Discrete GPCR-triggered endocytic modes enable β-arrestins to flexibly regulate cell signaling
Figures
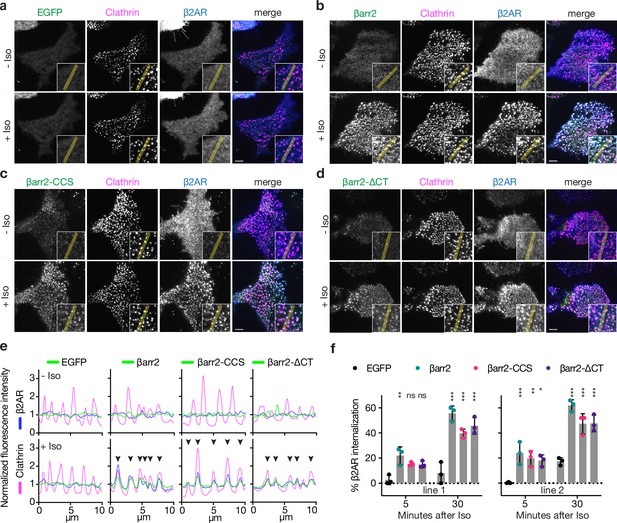
Known endocytic motifs in βarr2 are dispensable for β2-adrenergic receptor (β2AR) clustering and endocytosis.
Representative live-cell total internal reflection fluorescence (TIRF) microscopy images of βarr1/2 double knockout HEK293 cells coexpressing clathrin-light-chain-DsRed (magenta) and FLAG-tagged β2AR (blue) with either EGFP (a), βarr2-EGFP (b), β arr2-CCS-EGFP (c), or βarr2-ΔCT-EGFP (d) (all in green) and pre- and post-stimulation with 10 μM isoproterenol (Iso). Scale bars are 5 μm. (e) Representative fluorescence intensity profiles from line scans shown in insets from a to d. Chevrons indicate colocalization. (f) Percent internalization of FLAG-tagged β2AR coexpressed with either EGFP (black), wild-type βarr2-EGFP (green), βarr2-CCS-EGFP (pink), or βarr2-ΔCT-EGFP in two clonal lines of βarr1/2 DKO HEK293 cells at 5- and 30-min post-stimulation with 10 μM isoproterenol (Iso). Data shown as mean ± standard deviation (SD) for n = 3 independent experiments. Significance was determined by two-way analysis of variance (ANOVA) (df = 3, F = 24.48) with Tukey’s multiple comparisons test against the negative control (EGFP) for each time point (ns p ≥ 0.05, *p < 0.05, **p < 0.01, ***p < 0.001). Each dot is an average of three technical replicates. All data shown are from three independent experiments.
-
Figure 1—source data 1
Representative gating for all flow cytometry-based internalization assays.
P1 gate corresponds to cells. P2 gate corresponds to single cells. P3 gate corresponds to EGFP-positive single cells. M1-647 fluorescence measurements were carried out on the P3 population.
- https://cdn.elifesciences.org/articles/81563/elife-81563-fig1-data1-v2.zip
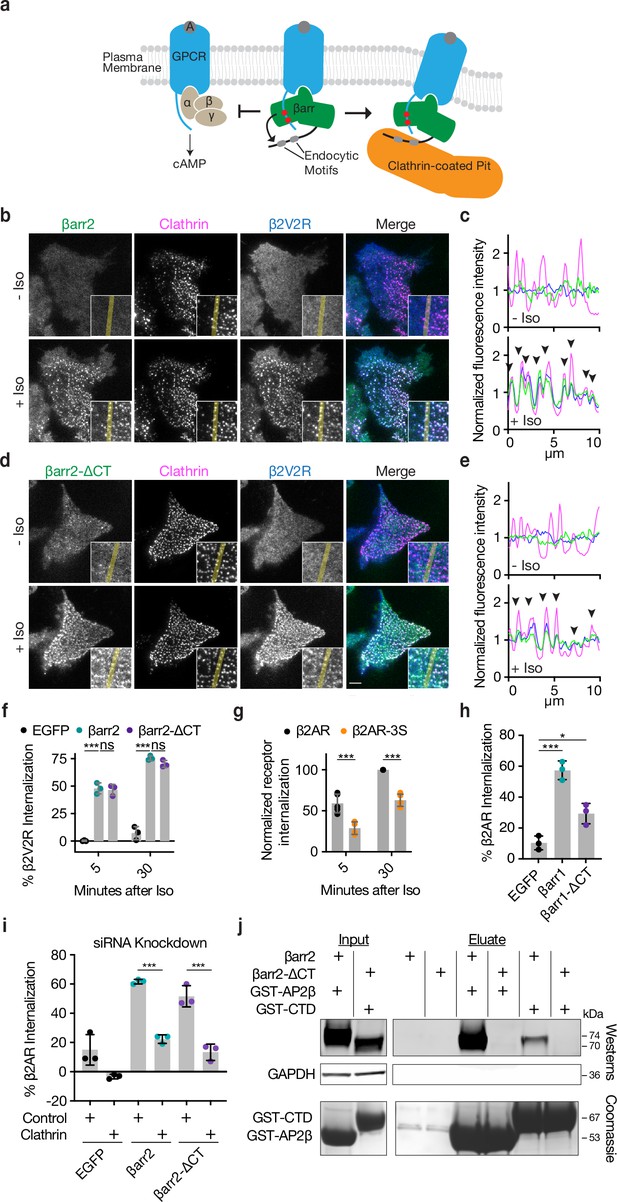
βarr2 and βarr1 C-terminus (CT) are dispensable for G-protein-coupled receptor (GPCR) internalization and β2-adrenergic receptor (β2AR) phospho-sites are required for efficient internalization.
