Generation of a transparent killifish line through multiplex CRISPR/Cas9mediated gene inactivation
Figures
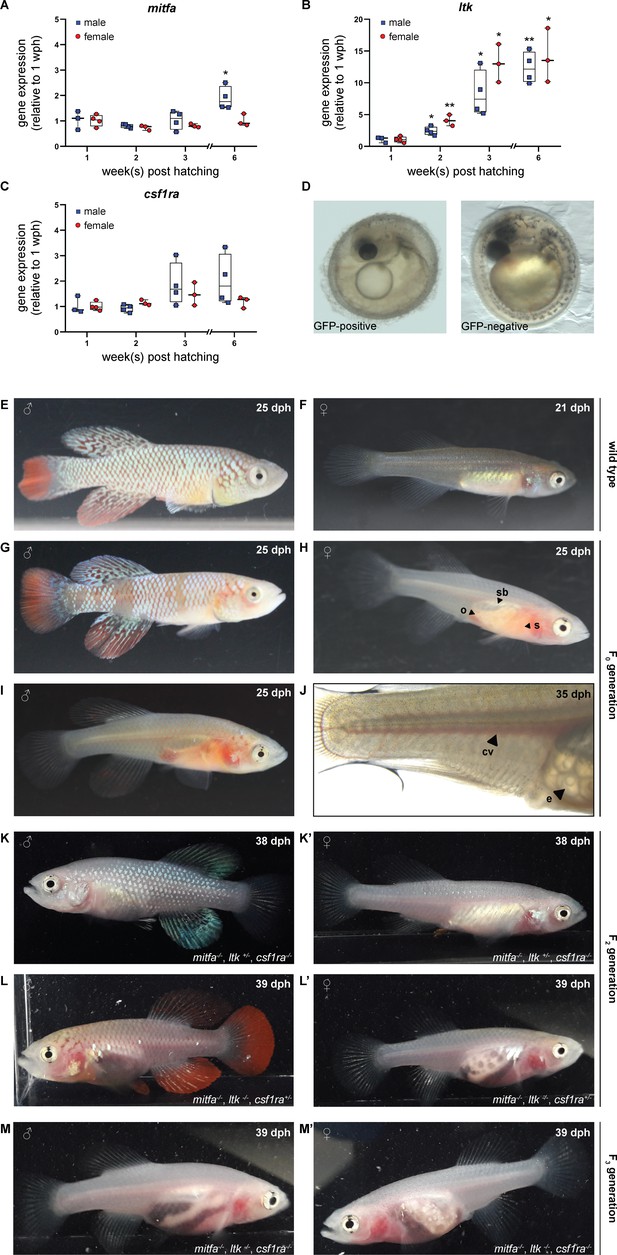
Simultaneous inactivation of genes involved in body pigmentation.
(A–C) Gene expression analysis of mitfa (A), ltk (B), and csf1ra (C) in the skin of male and female wild-type fish via quantitative real-time PCR. The expression of mitfa was only significantly up-regulated in male fish at the age of 6 weeks post-hatching (wph) (p: 0.048). For both sexes, the expression of ltk increased significantly over time (pmale, 2wph: 0.028, pmale, 3wph: 0.026, pmale, 6wph: 0.002; pfemale, 2wph: 0.002, pfemale, 3wph: 0.019, pfemale, 6wph: 0.033), whereas no significant differences were observed for csf1ra. Expression levels were normalized to the expression at 1 wph in the respective sex. Rpl13a was used as housekeeping gene (nmale_1wph = 3, nmale_2wph = 4, nmale_3wph = 4, nmale_6wph = 4; nfemale_1wph = 3, nfemale_2wph = 4, nfemale_3wph = 4, nfemale_6wph = 4). Relative gene expression was calculated using the ΔΔCT method (Pfaffl et al., 2002). Student’s or Welch’s t-tests were computed to determine significant changes in gene expression. Horizontal line represents median. Whiskers show min. to max. values. (D) Phenotypical analysis of F0 embryos revealed a reduction of melanophores in GFP-positive compared to GFP-negative embryos. (E,F) Male (E) and female (F) wild-type N. furzeri. (G–J) Males and females of the F0 generation, display a mosaic loss of body pigmentation. Note that almost completely transparent individuals can be observed in (H–J), allowing a view on inner organs (o: ovary, s: stomach, sb: swim bladder). (J) Microscopic analysis of a female F0 fish with a view on individual eggs within the ovary and blood vessels (cv: cardinal vein, e: egg). (K,K’) Male (K) and female (K’) fish at the age of 38 days post-hatching (dph) with the genotype mitfa-/-,ltk+/-,csf1ra-/- showed a lack of melanophores and xanthophores, whereas iridophores were present. (L,L’) A lack of melanophores and iridophores was observed in male (L) and female (L’) N. furzeri with the genotype mitfa-/-, ltk-/-, csf1ra+/-. Despite a homozygous mutation in ltk (ltk-/-) individual scales with iridophores were detected in fish of both sexes. (M,M’) The presence of homozygous mutations in all the three genes mitfa, ltk, and csf1ra resulted in the loss of body pigmentation in males (M) and females (M’) allowing a view on inner organs.
