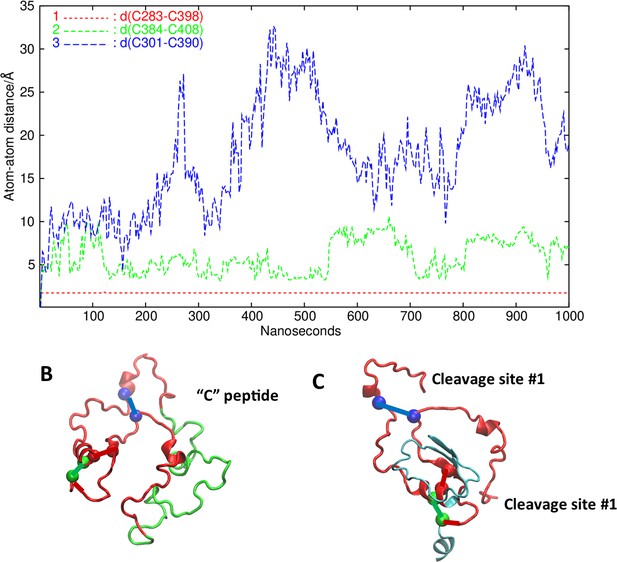
Analysis of putative disulfide bonds.
(A) Time evolution of the three cysteine-cysteine distances (in Å) in the linker region (LR). C283-C398: red, C384-C408: green, and C301-C390: blue. (B) The LR backbone (red and green) and the S atoms of the cysteines, colored to match the corresponding graph color. The putative pairs are connected by a line. (C) Here, we have left the cysteine pairs connected but taken away the 50 amino acid (AA) insert in the LR and show the reported cleavage sites. For giving context, a small part of the leucine-rich domain (LRD) and the transmembrane domain (TMD) are also shown in gray/blue.
Tables
Changes in helix length and radius.
| Helix # | 1 | 2 | 3 | 4 | 5 | 6.1 | 6.2 | 7.1 | 7.2 | 8 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Length: | –0.6 | S | –1.3 | S | –4.4 | S | 1.2 | S | 0.1 | n | 0.0 | n | 0.5 | S | 0.5 | n | –1.4 | S | –1.6 | S |
| Radius: | –0.5 | S | –0.5 | S | –3.7 | S | 0.4 | n | 0.2 | n | 0.0 | n | 0.2 | S | 0.5 | S | –0.4 | S | –0.4 | S |
Helix-helix distance changes.
| Helix# | 1 | 2 | 3 | 4 | 5 | 6.1 | 6.2 | 7.1 | 7.2 | 8 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.0 | n | –0.4 | S | 0.1 | n | –0.1 | n | 2.2 | S | 1.5 | S | 1.9 | n | –5.0 | S | 0.2 | n | –0.4 | n |
| 2 | 0.1 | n | 0.0 | n | 0.4 | S | –1.6 | S | 1.3 | S | –0.9 | S | –0.4 | n | 3.8 | S | 0.8 | n | –5.4 | S |
| 3 | 1.1 | S | 0.7 | S | 0.0 | n | 0.1 | n | –0.4 | n | –1.0 | S | –1.1 | n | 2.3 | n | 7.0 | n | –10.1 | S |
| 4 | 0.5 | n | 0.3 | S | 0.2 | S | 0.0 | n | 0.7 | S | 3.8 | S | –1.3 | S | 8.2 | S | 8.5 | S | –7.2 | S |
| 5 | 2.8 | S | 1.2 | S | –0.2 | n | –0.2 | n | 0.0 | n | –2.0 | S | 0.0 | n | 2.5 | S | 3.2 | n | –11.2 | S |
| 6.1 | 2.7 | S | 0.5 | n | –1.2 | S | –1.1 | S | 0.1 | n | 0.0 | n | –0.8 | S | –1.1 | S | –3.6 | n | –6.9 | S |
| 6.2 | 1.0 | S | –0.8 | S | –1.5 | S | –1.0 | S | 0.1 | n | 0.0 | n | 0.0 | n | –0.3 | n | –2.1 | S | 1.3 | n |
| 7.1 | 2.0 | S | 1.2 | S | 1.2 | S | 1.4 | S | 1.6 | S | 0.7 | S | –0.5 | S | 0.0 | n | 1.0 | n | –7.3 | S |
| 7.2 | 1.9 | S | –2.3 | S | –2.6 | S | –3.7 | S | –0.1 | n | 0.5 | n | 0.2 | n | 1.6 | S | 0.0 | n | –0.9 | S |
| 8 | 1.8 | n | –0.5 | n | –0.8 | S | –2.6 | S | 1.2 | S | 1.9 | S | 0.8 | S | 2.4 | S | 0.3 | n | 0.0 | n |
-
Upper triangle shows the change in the closest approach of the helix axes; the lower triangle shows the change in the distance between the helix centers. Positive number indicates an increase with respect to the starting structure. The characters ‘S’ and ‘n’ indicate that the reference value is within or outside the range of the representative structure values, respectively. The labels of the proline-separated segments of helices 6 and 7 have 0.1 and 0.2 added.
Helix-helix angle changes.
| Helix # | 2 | 3 | 4 | 5 | 6.1 | 6.2 | 7.1 | 7.2 | 8 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.9 | n | 1.4 | S | –7.8 | S | 6.9 | S | –10.8 | S | –8.8 | S | 16.4 | S | –27.8 | S | 11.8 | n |
| 2 | –0.3 | n | –1.7 | S | 0.2 | n | –0.6 | n | –0.8 | n | –8.6 | S | 34.8 | S | –5.8 | n | ||
| 3 | 2.4 | S | –2.3 | S | 7.5 | S | 1.6 | n | 6.6 | n | –24.2 | S | 1.0 | n | ||||
| 4 | –0.2 | n | 5.3 | S | 1.1 | n | –3.3 | n | 34.3 | S | –4.1 | n | ||||||
| 5 | 4.1 | S | –0.9 | n | 4.5 | n | –24.5 | S | 0.1 | n | ||||||||
| 6.1 | 5.5 | S | –9.0 | n | 36.1 | S | –7.4 | S | ||||||||||
| 6.2 | 4.1 | n | 0 8.9 | S | 2.9 | n | ||||||||||||
| 7.1 | –31.9 | S | 0.5 | n | ||||||||||||||
| 7.2 | –10.7 | S |
-
The characters ‘S’ and ‘n’ indicate that the reference value is within or outside the range of the representative structure values, respectively. The labels of the proline-separated segments of helices 6 and 7 have 0.1 and 0.2 added.