(a) Canonical model of β-arrestin desensitization and endocytosis of GPCRs. Representative live-cell total internal reflection fluorescence (TIRF) microscopy images of βarr2-EGFP (green) (b) or βarr2-ΔCT-EGFP (green) (d) with FLAG-tagged β2V2R (blue) and clathrin-light-chain (magenta) pre- and post-stimulation with 10 μM isoproterenol (Iso). Scale bars represent 5 μm. Insets correspond to the central area of each cell. (c, e) Normalized fluorescence intensity profiles from yellow lines shown in insets from panels (b) and (d) with colors corresponding to the image labels. (f) Percent internalization of FLAG-tagged β2V2R coexpressed with either EGFP (black), βarr2-EGFP (green), or βarr2-ΔCT-EGFP (purple) after 5- or 30-min treatment with 10 μΜ Iso. (g) Normalized internalization of FLAG-tagged β2AR (black) or its phosphorylation site mutant, β2AR-3S (orange), after 5- or 30-min treatment with 10 μM Iso, and coexpressed with βarr2-EGFP. (h) Percent internalization of FLAG-β2AR after 30 min of stimulation with 10 μM Iso, coexpressed with either EGFP (black), βarr1-EGFP (green), or βarr1-ΔCT-EGFP. (i). Percent internalization of FLAG-β2AR after 30 min of treatment with 10 μM Iso when coexpressed with the indicated βarr2 construct after siRNA knockdown of clathrin heavy chain or treatment with control siRNA. (j) Representative western blots and a Coomassie stained sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE) gel of GST-based pull-downs using purified GST-CTD (clathrin terminal domain, residues 1–363) or GST-AP2β (β-appendage of AP2, residues 701–937) and βarr1/2 DKO lysate from cells expressing either βarr2-EGFP or βarr2-ΔCT-EGFP. GAPDH was used as a loading control for cell lysate. See Figure 1—figure supplement 2 for uncropped images and Figure 1—figure supplement 1—source data 1 for raw images. For (f–i), data are shown as mean ± standard deviation of n = 3 independent experiments with significance determined by either two-way analysis of variance (ANOVA) (df = 2, F = 15.3) or (df = 1, F = 63.82) with Dunnett’s test for multiple comparisons (f, g, respectively), one-way ANOVA (df = 2, F = 49.81) with Tukey’s test for multiple comparisons (h), or one-way ANOVA (df = 5, F = 54.49) with Sidak’s test for multiple comparisons (i) (ns p ≥ 0.05, *p < 0.05, ***p < 0.001). All data shown are from three independent experiments.
-
Figure 1—figure supplement 1—source data 1
Source data for the quantifications graphed in Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/81563/elife-81563-fig1-figsupp1-data1-v2.zip
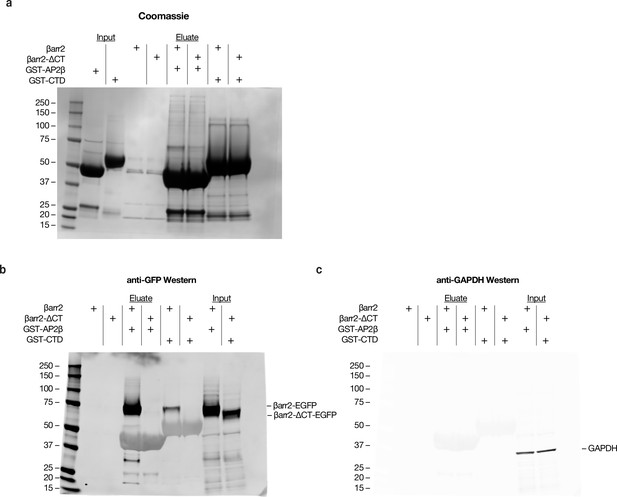
Representative unprocessed images of Coomassie stained sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE) gel and western blots.
(a) Uncropped Coomassie stained SDS/PAGE gel of GST-AP2β and GST-CTD showing input of both proteins and glutathione resin eluate (see Methods). (b) Anti-GFP western blot showing pull-down of βarr2-EGFP but not of βarr2-ΔCT-EGFP. (c) Anti-GAPDH western blot showing similar protein loading of βarr2-EGFP and βarr2-ΔCT-EGFP containing cell lysates. For (b, c), the large dimmer bands in eluate samples with molecular weights between 37 and 50 kDa are non-specifically labeled GST-AP2β and GST-CTD. See Figure 1—figure supplement 1j for cropped images and Figure 1—figure supplement 1—source data 1 for raw images. All data shown are representative of three independent experiments.