-
Figure 1—source data 1
Gene expression data and N. furzeri images.
- https://cdn.elifesciences.org/articles/81549/elife-81549-fig1-data1-v3.zip
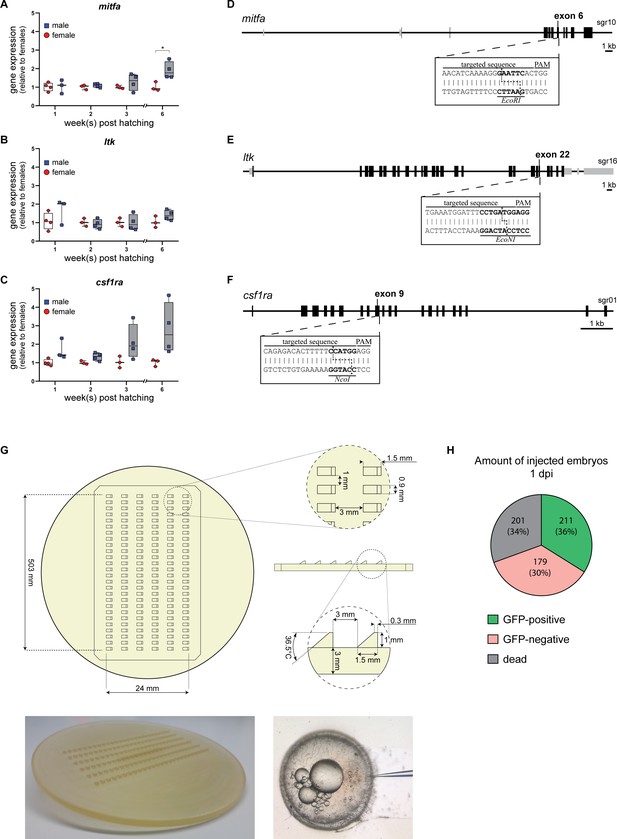
Targeting the mitfa, ltk, and csf1ra loci in N. furzeri.
(A–C) Comparison of the mitfa (A), ltk (B), and csf1ra (C) expression in the skin of male and female wild-type fish at 1 (nmale = 3, nfemale = 4), 2 (nmale = 4, nfemale = 3), 3 (nmale = 4, nfemale = 3) and 6 wph (nmale =4, nfemale = 3) using qRT-PCR. A significant change in gene expression between females and males was detected for mitfa at 6 wph (p: 0.031). Expression levels were normalized to the ones in females at the respective age. Rpl13a was used as a housekeeping gene. Relative gene expression was calculated using the ΔΔCT method. Student’s or Welch’s t-tests were computed to determine significant changes in gene expression. Horizontal line represents median. Whiskers show min. to max. values. (D,E,F) Schematic representation of the mitfa (D), ltk (E), and csf1ra (F) loci in N. furzeri. One sgRNA was designed per gene targeting a sequence part in exon 6 of mitfa, exon 22 of ltk, and exon 9 of csf1ra. Recognition sites for restriction enzymes (mitfa: EcoRI, ltk: EcoNI, csf1ra: NcoI) directly upstream of the respective PAM sequence were used to check for the presence of mutations. (G) Injection mold stamp designed for the preparation of plates for microinjections. Using this mold, single slots were generated on an agarose plate that stabilizes and holds freshly fertilized N. furzeri eggs for microinjections (Picture showing oocyte injection was kindly provided by Annekatrin Richter, FLI Jena). (H) One day after injection (dpi), injected eggs (n=591) were screened for the presence of a GFP signal. 36% of injected eggs were GFP-positive (n=211), 30% were GFP-negative (n=179), and 34% were dead (n=201).
-
Figure 1—figure supplement 1—source data 1
Gene expression data, genomic loci and microinjection.
- https://cdn.elifesciences.org/articles/81549/elife-81549-fig1-figsupp1-data1-v3.zip
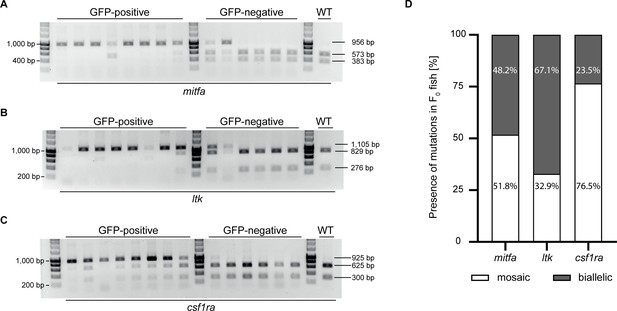
Analyzing the mitfa, ltk, and csf1ra loci upon simultaneous injection of three single guide RNAs (sgRNAs).
