Noncovalent antibody catenation on a target surface greatly increases the antigen-binding avidity
Figures
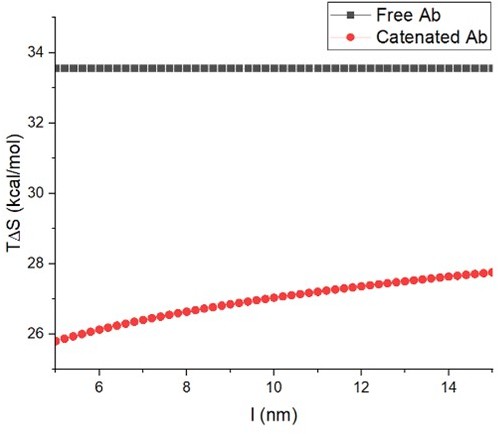
The concept of antibody catenation on a target surface by fusion of a catenator.
(A) Molecular model for catenator-fused antibodies. A flexible linker (Gly-Gly-Ser) between Fc and the catenator and the hinge segment between Fc and Fab were modeled by using the ROSETTA software. The catenator is an α-helical hairpin that forms four-helix anti-parallel coiled coils (PDB entry: 1ROP). The structure of Fc was derived from the IgG1 antibody (PDB entry: 1IGY) and that of Fab from an antibody against the receptor-binding domain of the SARS-CoV-2 spike protein (PDB entry: 6XE1). (B) Decreased dissociation by antibody catenation. Pairs of catAb-antigen complexes adjacent to each other can be catenated, and the catAb molecules are increasingly harder to dissociate from each other with increased catenation. The effective antigen-binding avidity would increase owing to a decreased off rate of catAb.

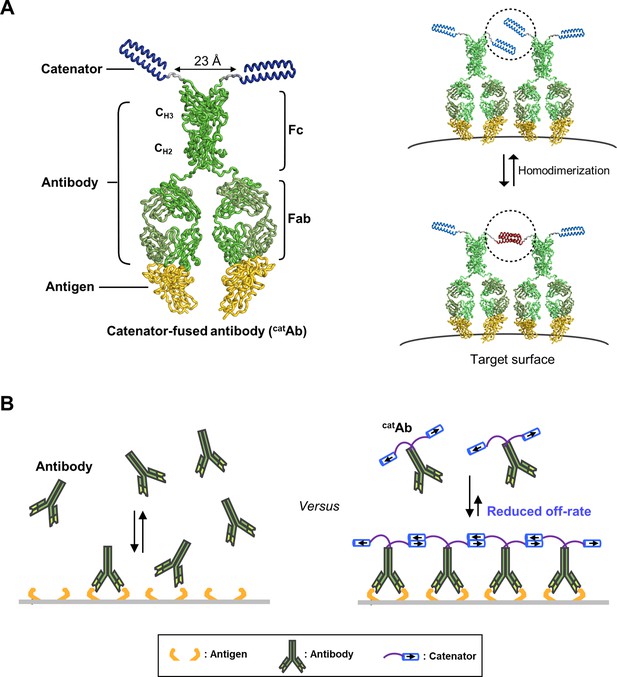
ABM for simulating the binding dynamics of a catenator-fused antibody.
(A) (Left) Each binding site is composed of two antigen molecules (2Ag). (Right) The gray circles indicate the sphere sampled by the catenator, and Voverlap is the overlapping volume between the adjacent spheres. Catenation between two catAb molecules is possible only in Voverlap. (B) The three rules of the ABM model. (Left) catAb-2Ag binding occurs with a relative likelihood, [catAb]/KD. (Middle) The catenation between adjacent catAb-2Ag complexes occurs with an indicated relative likelihood, f(d)/(KD)Catenator, determined by (KD)Catenator and the inter-complex distance d. (Right) It was assumed that catAb molecules that are catenated do not dissociate from the surface. (C) The simulation requires specification of the parameters for the binding site, antibody and catenator. Through the MCMC sampling, the state of binding sites on the target surface is iteratively updated with the ABM rules and eventually sampled. A sufficient number of sampling results are collected to quantify the binding occupancy and the effective dissociation constant.