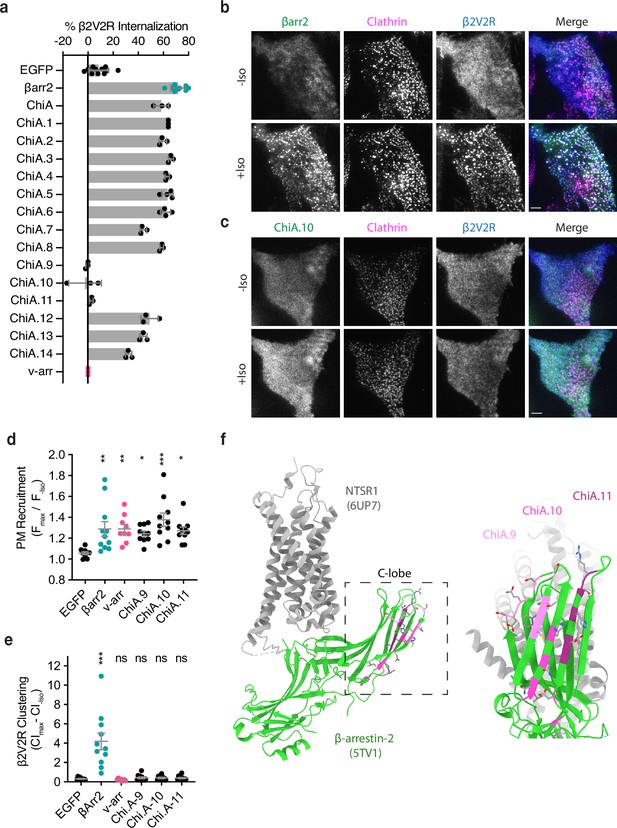
Identification of the βarr2 C-lobe base (CLB).
(a) Percent internalization of β2V2R after 30 min of 10 μM isoproterenol stimulation in βarr1/2 DKO HEK293s coexpressing the indicated construct (n ≥ 3 independent experiments, line is mean, error bars are standard deviation, each dot is an average of three technical replicates). Representative total internal reflection fluorescence (TIRF) microscopy images of cells expressing β2-adrenergic receptor (β2AR) (blue) and clathrin-light-chain-dsRed (magenta) with either wild-type βarr2-EGFP (b) or an example of one of the three internalization-defective chimeras, ChiA.10-EGFP (c) pre- and post-stimulation with 10 μM isoproterenol (Iso). Scale bars are 5 μm. (d) Plasma membrane recruitment of the indicated EGFP-tagged proteins (see Methods) in response to stimulation with 10 μM isoproterenol. (e) Maximum clustering index (CI, see Methods) of plasma membrane β2V2R after treatment with 10 μM Iso. For (d, e), each dot represents an individual cell. Data are shown as mean ± standard error of the mean (SEM) (n ≥ 9 cells). Significance was determined by ordinary one-way analysis of variance (ANOVA) (df = 5 for both, F = 22.21 and 4.531, respectively) with Dunnett’s multiple comparison test against negative control (EGFP) (ns p ≥ 0.05, *p < 0.05, **p < 0.01, ***p < 0.001). (f) Location of mutations unique to ChiA.9–11 (shades of pink and purple) in an active state structure of β-arrestin-2 (5TV1, green) (Chen et al., 2017) fit to the NTSR1/βarr1 structure (6UP7, gray) (Huang et al., 2020) (βarr1 not shown) and the same model rotated and zoomed to the cytoplasmic face of the C-lobe. All data shown are from at least three independent experiments.
-
Figure 2—source data 1
Source data for quantifications graphed in Figure 2.
- https://cdn.elifesciences.org/articles/81563/elife-81563-fig2-data1-v2.zip
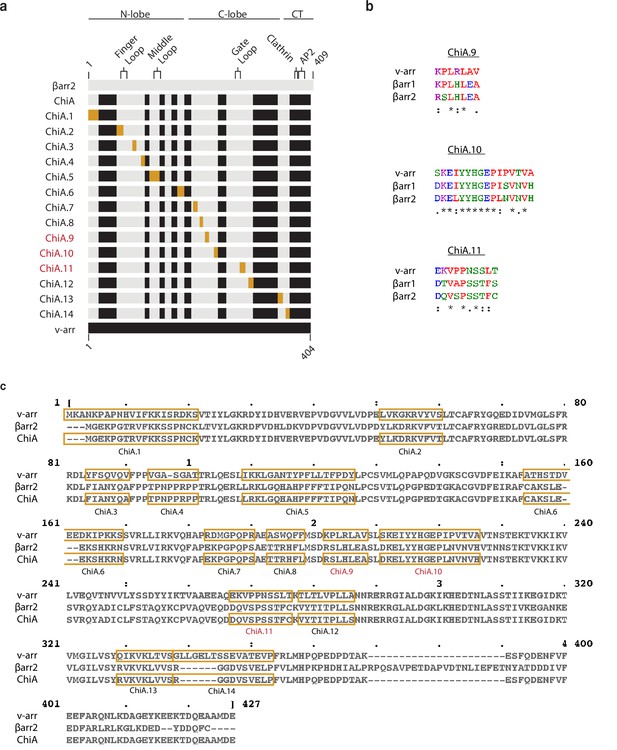
Diagram of chimeras and sequence alignments of arrestins.