(A–C) Restriction enzyme digests to analyze the presence of mutations in mitfa (A), ltk (B), and csf1ra (C) of eight randomly selected GFP-positive and six randomly selected GFP-negative embryos as well as a wild-type control. Non-cleaved products (mitfa: 956 bp, ltk: 1.105 bp, csf1ra: 926 bp) indicated the presence of a mutation in the respective locus. The occurrence of two smaller products (mitfa: 383 bp +573 bp, ltk: 276 bp +829 bp, csf1ra: 300 bp +625 bp) indicated the presence of the wild-type sequence. (D) Based on restriction enzyme digests, the occurrence of mutations in the mitfa, ltk, and csf1ra locus of 85 F0 fish were analyzed. All fish (85/85) carried a mutation in mitfa (mosaic: 51.8%; biallelic: 48.2%), ltk (mosaic: 32.9%; biallelic: 67.1%), and csf1ra (mosaic: 76.5%; biallelic: 23.5%).
-
Figure 1—figure supplement 2—source data 1
Control digest and mutation frequencies in F0 fish.
- https://cdn.elifesciences.org/articles/81549/elife-81549-fig1-figsupp2-data1-v3.zip
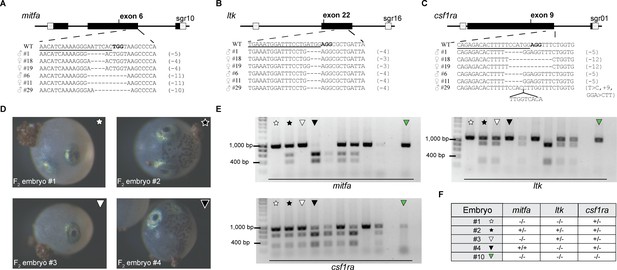
Analyzing the mitfa, ltk, and csf1ra loci in animals of the F1 and F2 generation.
(A–C) Via sequencing, indel mutations were identified in mitfa (A), ltk (B), and csf1ra (C) of F1 fish, originating from an outcross of F0 animals with wild-type fish. CLUSTALW was employed for sequence alignments. Each line represents one fish. Deletions are indicated by dashed lines. The sequence part targeted by the respective single guide RNA (sgRNA) is underlined. (D) Phenotypic analysis of F2 embryos, resulting from an incross of two triple heterozygous (mitfa+/-, ltk+/-, csf1ra+/-) fish. The absence of melanophores was detected in embryos #1 and #3. In contrast, melanophores were present in embryos #2 and #4. (E) Restriction enzyme digests to analyze the presence of mutations in mitfa, ltk, and csf1ra of 10 randomly selected F2 embryos. Non-cleaved products (mitfa: 956 bp, ltk: 1.105 bp, csf1ra: 926 bp) indicated the presence of a mutation in the respective locus. Occurrence of two smaller products (mitfa: 383 bp +573 bp, ltk: 276 bp +829 bp, csf1ra: 300 bp +626 bp) indicated the presence of the wild-type sequence. (F) Assessment of genotypes based on the results from the restriction enzyme digests. Embryo #10 showed a homozygous mutation in all three analyzed loci (mitfa-/-, ltk-/-, csf1ra-/-).
-
Figure 1—figure supplement 3—source data 1
Sequencing results, images of F2 embryos and control digests.
- https://cdn.elifesciences.org/articles/81549/elife-81549-fig1-figsupp3-data1-v3.zip
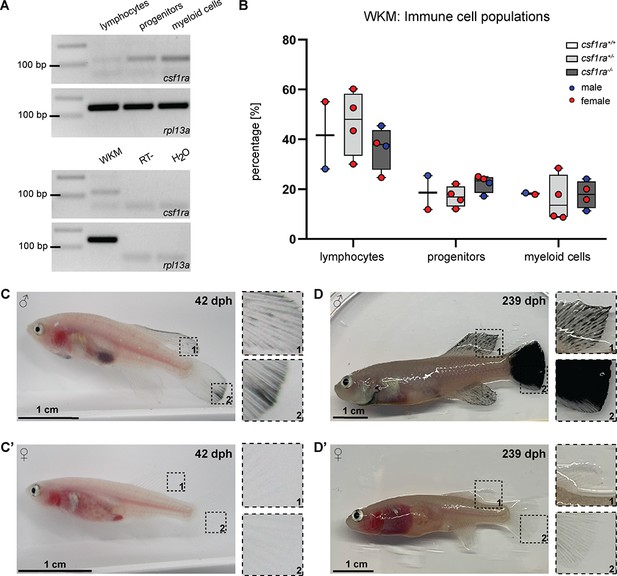
Characterization of klara.
