Asymmetric retinal direction tuning predicts optokinetic eye movements across stimulus conditions
Figures
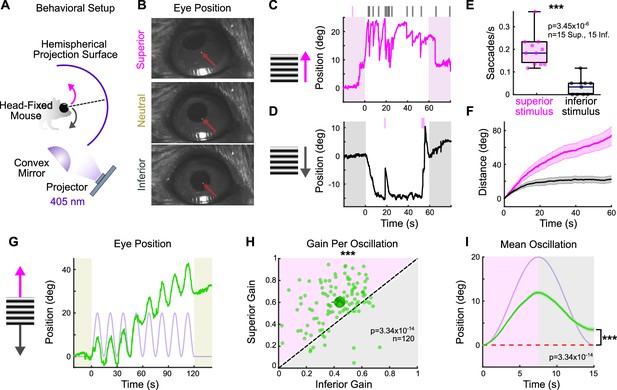
The superior and inferior optokinetic reflex (OKR) are asymmetric in adult mice.
(A) Schematic of behavioral setup to elicit the vertical OKR. The mouse is situated so that one eye is centered in a hemisphere. Stimuli are projected onto the hemisphere’s concave surface via reflection off of a convex mirror. Eye movements are tracked using an infrared-sensitive camera and a corneal reflection (see ‘Materials and methods’). (B) Example video frames demonstrating that the eye traverses between superior, neutral, and inferior positions in the presence of vertically drifting sinusoidal gratings. Red arrows mark the infrared corneal reflection. (C, D) Example of OKR in response to full contrast (C) superior and (D) inferior unidirectional drifting gratings (10°/s). For each epoch, a continuous 60 s stimulus was flanked by 20 s of a static grating (shaded regions). Ticks above the plots mark the time of fast nystagmus either in the superior (magenta) or inferior (gray) direction. Examples from one animal. (E) Rate of vertical fast nystagmus for superior and inferior stimuli on each epoch for N = 5 mice. Horizontal line represents median, box boundaries are the interquartile range (IQR), whiskers represent most extreme observation within 1.5× IQR. (F) Cumulative vertical distance traveled during slow nystagmus in response to superior (magenta) and inferior (gray) drifting gratings (mean ± SEM). (G) Example of OKR in response to a vertically oscillating sinusoidal grating. The eye position is shown in green, and the stimulus position is shown in lavender. Saccades (‘fast nystagmuses’) have been removed to reveal the asymmetry between superior and inferior OKR. For each epoch, animals viewed eight oscillation cycles lasting a total of 120 s, flanked by 20 s of a static grating (shaded regions). (H) Average gain of slow nystagmus during the superior versus inferior stage of individual oscillations. Each small dot is a single oscillation. The region of magenta (or gray) indicates that gain was greater for the superior (or inferior) stage of the oscillation. Points that fall on the line indicate equivalent gain for both stimulus directions. Large dot and whiskers represent univariate medians and 95% confidence intervals (via bootstrapping), respectively. Significance value indicates whether the points tend to fall unevenly on one side of the unity line (two-sided signed-rank). (I) Eye position (green) and stimulus position (lavender) averaged across all oscillations and all animals (mean ± SEM). Starting eye position is normalized to 0° at cycle onset. The average ending eye position is displaced in the superior direction (two-sided signed-rank). N = 5 mice for all experiments; n = number of trials. *p<0.05, **p<0.01, ***p<0.001.
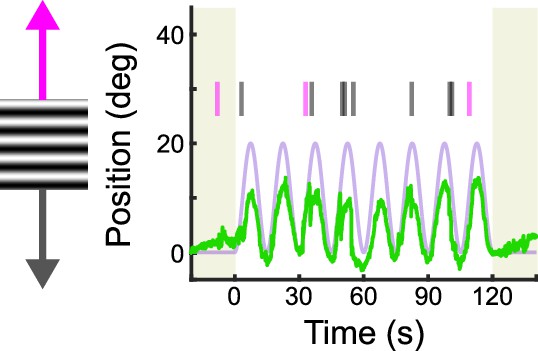
Example of sinusoidal vertical optokinetic reflex (OKR) before saccade removal.
Eye position (green) is plotted across time as a full-field grating oscillates vertically (lavender). The eye trace includes saccades (i.e., ‘fast nystagmuses,’ as indicated by tick marks: magenta for superior, gray for inferior). Saccades tend to occur in the opposite direction as the ‘slow nystagmus’ and do not facilitate image stabilization. However, saccades are necessary to keep the eye centered in its orbit. Removing these resetting saccades from the eye trace isolates the slow nystagmus component and reveals the asymmetry between superior and inferior OKR (Figure 1G).