(a) Diagram of βarr2 (gray) and visual arrestin (black) sequences. Visual arrestin sequences swapped into ChiA are gold to make ChiA.1–14. Major structural landmarks are labeled for βarr2. (b) Multiple sequence alignment of visual arrestin (v-arr), βarr1, and βarr2 of regions that are necessary for CT-independent endocytic activity of ChiA.9–11. “*” indicates a single, fully conserved residue, “:” indicates conservation between residues with strongly similar properties, “.” indicates conservation between residues with weakly similar properties. (c) Multiple sequence alignment of visual arrestin (v-arr), βarr2, and ChiA. Gold boxes in the v-arr sequence replace gold boxes in the ChiA sequence to make the indicated chimera. ChiA.9, 10, and 11 (red) abolished internalization of the β2V2R.
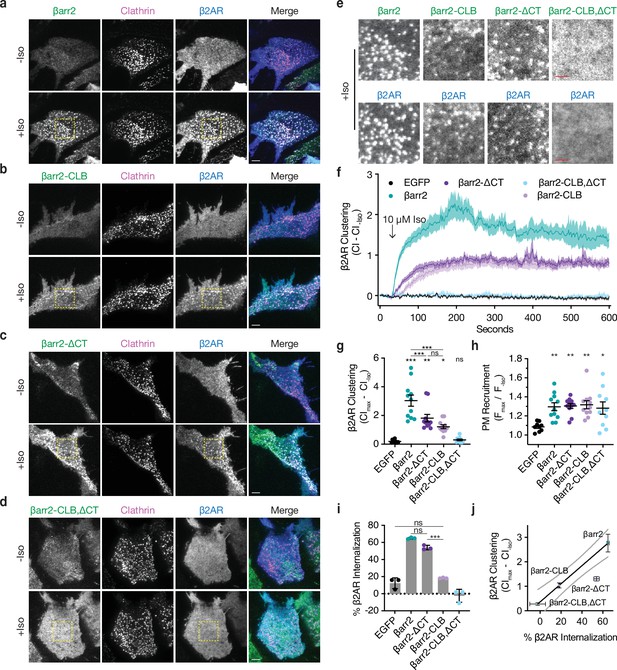
βarr2 C-terminus (CT) is not sufficient for β2-adrenergic receptor (β2AR) internalization.
Representative live-cell total internal reflection fluorescence (TIRF) microscopy images of βarr1/2 double knockout HEK293s coexpressing clathrin-light-chain-DsRed (magenta) and FLAG-tagged β2AR (blue) with either EGFP-tagged βarr2 (a), β arr2-CLB (b), βarr2-ΔCT (c), or βarr2-CLB,ΔCT (d) (all in green) pre- and post-stimulation with 10 μM isoproterenol (Iso). Scale bars represent 5 μm. EGFP condition not shown (see Figure 1a for example). (e) Zoomed images corresponding to dashed boxes in panels a–d for βarr2 and β2AR images. Scale bars (red) represent 2.5 μm. (f) β2AR clustering index (CI, see Methods) pre- and post-stimulation with 10 μM Iso over 10 min. (g) Max plasma membrane recruitment of the indicated EGFP-tagged proteins in response to treatment with 10 μM Iso. (h) Max clustering index (CI) of β2AR calculated from within the first 300 s of (f) and normalized to clustering index prior to Iso treatment. For (f–h), data shown as mean ± standard error of the mean (SEM) (n ≥ 9 cells, represented as dots in g and h). (i) Internalization of β2AR when coexpressed with the indicated EGFP-tagged proteins (n = 3, each dot is an average of three technical replicates) in βarr1/2 DKO HEK293 cells. (j) Correlation between β2AR clustering and internalization. Solid line is a simple linear regression fit to βarr2 and βarr2-CLB,ΔCT (R2 = 0.69, dashed lines = 95% CI, vertical error = SEM, and horizontal error = std. dev.). For (g–i), significance was determined by ordinary one-way analysis of variance (ANOVA) (df = 4 for all, F = 21.32, 4.828, and 117.6, respectively) with Tukey’s test for multiple comparisons (ns p ≥ 0.05, *p < 0.05, **p < 0.01, ***p < 0.001). All data shown are from at least three independent experiments.
-
Figure 3—source data 1
Source data for the figures displayed in Figure 3.