(A) Expression of csf1ra was analyzed via RT-PCR using cDNA from FACS-sorted populations of lymphocytes, progenitors, and myeloid cells obtained from the whole kidney marrow (WKM) of a wild-type N. furzeri. Csf1ra was detected in all subpopulations, most strongly in myeloid cells. As a negative control, an RT-sample (no reverse transcriptase during cDNA synthesis) was used to exclude contaminations with genomic DNA. As loading control, rpl13a was used. (B) Comparison of cell numbers in the different subpopulations of the WKM of fish with the following genotypes: mitfa-/-, ltk-/-, csf1ra+/+ (n=2), mitfa-/-, ltk-/-, csf1ra+/- (n=4), and mitfa-/-, ltk-/-, csf1ra-/- (n=4). One-way ANOVA followed by Tukey’s post hoc test did not reveal any significant differences. Horizontal line represents median. Whiskers show min. to max. value. (C,C’) Male klara fish (C) showed an appearance of melanophores on fin appendages, which was not observed in females (C’). (D,D’) Occurrence of melanophores intensified with age resulting in black fins in male fish (D). In female klara animals (D’) black fins were not observed.
-
Figure 2—source data 1
PCR, FACS analysis of the WKM and images of klara fish.
- https://cdn.elifesciences.org/articles/81549/elife-81549-fig2-data1-v3.zip
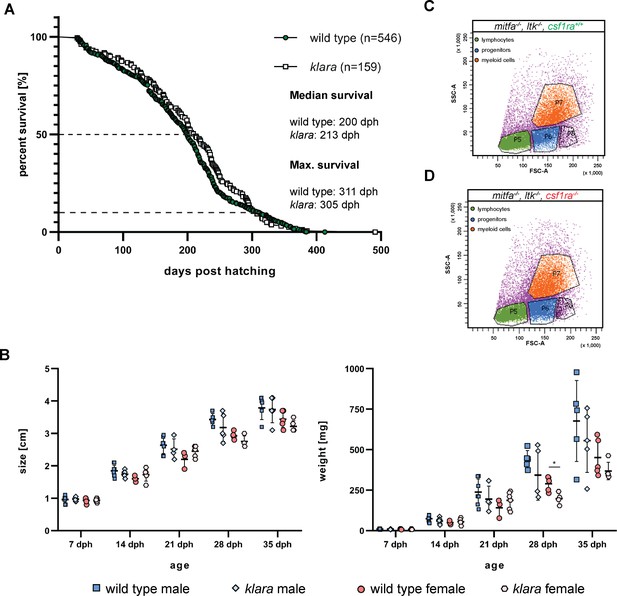
Characterization of klara fish.
(A) The lifespan of wild-type (MZCS-08/122, n=546) and klara (n=159) fish did not differ (p: 0.1353). The median lifespan (50% survival) was 200 days for wild-type fish and 213 days for klara animals. The maximum lifespan (10% survival) was 311 days in wild-types and 305 days in klara. Lifespan curves were analyzed via a Log-rank test for statistical differences. (B) Analysis of body size and weight in male and female wild-type and klara fish at the age of 7 (nWT_male = 10, nWT_female = 5, nklara_male = 7, nklara_female = 9), 14 (nWT_male = 9, nWT_female = 3, nklara_male = 7, nklara_female = 5), 21 (nWT_male = 7, nWT_female = 3, nklara_male = 4, nklara_female = 6), 28 (nWT_male = 5, nWT_female = 5, nklara_male = 5, nklara_female = 4) and 35 (nWT_male = 5, nWT_female = 5, nklara_male = 5, nklara_female = 5) days post-hatching (dph). Within one age cohort, the body size and weight of wild-types and klara fish were only compared between animals of the same sex via Student’s or Welch’s t-test. Regarding size, there was no difference between wild-types and klara fish. Among male fish, there was no difference between wild-type and klara fish. Only at the age of 28 dph, female wild-type fish were significantly heavier than klara females (p: 0.014). Error bars represent standard deviation. (C,D) Forward scatter (FSC-A) versus side scatter (SSC-A) plot of the whole kidney marrow (WKM) of a mitfa-/-, ltk-/-, csf1ra+/+ fish (C) and a mitfa-/-, ltk-/-, csf1ra-/- fish (D).
-
Figure 2—figure supplement 1—source data 1
Lifespan data, size and weight measurements and gating strategy.