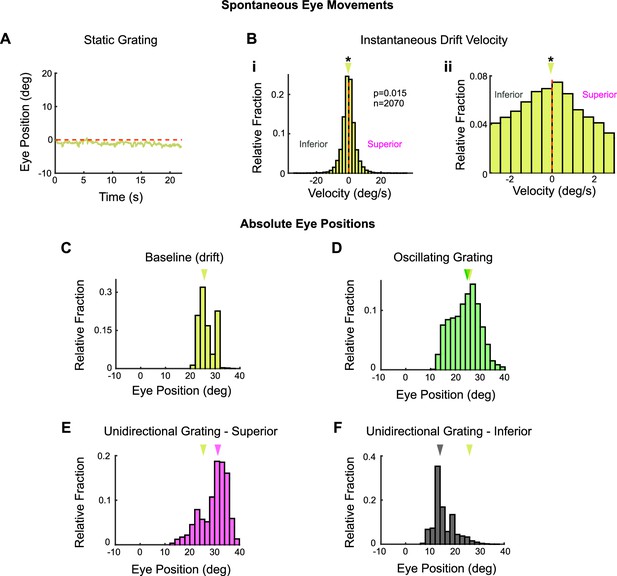
Baseline vertical eye movements in head-fixed mice (see also Figure 8—figure supplement 4).
Vertical eye movements were measured in response to static gratings to calculate eye drifts for baseline subtraction. (A) Example raw eye trace over 22 s of a static grating. The calculated position of the eye drifts downward over time, which could reflect true eye movements or a calibration error in our recording configuration. These two possibilities cannot be disambiguated (see ‘Materials and methods’). The magnitude of eye position drift during static gratings is approximately 18-fold less than the magnitude of the eye movements elicited by high-contrast drifting gratings. (B) Distribution of instantaneous eye velocity across N = 5 animals for the 20 s prior to the onset of all drifting grating stimuli (unidirectional and oscillating gratings at high [full] and low [20% relative] contrasts) used to evoke the optokinetic reflex (OKR). On average, there is a slight bias toward inferior (i.e., downward/negative) eye velocities during this baseline period, with a median velocity of –0.0787°/s. (Bi) Full distribution. (Bii) Same data, zoomed in on 0° to reveal the inferior bias. (C–F) Absolute vertical position of the eye without drift correction (C) prior to stimulus onset (when the drift was calculated as in [B], includes data from high (full) and low (20% relative) contrast, oscillating and unidirectional experiments), and during (D) high-contrast oscillating gratings, (E) superior unidirectional gratings and (F) inferior unidirectional gratings. Absolute eye position is similar to that measured during baseline only for oscillating gratings. The eye moves to more extreme positions during unidirectional stimuli. For this reason, the baseline subtraction was only applied to eye movements measured in response to oscillating gratings. For all histograms, arrows mark the median of the distribution. Yellow arrow in (D–F) marks the median of the distribution shown in (C). See Figure 8—figure supplement 4 for further data on eye drift in response to low-contrast gratings. *p<0.05, **p<0.01, ***p<0.001.
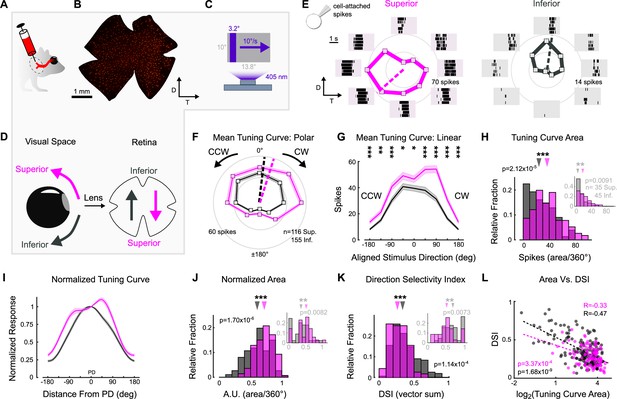
Superior and Inferior ON direction-selective retinal ganglion cells (oDSGCs) have asymmetric spike tuning curves.