- https://cdn.elifesciences.org/articles/81563/elife-81563-fig3-data1-v2.zip
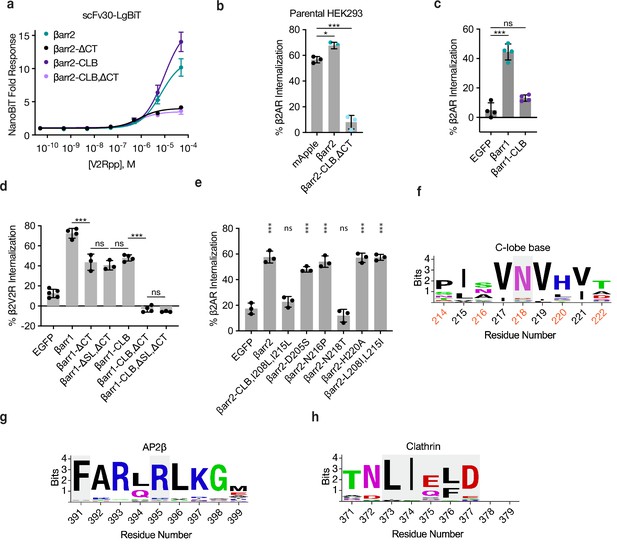
βarr2 and mutants bind scFv30, βarr2-CLB,ΔCT acts as a dominant negative, CLB is required for β2-adrenergic receptor (β2AR) internalization in βarr1, and mutating a conserved residue (N218) in βarr2 abolished β2AR internalization.
(a) NanoBiT complementation of scFv30-LgBiT and βarr2-SmBiT (or mutants) in βarr1/2 DKO cell lysate incubated with the indicated concentrations of V2Rpp. Dose–response curves were generated with three-parameter nonlinear fit (R2 = 0.98-0.99). logEC50 ± 95% CI values are −5.2 ± 0.1, −6.1 ± 0.4, −5.0 ± 0.1, and −6.1 ± 0.5 for wild type, ΔCT, CLB, and double mutant, respectively. Data shown as mean ± standard error of the mean (SEM). (b) Percent β2AR internalization in parental cells when co-expressed with either mApple, βarr2-mApple, or βarr2-CLB,ΔCT-mApple mutant after 30 min of treatment with 10 μM isoproterenol. (c) Internalization of β2AR after 30 min of stimulation with 10 μM isoproterenol when coexpressed with either EGFP, EGFP-tagged βarr1, or the βarr1 CLB mutant (D204S, S215P, N217T, and H219A) in βarr1/2 DKO HEK293s. (d) Percent internalization of β2V2R in βarr1/2 DKO cells when coexpressed with βarr1 or βarr1 constructs with the clathrin-binding splice loop removed (334-LLGDLASS-341). (e) Internalization of FLAG-tagged β2AR coexpressed with the indicated construct after 30 min of stimulation with 10 μM isoproterenol. Data shown as mean ± standard deviation. For (b–e), significance determined by ordinary one-way analysis of variance (ANOVA) (df = 4, F = 121.5) with Sidak’s multiple comparison test (b), ordinary one-way ANOVA (df = 2, F = 88.87) with Tukey’s multiple comparison test (c), ordinary one-way ANOVA (df = 19, F = 161.7) with Tukey’s multiple comparison test (d), one-way ANOVA (df = 8, F = 75.64) with Dunnett’s multiple comparison test against the negative control (EGFP) (e). All data shown are from at least three independent experiments (n ≥ 3, ns p ≥ 0.05, * p < 0.05, ***p < 0.001). Logos for sequence from both βarr1 and βarr2 around N218 in the βarr2 CLB (f), AP2β-binding site (g), and clathrin-binding box (gray) (h). Solvent exposed residues (CLB only) are numbered in orange. Amino acids that are critical to function or binding are colored in gray.
-
Figure 3—figure supplement 1—source data 1
Source data for results graphed in Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/81563/elife-81563-fig3-figsupp1-data1-v2.zip
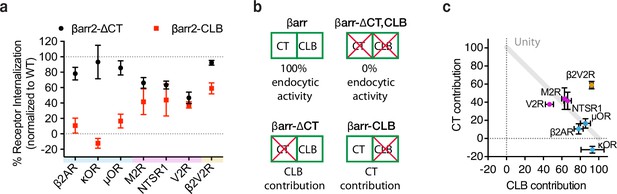
G-protein-coupled receptors (GPCRs) selectivity utilizes the βarr2 CLB and C-terminus (CT) for endocytosis.
(a) Internalization of the CT (black) and CLB (red) mutants normalized to wild-type βarr2 for each receptor after 30 min of agonist (see Figure 4—figure supplement 1). Each dot is the mean of three independent experiments ± standard deviation. Shading indicates whether receptors are naturally occurring ‘class a’ (blue), ‘class b’ (magenta), or ‘engineered class b’ (gold). (b) Schematic summarizing the conceptual basis for estimating contributions of the CT and CLB. Contribution of each determinant within βarr2 is defined by subtracting internalization measured in the negative control (EGFP) from βarr2, βarr2-ΔCT, βarr2-CLB, and dividing the resulting values by control (EGFP) subtracted wild-type (βarr2) value. (c) Contribution to total endocytic activity of each determinant plotted as x and y coordinates for each receptor from panel (a). Unity is defined as 100% endocytic activity when individual activities are summed. Dot color corresponds to the typology described for panel (a). All data shown are from three independent experiments that were performed in βarr1/2 DKO HEK293 cells.
-
Figure 4—source data 1
Source data for results graphed in Figure 4.