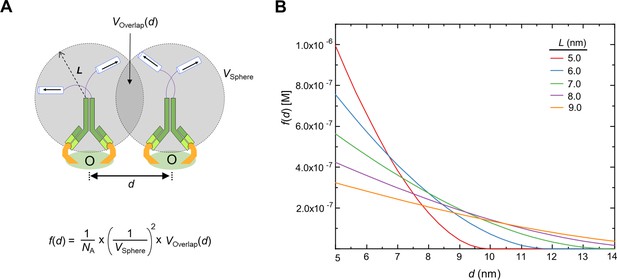
Calculation of f(d) using uniform local density approximation.
(A) Definition of parameters and the equation of f(d). The forward catenation rate at which two catenators dimerize is proportional to the volumetric overlap (V(d)) between the effective concentration of the catenator, which is assumed to be uniformly distributed over a sphere defined by the reach length (L). V(d) depends on the distance (d) between the two adjacent catAb-2Ag complexes as well as the reach length. (B) Plot of f(d) as a function of d calculated for the indicated reach length (L).
-
Figure 2—figure supplement 1—source data 1
Simulation data for catenation enhancement by binding sites interval and linker length in Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/81646/elife-81646-fig2-figsupp1-data1-v1.xlsx
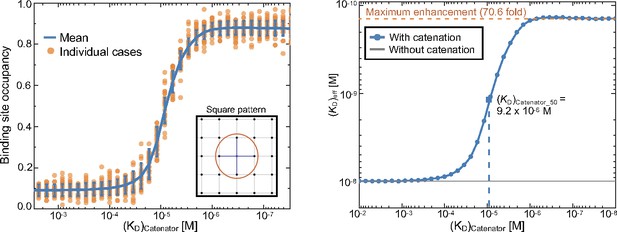
Simulations of the binding site occupancy and (KD)eff in response to (KD)catenator.
(Left) Binding site occupancy. The simulations were carried out for a square array of the binding sites. The values for a set of variables were KD = 10–8 M, [catAb]=10–9 M, reach length = 7 nm, spacing between the binding sites = 12 nm and the number of total binding sites = 98. The mean value and standard deviations of 1024 MCMC simulations for each (KD)catenator value are shown in blue, and the data are shown as a scatter plot of representative runs (orange). (Right) The effective dissociation constant. The data shown on the left were converted into the (KD)eff values. The dashed line represents the KD value for the same antibody without a catenator. The maximum fold enhancement of the effective binding avidity, which is equivalent to the reduction of (KD)eff, is 70.6.
-
Figure 3—source data 1
Simulation data for Figure 3.
- https://cdn.elifesciences.org/articles/81646/elife-81646-fig3-data1-v1.xlsx
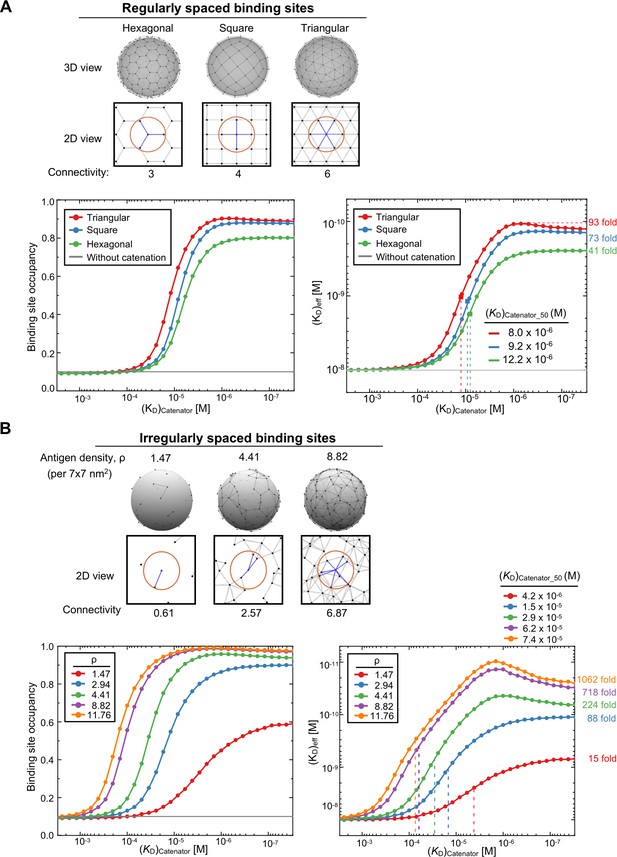
Simulations for different arrays of the binding sites.