- https://cdn.elifesciences.org/articles/81549/elife-81549-fig2-figsupp1-data1-v3.zip
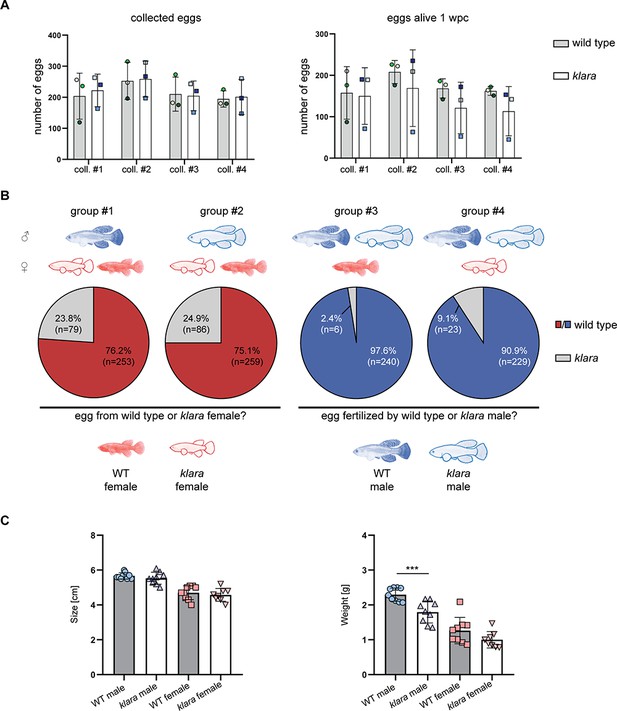
Pigmented fish are preferred mating partners.
(A) For the analysis of egg quantity, eggs were collected once per week for four weeks (coll. #1–4) from wild-type (n=3 tanks with one male and two females each) and klara (n=3 tanks with one male and two females each) fish. To assess egg quality, the number of alive eggs stored on coconut coir plates was determined one-week post-collection (wpc). Student's or Welch's t-tests were computed revealing no significant differences regarding egg quantity and quality. Error bars represent standard deviation. (B) Collected eggs obtained from breeding groups with a wild-type female and a klara female and either a wild-type male (group #1) or a klara male (group #2) were genotyped via HRMA. As a mating partner, male fish preferred wild-type females (group #1: 76.2%; group #2: 75.1%) over klara females (group #1: 23.8%; group #2: 24.9%). In the presence of a wild-type male and a klara male and either a wild-type female (group #3) or a klara female (group #4) the majority of analyzed eggs were fertilized by the wild-type male (group #3: 97.6%; group #4: 90.9%). (C) Within the same sex, wild-type and klara fish did not differ in size(nWT_male = 9 nWT_female = 9, nklara_male = 9, nklara_female = 8). Wild-type males were significantly heavier than klara males (p: 0.0009). Student’s or Welch’s t-tests were computed to determine differences in size or weight. Error bars represent standard deviation.
-
Figure 3—source data 1
Egg quantity and quality data, genotyping of eggs and size and weight measurements.
- https://cdn.elifesciences.org/articles/81549/elife-81549-fig3-data1-v3.zip
Mating of wild-type N. furzeri animals.
Mating of N. furzeri klara animals.
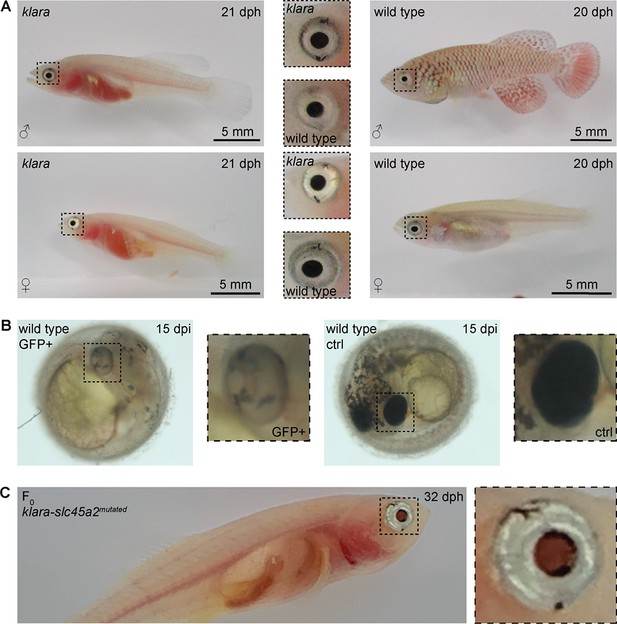
Partial loss of eye pigmentation.