- https://cdn.elifesciences.org/articles/81563/elife-81563-fig4-data1-v2.zip
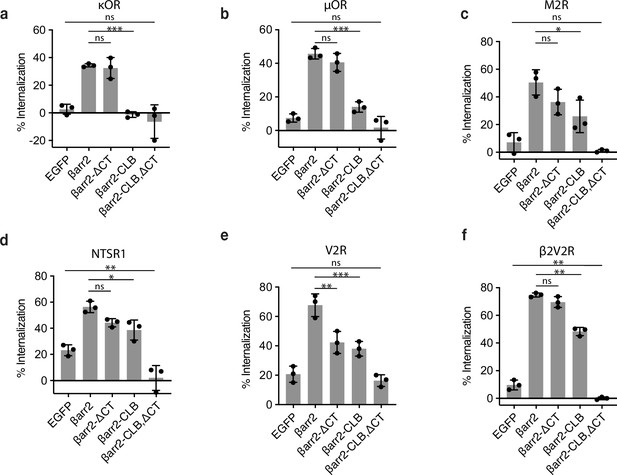
Internalization of G-protein-coupled receptors (GPCRs) coexpressed with βarr2 wild type or mutants.
Percent internalization of the indicated receptor after 30 min of stimulation with either 10 μM dynorphin A-17 (a), 10 μM DAMGO (b), 10 μM carbachol (c), 10 μM neurotensin (d), 1 μM arginine vasopressin (e), or 10 μM isoproterenol (f) in βarr1/2 DKO HEK293 cells. Significance was determined by ordinary one-way analysis of variance (ANOVA) (df = 4 for all, F = 436.6, 33.13, 17.52, 33.88, 25.55, and 60.51, respectively) (ns p ≥ 0.05, *p < 0.05, ** p < 0.01, ***p < 0.001). Data shown as mean ± standard deviation of three independent experiments.
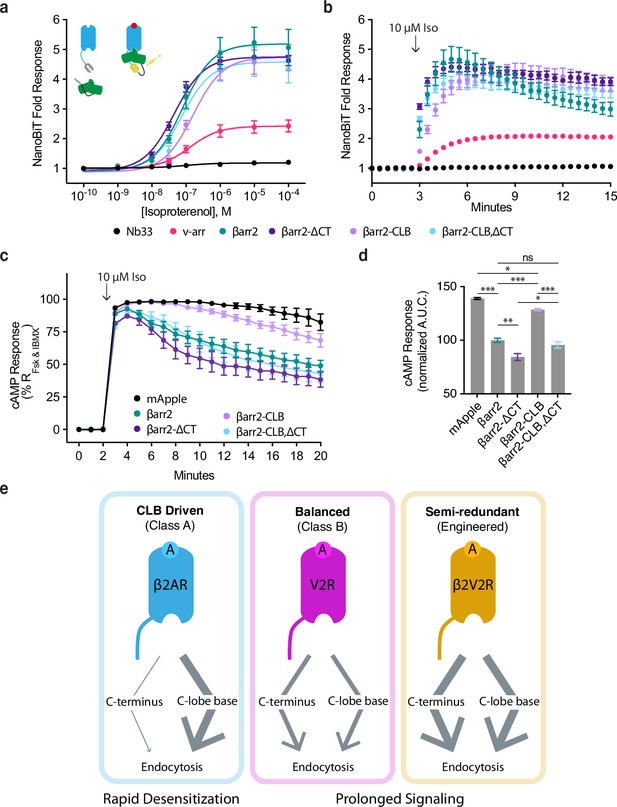
CLB and C-terminus (CT) determinants reveal two allosteric paths from G-protein-coupled receptors (GPCRs) to the endocytic network.
Direct NanoBiT luciferase complementation of β2-adrenergic receptor (β2AR)-LgBiT and SmBiT-tagged: Nb33 (a μOR receptor-specific nanobody, black), visual arrestin (pink), wild-type β-arrestin-2 (green), CT mutant (dark purple), CLB mutant (light purple), or double mutant (cyan) measured as an end point across a range of isoproterenol (Iso) concentrations (a) and kinetically (b) pre- and post-stimulation with 10 μM Iso. Dose–response curves were generated with three-parameter nonlinear fit (R2 = 0.94–0.99). (c) Endogenous β2AR cAMP response after stimulation with 10 μM Iso measured by a genetically encoded fluorescent cAMP biosensor, cADDis, and normalized to the response elicited by simultaneous treatment with 10 μM forskolin (Fsk) and 300 μM 3-isobutyl-1-methylxanthine (IBMX). (d) Area under the curve calculated from panel (c). All data are shown as mean ± standard error of the mean (SEM). from three independent experiments performed in βarr1/2 DKO HEK293 cells. Significance was determined by an ordinary one-way analysis of variance (ANOVA) (df = 4, F = 112.1) with Tukey’s multiple comparisons test. ns p ≥ 0.05, *p < 0.05, ** p < 0.01, ***p < 0.001. (e) Diagram of proposed model involving two differentially utilized allosteric paths from GPCRs through β-arrestins to promote endocytosis. Class A GPCRs (blue), exemplified by the β2AR, primarily utilize the CLB to drive endocytosis while Class B GPCRs, exemplified by V2R (magenta) and β2V2R (gold) utilize both determinants. Arrows represent the proposed allosteric paths linking the GPCR/β-arrestin interface to the β-arrestin/clathrin-coated pit (CCP) interface, explaining how the CLB-dependent endocytic mode is coupled to rapid desensitization of receptor signaling while the CT-dependent mode enables prolonged signaling.