(A) Comparison for regularly distributed binding sites. Three different regular arrays of the binding sites are shown at the top. The black dots represent the binding sites and the gray lines the connectable pairs by the catenators. The red circles and the blue lines represent the maximum range of catenation and the connectivity number, respectively, for a given binding site. Binding site occupancy and (KD)eff in response to (KD)catenator are shown at the bottom. 1024 trials were sampled for each (KD)catenator value and the results are plotted. The variables were KD = 10–8 M, [catAb]=10–9 M, reach length = 7 nm, spacing between the binding sites = 12 nm, and the number of total binding sites were 98 for the square array and 102 for hexagonal and triangular array, respectively. The numbers on the right are the maximum fold enhancement of the effective binding avidity for each array. (B) Comparison for randomly distributed binding sites. Three random arrays of the binding sites with different binding site densities (ρ) are shown at the top. The surface area for the simulation was 5760 nm2. The simulation conditions were the same as in (A). Binding site occupancy and (KD)eff in response to (KD)catenator are plotted as in (A).
-
Figure 4—source data 1
Simulation data for regularly spaced binding sites in Figure 4.
- https://cdn.elifesciences.org/articles/81646/elife-81646-fig4-data1-v1.xlsx
-
Figure 4—source data 2
Simulation data for irregularly spaced binding sites in Figure 4.
- https://cdn.elifesciences.org/articles/81646/elife-81646-fig4-data2-v1.xlsx
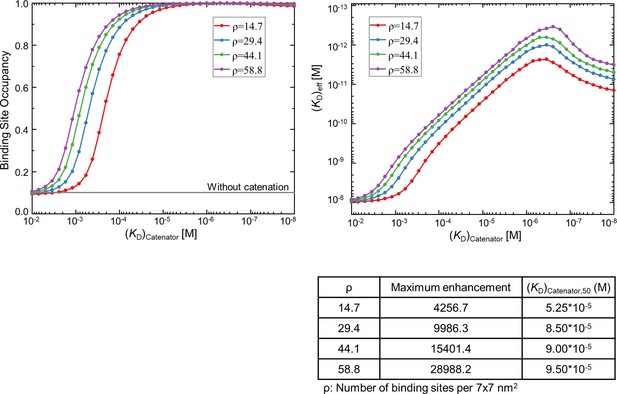
Simulations for randomly distributed, high-density binding sites.
1024 trials were sampled for each (KD)catenator value at the indicated density and the results are plotted. The variables were KD = 10–8 M, [catAb]=10–9 M, reach length = 7 nm, spacing between the binding sites = 12 nm, and the surface area = 5760 nm2. The maximum fold enhancement of the effective binding avidity and (KD)catenator,50 are tabulated at the bottom.
-
Figure 4—figure supplement 1—source data 1
Simulation data for binding sites with high density in Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/81646/elife-81646-fig4-figsupp1-data1-v1.xlsx
-
Figure 4—figure supplement 1—source data 2
Values of the simulation data for binding sites with high density in Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/81646/elife-81646-fig4-figsupp1-data2-v1.xlsx
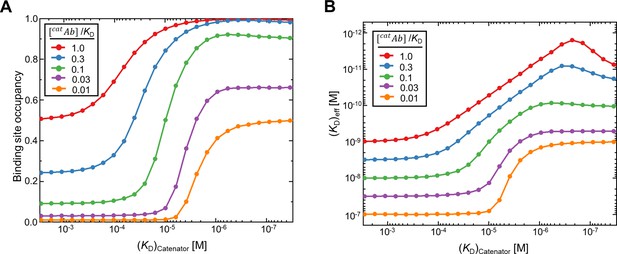
Influence of the likelihood of intrinsic antigen binding ([catAb]/KD) on binding site occupancy and (KD)eff.
(A) The binding occupancy and (B) the effective dissociation constant (KD)eff in response to [catAb]/KD for [catAb]/KD = 1.0, 0.3, 0.1, 0.03, 0.01, where [catAb] was fixed to 10–9 M and KD was varied from 10–8 to 10–6. The simulations were carried out with a square array of the binding sites as in Figure 3. The set values for the variable parameters were reach length = 7 nm, spacing between the binding sites = 12 nm and the number of total binding sites = 98. The mean binding occupancy of 1024 MCMC simulations was plotted. (KD)catenator was varied from 1 mM to 10 nM. The antibody binding avidity is substantially enhanced across a broad range of [catAb]/KD.