(A) Despite the inactivation of three genes involved in pigmentation, the eye pigmentation did not differ between klara and wild-type fish. (B) Targeting the slc45a2 locus in wild-types resulted in a reduction of melanophores in the eye, but also on the whole body of injected embryos compared to wild-type controls. (C) A loss of melanophores in the retinal pigmented epithelium was observed in F0 fish upon a microinjection of the sgRNA targeting slc45a2 in klara embryos. A silver-pigmented ring around the eye was still present. (ctrl: control; dpi: days post-injection; dph: days post-hatching).
-
Figure 4—source data 1
Images of embryos and fish.
- https://cdn.elifesciences.org/articles/81549/elife-81549-fig4-data1-v3.zip
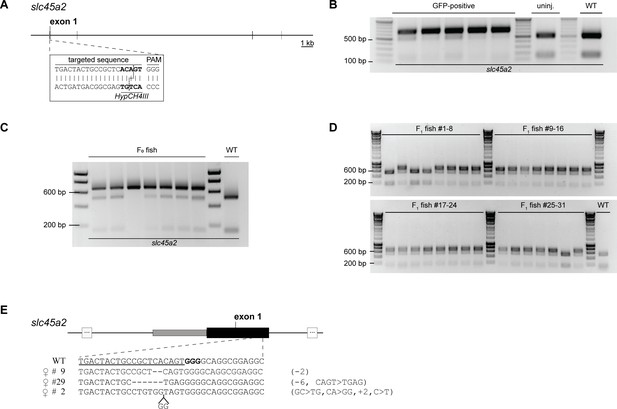
Inactivation of the slc45a2 locus.
(A) Schematic representation of the slc45a2 locus in N. furzeri. A sgRNA was designed to target a sequence part in exon 1. The HypCH4III recognition site upstream of the PAM sequence allowed checking for the presence of mutations via restriction enzyme digest. (B) All GFP-positive embryos (5/5) from microinjections of the slc45a2 sgRNA into wild-type oocytes showed the presence of a non-cleaved fragment (652 bp) indicating the presence of a mutation. Cleaved fragments (152 bp +500 bp) were detected in uninjected and wild-type control samples. (C) Analyzing seven hatched F0 animals via restriction enzyme digest revealed the presence of mutations in all (7/7) fish indicated by the non-cleaved product. One fish (#3) did not show the additional cleaved fragments (152 bp +500 bp) indicating a biallelic mutation in slc45a2. (D) Mutations in slc45a2 were present in 27/31 F1 fish originating from the outcross of an F0 animal with a wild-type fish. (E) Various indel mutations were identified in F1 fish via sequencing. CLUSTALW was used for sequence alignments, in which each line represents one fish. Deletions are indicated by dashed lines. The sequence part targeted by the used sgRNA is underlined.
-
Figure 4—figure supplement 1—source data 1
slc45a2 locus, control digests and sequencing results.
- https://cdn.elifesciences.org/articles/81549/elife-81549-fig4-figsupp1-data1-v3.zip
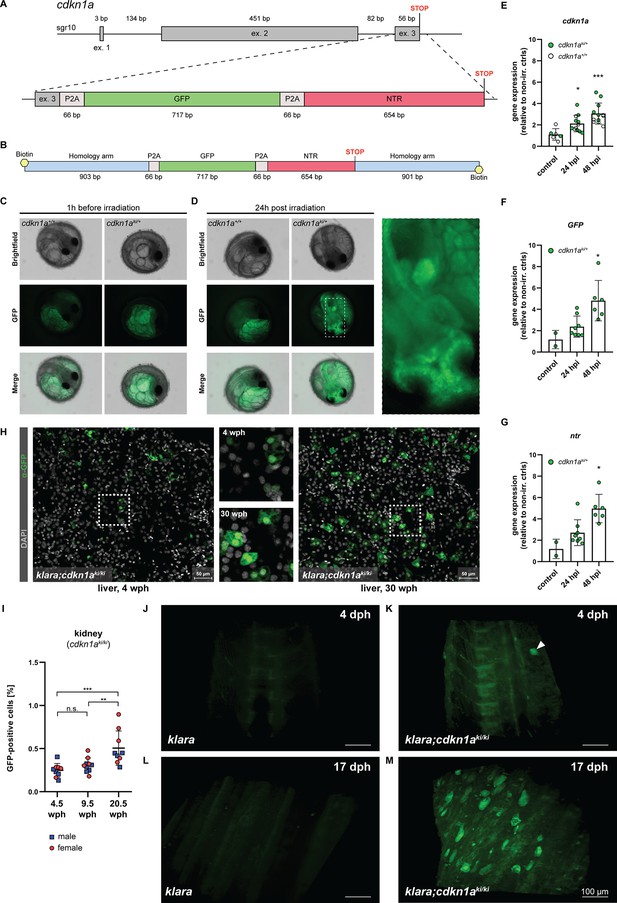
CRISPR/Cas9-mediated insertion of a reporter construct.