-
Figure 5—source data 1
Source data for results graphed in Figure 5.
- https://cdn.elifesciences.org/articles/81563/elife-81563-fig5-data1-v2.zip
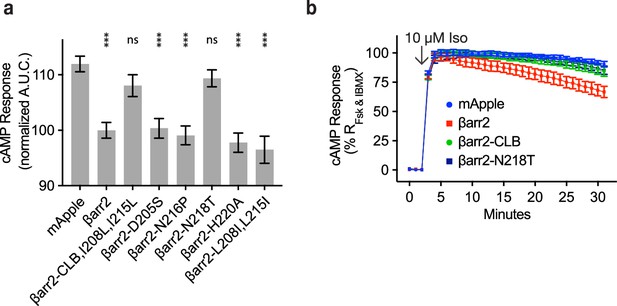
N218 in βarr2 is required for endogenous β2-adrenergic receptor (β2AR) desensitization.
(a) Area under the curve (AUC) for the cAMP response elicited by endogenous β2AR after stimulation with 10 μM isoproterenol in βarr1/2 DKO HEK293s expressing the indicated construct and normalized to response in cells expressing wild-type βarr2. Data shown as mean ± standard error of the mean (SEM). Significance determined by ordinary one-way analysis of variance (ANOVA) (df = 9, F = 13.47) with Dunnett’s multiple comparison test against the negative control (EGFP). (b) Example kinetics of the cAMP response from endogenous β2AR after treatment with 10 μM Iso in βarr1/2 DKO HEK293 expressing the negative control (mApple, blue), βarr2-mApple (red), βarr2-CLB-mApple (green), and βarr2-N218T-mApple (dark blue). Data shown as mean ± standard deviation (n = 3). Kinetics for other mutants are not shown. All data shown are from three independent experiments. (ns p ≥ 0.05, *p < 0.05, ** p < 0.01, ***p < 0.001).
-
Figure 5—figure supplement 1—source data 1
Source data for results graphed in Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/81563/elife-81563-fig5-figsupp1-data1-v2.zip
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | Parental HEK293 | O’Hayre et al., 2017 (PMID: 28634209) | ||
| Cell line (Homo sapiens) | HEK293 βarr1/2 double knockout, line 1 | O’Hayre et al., 2017 (PMID: 28634209) | ||
| Cell line (Homo sapiens) | HEK293 βarr1/2 double knockout, line 2 | O’Hayre et al., 2017 (PMID: 28634209) | ||
| Recombinant DNA reagent | FLAG-β2AR (plasmid) | Cao et al., 1999 (PMID: 10499588) | ||
| Recombinant DNA reagent | FLAG-β2AR-3S (plasmid) | Hausdorff et al., 1991 (PMID: 1849641) | ||
| Recombinant DNA reagent | FLAG-V2R (plasmid) | Lefkowitz Laboratory, Duke University | ||
| Recombinant DNA reagent | FLAG-μOR (plasmid) | Tanowitz and von Zastrow, 2003 (PMID: 12939277) | ||
| Recombinant DNA reagent | FLAG-κOR (plasmid) | Chu et al., 1997 (PMID: 9341153) | ||
| Recombinant DNA reagent | FLAG-NTSR1 (plasmid) | Huang et al., 2020 (PMID: 31945771) | ||
| Recombinant DNA reagent | FLAG-β2V2R (plasmid) | Oakley et al., 1999 (PMID: 10542263) | ||
| Recombinant DNA reagent | FLAG-M2R (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | FLAG-β2AR-LgBiT (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | βarr2-EGFP (plasmid) | Barak et al., 1997 (PMID: 9346876) | ||
| Recombinant DNA reagent | βarr2-mApple (plasmid) | Eichel et al., 2016 (PMID: 26829388) | ||
| Recombinant DNA reagent | Clathrin-light-chain-dsRed (plasmid) | Merrifield et al., 2002 (PMID: 12198492) | ||
| Recombinant DNA reagent | βarr2-CCS-EGFP (plasmid) | Eichel et al., 2018 (PMID: 29720660) | ||
| Recombinant DNA reagent | βarr2-ΔCT-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | v-arr-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | ChiA-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | ChiA.1-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | ChiA.2-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | ChiA.3-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | ChiA.4-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | ChiA.5-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | ChiA.6-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | ChiA.7-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | ChiA.8-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | ChiA.9-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | ChiA.10-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | ChiA.11-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | ChiA.12-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | ChiA.13-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | ChiA.14-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | βarr2-CLB-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | βarr2-CLB,ΔCT-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | EGFP-Nb33-SmBiT (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | βarr2-SmBiT (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | βarr2-ΔCT-SmBiT (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | βarr2-CLB-SmBiT (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | βarr2-CLB,ΔCT-SmBiT (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | mApple (plasmid) | Steinbach et al., 2008 (PMID: 18454154) | ||
| Recombinant DNA reagent | EGFP (plasmid) | Clontech | Discontinued | |