-
Figure 4—figure supplement 2—source data 1
Simulation data for varying likelihood of intrinsic antigen binding in Figure 4—figure supplement 2.
- https://cdn.elifesciences.org/articles/81646/elife-81646-fig4-figsupp2-data1-v1.xlsx
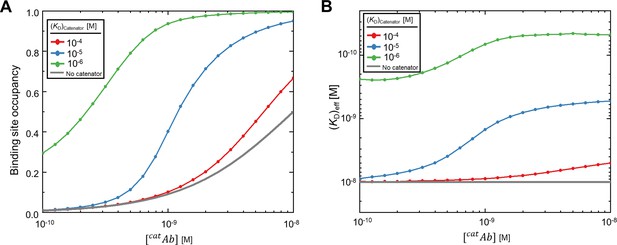
Influence of [catAb]/KD on binding site occupancy and (KD)eff.
(A) The binding occupancy and (B) the effective dissociation constant (KD)eff in response to [catAb] for (KD)catenator = 1, 10, 100 μM. KD was set to 10 nM, and [catAb] was varied from 0.1 to 10 nM. The simulations were carried out essentially the same as for Figure 2.
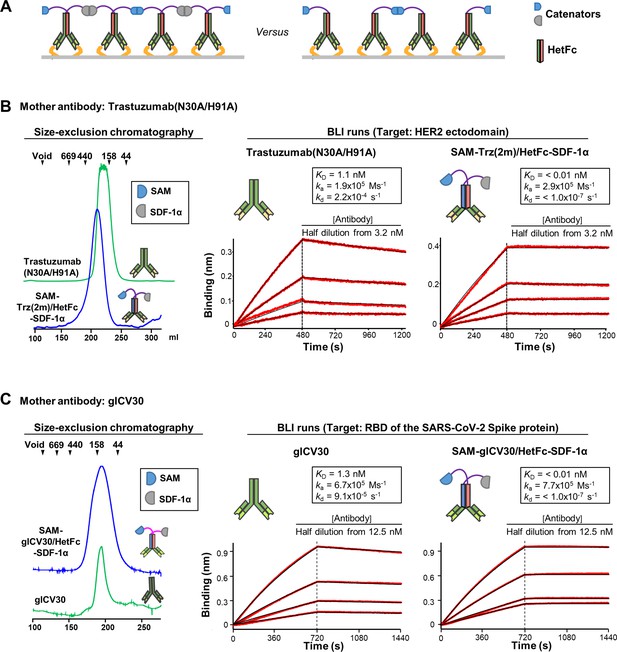
Catenation effect on the binding avidity.
(A) By employing a heterodimeric Fc (HetFc), an antibody fused to two different catenators (Left) or an antibody with one catenator arm (Right) can be generated. Only the former could form catenated antibodies on a surface where target antigens are abundant. (B) Elution profile and BLI analyses of Trastuzumab(N30A/H91A) and SAM-Trz(2 m)/HetFc-SDF-1α. Trz(2 m)/Het stands for Trastuzumab(N30A/H91A) with its homodimeric Fc replaced by a HetFc. The two heavy chains were fused to SAM or SDF-1α. The two antibodies were eluted from a size-exclusion column as if they were monomeric (Left). Elution positions of the size marker proteins are indicated by triangles. BLI runs for the two antibodies are shown (Right). The ectodomain of HER2 was immobilized on the biosensor tip. (C) Elution profile and BLI analyses of glCV30 and SAM-glCV30/HetFc-SDF-1α. The two antibodies were analyzed as described in (B). Commonly for all BLI runs, the experimental signals and fitted curves are shown in red and black, respectively. For curve fitting, 1:1 binding was assumed. The kinetic parameters are shown in the insets. ka, association rate constant; kd, dissociation rate constant. The experiments were performed in triplicates, and representative sensorgrams are shown.