(A) The cdkn1a locus in klara was targeted via CRISPR/Cas9 to insert a reporter construct consisting of a GFP and an NTR cassette, separated by P2A self-cleavage sites, allowing the detection and ablation of senescent cells. (B) A 5’-biotinylated, double-stranded donor template flanked by two ~0.9 kb flanking arms was used for the target-specific insertion. (C,D) Function of the reporter construct was tested by exposing embryos (cdkn1a+/+ and cdkn1aki/+) to a γ-irradiation dose of 10 Gy. Representative images taken before irradiation showed autofluorescence originating from the yolk, which was observed in embryos of both genotypes (C). 24 hr post-irradiation (hpi), the presence of GFP-positive cells was detected only in cdkn1aki/+ embryos (D). (E) Gene expression analysis of cdkn1a in embryos (klara;cdkn1a+/+ and klara;cdkn1aki/+) at 24 (n=12) and 48 hpi (n=12) compared to non-irradiated controls (n=6). Expression of cdkn1a increased significantly as revealed by Student's t-test (p24hpi: 0.011, p48hpi: 0.0004). (F,G) For the expression of GFP (F) and ntr (G) at 24 (n=8) and 48 hpi (n=6) compared to non-irradiated controls (n=2), klara;cdkn1a+/+ embryos were excluded due to the absence of the reporter construct. Expressions of GFP and ntr were significantly increased at 48 hpi (pGFP: 0.045, pntr: 0.006) compared to controls. For all qRT-PCRs, rpl13a was used as a housekeeping gene. Relative gene expression was calculated using the ΔΔCT method. Student’s t-tests were computed to determine significant changes in gene expression. Error bars represent standard deviation. (H) Detection of GFP-positive cells in the liver of a young (4 wph) and an old (30 wph) klara;cdkn1aki/ki fish. A higher number of GFP+ cells was detected in the old liver sample. Images are displayed as extended depth of focus projections. In the young liver tissue, small individual GFP-positive cells were detected, whereas in the old liver, a high prevalence of enlarged GFP-positive cells was identified (see zoom-in). (I) Analysis of kidney samples via flow cytometry from klara;cdkn1aki/ki fish at the age of 4.5 wph (nmale = 5; nfemale = 4), 9.5 wph (nmale = 5; nfemale = 5) and 20.5 wph (nmale = 4; nfemale = 5). A significantly higher proportion of GFP-positive cells was detected in 20.5 wph versus 4.5 wph (p: 0.0009) and 9.5 wph (p: 0.0084) fish. One-Way ANOVA followed by Tukey’s post hoc test was computed to analyze for differences in cell number. Horizontal line shows mean value. Error bars represent standard deviation. (J–M) Representative images from light sheet microscopy show the presence of GFP-positive cells in living klara;cdkn1aki/ki fish (K, M) at 4 dph (K) and 17 dph (M). The panel displays representative 3D reconstructions from stacks of 31 (J,L) and 372 (K,M) optical slices. Klara fish at the same ages (J,L), which do not have any GFP + cells, were used as controls. The image processing settings for the klara fish were adjusted to make the background fluorescence visible in order to create comparable image pairs. While a high number of GFP-positive cells was present in the dorsal fin area of klara;cdkn1aki/ki fish at the age of 17 dph (M), only one cell (marked by the white arrowhead) was detected in the dorsal developing fin of the younger klara;cdkn1aki/ki animal (K).
-
Figure 5—source data 1
cdkn1a locus, images of embryos, gene expression data, immunofluorescence stainings, FACS analysis and images obtained by light sheet microscopy.
- https://cdn.elifesciences.org/articles/81549/elife-81549-fig5-data1-v3.zip
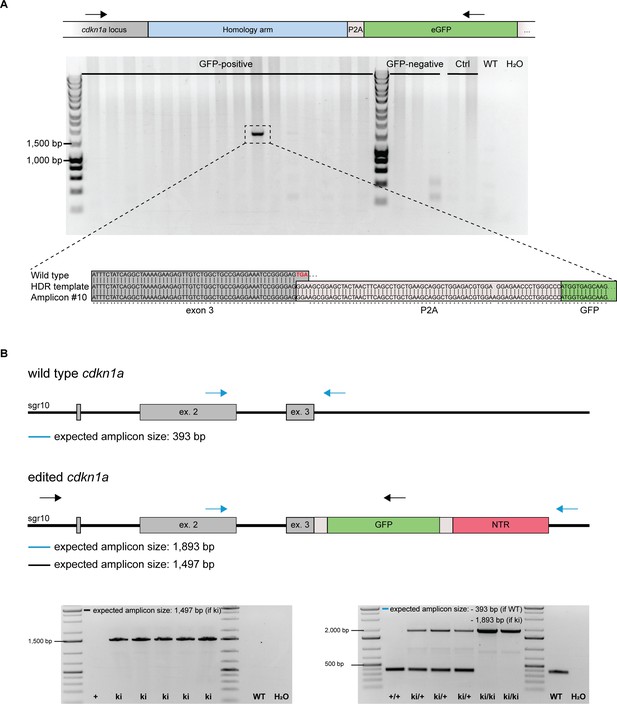
Analysis of embryos from the senescence reporter line.