| Recombinant DNA reagent | βarr2-ΔCT-mApple (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | βarr2-CLB-mApple (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | βarr2-CLB,ΔCT-mApple (plasmid) | This paper | See Materials and methods | |
| Commercial assay or kit | In-Fusion HD Cloning | Takara | 638920 | |
| Recombinant DNA reagent | βarr1-ΔCT-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | GST-AP2β (plasmid) | Jeff Benovic and Harvey McMahon | ||
| Recombinant DNA reagent | GST-CTD (plasmid) | Kang et al., 2009 (PMID: 19710023) | ||
| Strain, strain background (Escherichia coli) | BL21 DE3 | QB3 MacroLab UC Berkeley | ||
| Recombinant DNA reagent | scFv30-LgBiT (plasmid) | This paper | See Materials and methods | |
| Commercial assay, kit | Lipofectamine 2000 | Thermo Fisher Scientific | 11668019 | |
| Commercial assay, kit | Lipofectamine RNAi Max | Invitrogen | 13778075 | |
| Transfected construct (human) | siRNA, negative control | Qiagen | 1027281 | |
| Transfected construct (human) | siRNA, clathrin heavy chain | Qiagen | 5′- AAGCAATGAGCTGTTTGAAGA-3′ | |
| Peptide, recombinant protein | V2Rpp | Tufts University Core Facility | ARGRTPPSLGPQDESCTTASSSLAKDTSS (phosphorylated residues underlined) | |
| Recombinant DNA reagent | βarr1-CLB-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | βarr1-ΔSL,ΔCT-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | βarr1-CLB,ΔCT-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | βarr1-CLB,ΔSL,ΔCT-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | βarr2-CLB,I208L,I215L-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | βarr2-D205S-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | βarr2-N216P-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | βarr2-N218T-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | βarr2-H220A-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | βarr2-L208I,L215I-EGFP (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | βarr2-CLB,I208L,I215L-mApple (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | βarr2-D205S-mApple (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | βarr2-N216P-mApple (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | βarr2-N218T-mApple | This paper | See Materials and methods | |
| Recombinant DNA reagent | βarr2-H220A-mApple (plasmid) | This paper | See Materials and methods | |
| Recombinant DNA reagent | βarr2-L208I,L215I-mApple (plasmid) | This paper | See Materials and methods | |
| Commercial assay, kit | cADDis Green Upward | Montana Molecular | #U0200G | |
| Chemical compound, drug | Forskolin (Fsk) | Sigma-Aldrich | F6886 | |
| Chemical compound, drug | 300 μM 3-isobutyl-1-methylxanthine (IBMX) | Sigma-Aldrich | F5879 | |
| Chemical compound, drug | (−)-Isoproterenol hydrochloride | Sigma-Aldrich | I6504 | |
| Chemical compound, drug | DADLE, [D-Ala2, N-Me-Phe4, Gly5-ol]-Enkephalin acetate salt | Sigma-Aldrich | E7131 | |
| Chemical compound, drug | DAMGO, [D-Ala2, N-Me-Phe4, Gly5-ol]-Enkephalin acetate salt | Sigma-Aldrich | E7384 | |
| Chemical compound, drug | Carbamoylcholine chloride, ≥98% (titration), crystalline | Sigma-Aldrich | C4382 | |
| Chemical compound, drug | Neurotensin, ≥90% (HPLC) | Sigma-Aldrich | N6383 | |
| Chemical compound, drug | AVP [Arg8]-Vasopressin acetate salt | Sigma-Aldrich | V9879 | |
| Chemical compound, drug | Coelenterazine-H | Thermo Fisher Scientific | 50-995-840 | |
| Antibody | αGFP (mouse diclonal) | Roche | 11814460001 | 1:1000 |
| Antibody | αGAPDH (rabbit monoclonal) | Cell Signalling Technologies | 5174S | 1:1000 |
| Antibody | M1 anti-flag (mouse monoclonal) | Sigma-Aldrich | F-3040 | 1:1000 |
| Antibody | Donkey Anti-Mouse IgG Antibody, IRDye 680RD Conjugated – 0.5 mg (donkey polyclonal) | Li-cor Biosciences | 926-68072 | 1:3000 |
| Antibody | IRDye 800CW Donkey anti-Rabbit IgG (H+L), 0.5 mg (donkey polyclonal) | Li-cor Biosciences | 926-32213 | 1:3000 |
| Commercial assay, kit | Alexa Fluor 647 Protein Labeling Kit | Thermo Fisher Scientific | A20173 | |
| Software, algorithm | Prism | GraphPad | 9.0 | |
| Software, algorithm | ImageJ | https://imagej.net/downloads | 2.0.0-rc-54/1.51g | |
| Software, algorithm | Excel | Microsoft | 16.11.1 | |
| Software, algorithm | ChimeraX | UCSF Resource for Biocomputing, Visualization, and Informatics | 1.4 | |
| Software, algorithm | Illustrator CC | Adobe | 21.0.2 | |
| Software, algorithm | Python | Python Software Foundation | 3.7.4 |
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/81563/elife-81563-mdarchecklist1-v2.pdf
-
Supplementary file 1
List of primers used.
- https://cdn.elifesciences.org/articles/81563/elife-81563-supp1-v2.xlsx