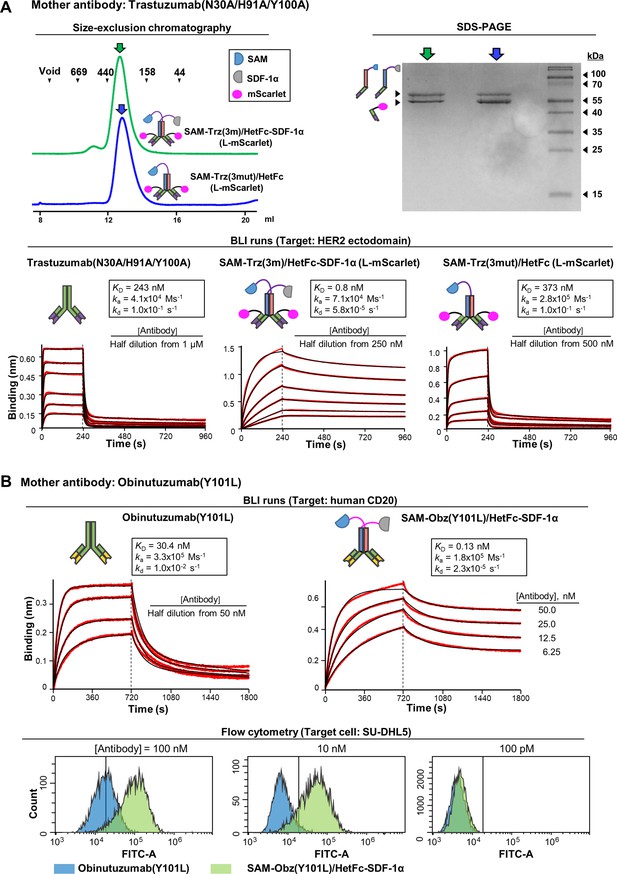
Catenation effects were observed for low-affinity mother antibodies.
(A) The triply mutated Trastuzumab(N30A/H91A/Y100A), the antibody with two catenator arms (SAM-Trz(3 m)/HetFc-SDF-1α) and the same antibody with one catenator arm (SAM-Trz(3 m)/HetFc) were prepared with their light chain (L) fused to mScarlet at the C-terminus. The two antibody-catenators were eluted from a size-exclusion column as if they were monomeric (Left). The SDS-PAGE of the peak fractions shows the correct sizes of the indicated chains (Right). BLI was performed similarly as described in Figure 5B. The two catenator-armed antibody exhibits a significant increase in the binding avidity, but the one catenator-armed antibody did not (KD values in the inset). (B) Obnutuzumab(Y101L) and SAM-Obz(Y101L)/HetFc-SDF-1α were prepared, and BLI was performed with biotinylated human CD20 immobilized on the biosensor tip (Top). Flow cytometric analyses of SU-DHL5-binding by the two antibodies were performed (Bottom). For this experiment, the two antibodies were labeled with muGFP at the C-terminus of the light chain. The fluorescent signal from muGFP was detected by using a 525/40 bandpass filter.
-
Figure 6—source data 1
SDS-PAGE result for SAM-Trastuzumab(N30A/H91A/Y100A)/HetFc-SDF1a and SAM-Trastuzumab(N30A/H91A/Y100A)/HetFc.
- https://cdn.elifesciences.org/articles/81646/elife-81646-fig6-data1-v1.zip
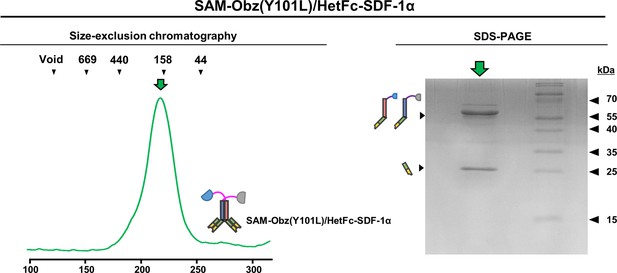
Purification of SAM-Obz(Y101L)/HetFc-SDF-1α.
Elution profile and SDS-PAGE analysis of SAM-Obz(Y101L)/HetFc-SDF-1α.
-
Figure 6—figure supplement 1—source data 1
SDS-PAGE result for SAM-Obinutuzumab(Y101L)/HetFc-SDF1a.