(A) Identification of the reporter constructs integration into the cdkn1a locus via PCR. A primer pair combination was selected in which one oligonucleotide binding site was located within the cdkn1a locus but outside of the sequence that was covered by the homology arm and the other one binding within the insert. In 1 out of 16 randomly selected GFP-positive embryos an amplicon indicating the insertion of the construct was detected. Proper insertion, i.e., lack of the endogenous STOP codon followed by the presence of the P2A-GFP-P2A-NTR sequence was verified via sequencing of the PCR amplicon. (B) Two primer pair combinations were used to verify the presence of the reporter constructs in the cdkn1a locus. Besides the primer pair combination described in (A) (black arrows; expected amplicon size: 1497 bp), the alternative primer pair (blue arrows) was used in addition to distinguish between cdkn1a+/+, cdkn1aki/+, and cdkn1aki/ki animals. In the presence of a wild-type allele, an amplicon at the size of 393 bp was expected. The insertion of the reporter cassette caused an increase in the fragment size to 1893 bp. In cdkn1aki/+ fish, both amplicons were detected.
-
Figure 5—figure supplement 1—source data 1
Genotyping PCRs and sequencing results.
- https://cdn.elifesciences.org/articles/81549/elife-81549-fig5-figsupp1-data1-v3.zip
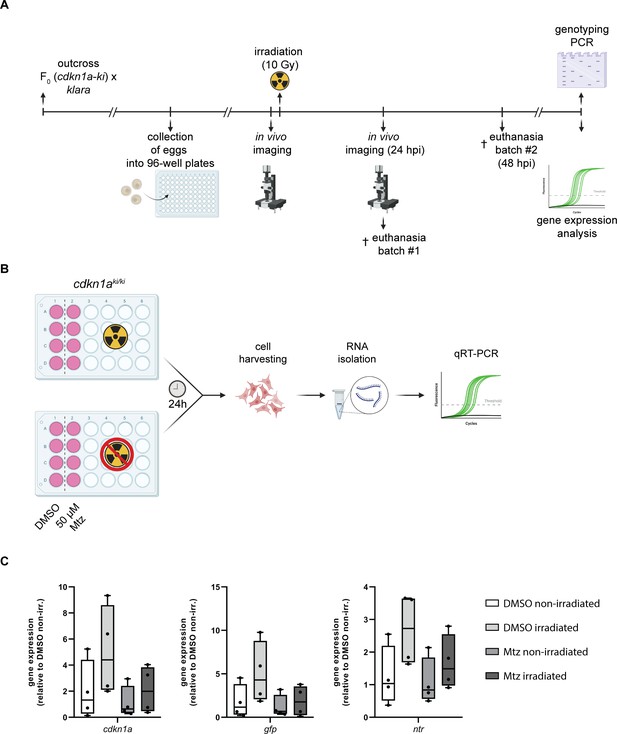
Characterization of the senescence reporter line.
(A) Workflow of the reporter construct activity test via γ-irradiation (10 Gy) in embryos (klara;cdkn1a+/+ and klara;cdkn1aki/+). The image was created with biorender.com. (B) Experimental setup to test the NTR/Mtz system in primary cells (fibroblasts) isolated from a klara;cdkn1aki/ki fish. The image was created with BioRender. (C) The expression of cdkn1a, gfp, and ntr was analyzed in irradiated and non-irradiated cells upon Mtz treatment (50 µM). For all qRT-PCRs, rpl13a was used as a housekeeping gene. Gene expression (relative to non-irradiated, DMSO-treated cells) was calculated using the ΔΔCT method. One-way ANOVA followed by Tukey’s post hoc test did not reveal any significant differences in gene expression. Horizontal line represents median. Whiskers show min. to max. values.
-
Figure 5—figure supplement 2—source data 1
Schematic presentation of workflows and gene expression data.
- https://cdn.elifesciences.org/articles/81549/elife-81549-fig5-figsupp2-data1-v3.zip
This movie represents the z-stacks of optical slices from which the 3D projections of Figure 5K were generated.
Microscopy has been done using a light sheet microscope and a 4-day-old klara;cdkn1aki/ki animal.
This movie represents the z-stacks of optical slices from which the 3D projections of Figure 5M were generated.
Microscopy has been done using a light sheet microscope and a 17-day-old klara;cdkn1aki/ki animal.