- https://cdn.elifesciences.org/articles/81646/elife-81646-fig6-figsupp1-data1-v1.zip
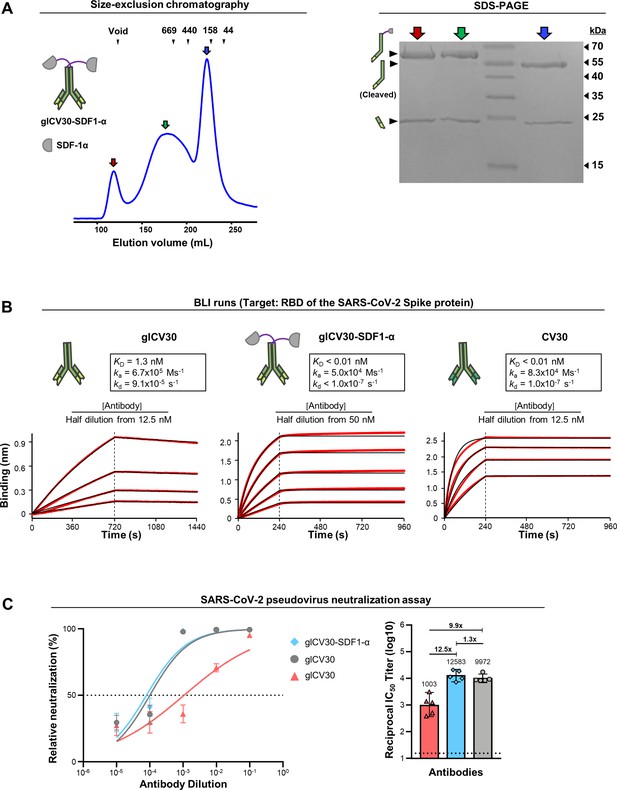
Analyses of glCV30-SDF-1α.
(A) Elution profile and SDS-PAGE analysis of glCV30-SDF-1α. (B) The BLI runs shown on the left of Figure 5C are reused here for easy comparison with those for glCV30-SDF-1α. The KD values could not be accurately determined due to instrumental insensitivity (KD <10 pM). The affinity-matured version, CV30, was analyzed and its BLI sensorgrams are shown on the right. (C) Neutralization of VSV virus pseudotyped with the SARS-CoV-2 spike protein. Each of the three antibodies was serially diluted and added to the rVSV-ΔG-Luc bearing the SARS-CoV-2 Spike protein of the Wuhan-Hu-1 strain. The mixture was incubated with HEK293T-hACE2 cells and the luciferase activity was measured. The geometric mean titer (GMT) values with a 95% confidence interval are shown on the right, which corresponds to the IC50 values of 0.25, 3.22 and 0.21 μg/ml of CV30, glCV30 and glCV30-SDF-1α, respectively.
-
Figure 7—source data 1
SDS-PAGE result for glCV30-SDF-1α.
- https://cdn.elifesciences.org/articles/81646/elife-81646-fig7-data1-v1.zip
Tables
Simulation specifications.
The definition and values of the parameters used in the presented simulations are tabulated.
| Parameters | Description | Values |
|---|---|---|
| Specification of catAb | ||
| KD | Dissociation constant of antibody | 10 nM |
| (KD)catenator | Dissociation constant of catenator | 10 nM-10 mM |
| [catAb] | Antibody concentration | 1 nM |
| l | Length of the flexible linker | 6 nm |
| c | Length of the catenator | 2 nm |
| L | Reach length (l+c/2) | 7 nm |
| Specification of the target surface | ||
| Ntotal_binding_sites (in Figures 3 and 4) | Number of antibody-binding sites | 98–102 |
| Connectivity number (in Figures 3 and 4) | Number of possible catenation | 3 (Hexagonal) 4 (Square) 6 (Triangular) |
| d (in Figures 3 and 4) | Distance between adjacent binding sites | 12 nm |
| Lsurface (in Figure 4) | Surface area of the target surface | 40 nm2 |
| Binding site density (in Figure 4) | Surface density of the binding sites | 1.47–11.76 (per 7x7 nm2) |
| Specification of simulation | ||
| Updates/MCMC step | Number of updates in one MCMC step | 30,000–100,000 |
| Sampling size | Number of sampling for a parameter set | 1024 |
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/81646/elife-81646-mdarchecklist1-v1.docx
-
Source code 1
Github repository to generate simulation data for Figures 3 and 4.
- https://cdn.elifesciences.org/articles/81646/elife-81646-code1-v1.zip