(A) Schematic illustrating unilateral bead injections into medial terminal nucleus (MTN) to retrogradely label ganglion cells in the contralateral retina. (B) Flat-mount retina with retrogradely labeled, MTN-projecting retinal ganglion cells. (C) Drifting bar stimulus (3.2° × limiting projector dimension, 10°/s, 2.4 × 104 S-cone photoisomerization/s). (D) Definitions of superior (magenta) and inferior (gray) motion in visual space and on the retina. Directions are inverted by the lens. (E) Cell-attached spikes from labeled, MTN-projecting retinal ganglion cells in a flat-mount retina in response to a bar drifting in eight directions. Spike responses and average tuning curves from example Superior (left, magenta) and Inferior (right, gray) oDSGCs. Mean spike counts are presented as the distance from the origin, marked by concentric circles. Numbers on circles indicate spike counts. Dashed lines represent the preferred direction of each cell, calculated as the direction of the vector sum of all responses. Coordinates are in retinal space. (F, G) Population tuning curves across all Superior and Inferior oDSGCs (mean ± SEM). (F) Polar plots (as in [E]) aligned by rotating the tuning curves of Superior cells by 180°. (G) Linear representation of the same data (referred to as the ‘linear tuning curve’). CW: clockwise, nasal for Superior oDSGCs, temporal for Inferior oDSGCs; CCW: counterclockwise, temporal for Superior oDSGCs, nasal for Inferior oDSGCs. 0° represents directly superior/inferior motion. (H) Histograms of the area under the curve of the linear tuning curve of every cell. Inset shows the same metric for a stimulus at 20% relative contrast. (I) Population mean (± SEM) normalized tuning curves – computed by normalizing and aligning (at 0°) the response of each cell to its response in the preferred direction. (J) Histograms of the area under the curve of the normalized tuning curve (as in [I]) of every cell (referred to as ‘normalized area’). A larger normalized area indicates a wider tuning curve. Inset shows the same metric for a stimulus at 20% relative contrast. (K) Histograms of the direction selectivity index (DSI, vector sum, see ‘Materials and methods’) of every cell. Inset shows the same metric for a stimulus at 20% relative contrast. (L) Linear tuning curve area (as in [H]) and direction selectivity index (as in [K]) were correlated on a cell-by-cell basis for both Superior and Inferior oDSGCs. Dashed lines are least squares linear regressions, R and p values are Spearman’s rank correlation coefficient and associated two-sided significance value, respectively. For all histograms, medians of Superior (magenta) and Inferior (gray) oDSGC distributions are indicated by arrows. *p<0.05, **p<0.01, ***p<0.001.
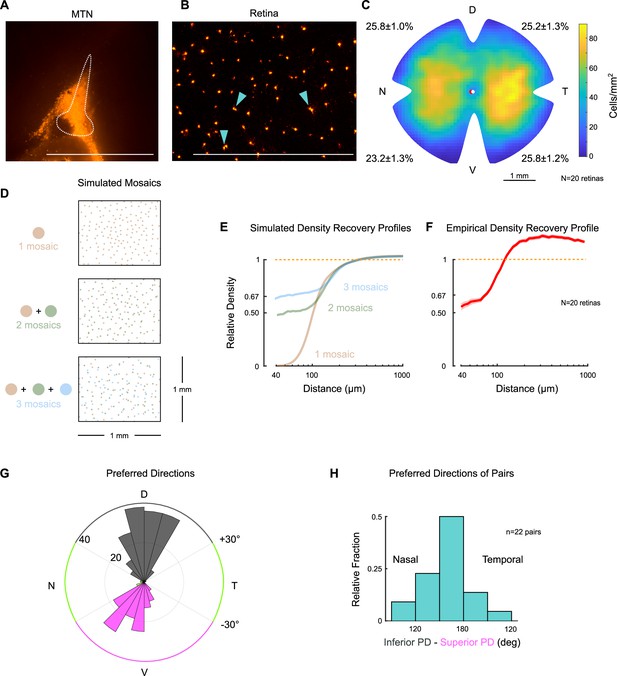
Two retinal ganglion cell types project to the medial terminal nucleus.
(A) Sagittal section of medial terminal nucleus (MTN) following injection of fluorescent retrobeads (scale bar = 1mm). Dotted line outlines MTN. (B) Retrogradely labeled retinal ganglion cell somas in a flat-mount retina after contralateral MTN injection. Arrowheads point to examples where labeled cells form ‘pairs’ (i.e., are within 30 µm of each other; scale bar = 1 mm), as described previously (Yonehara et al., 2008). (C) Density heatmap of retrogradely labeled MTN-projecting retinal ganglion cells across the contralateral retina. Numbers around the perimeter indicate the percentage of cells found in each quadrant (mean ± SEM). D, T, V, and N denote dorsal, temporal, ventral, and nasal directions, respectively, on the retina. Pairwise comparisons of the average number of cells per quadrant did not reveal any significant differences. However, comparing across halves showed that densities were marginally greater in dorsal compared to ventral retina (p=0.047), and in temporal compared to nasal retina (p=0.021). (D) Simulations of retinal ganglion cell populations that consist of one (top), two (middle), and three (bottom) mosaics. Each simulation contains approximately the same number of total cells. When only one mosaic is present (top), cells obey exclusion zones and do not cluster next to each other. When more than one mosaic is present, cells do not respect the exclusion zones of other cell types and ‘pairs’ (middle) and ‘trios’ (bottom) begin to form. Retinal ganglion cells of distinct types are well known to tile the retina in separate mosaics. (E) Mean density recovery profiles (DRPs) for simulated retinal ganglion cell populations of one (tan), two (green), and three (blue) mosaics (n = 30 repetitions each). Only single mosaics exhibit a complete exclusion zone. The DRP of two mosaics converges to 50% of its average density (dashed line) as distance approaches 0, and the DRP of three mosaics converges to 67% of its average density. More generally, a spatial distribution of ganglion cells will converge to of its average density, where n is the total number of mosaics (Cook and Podugolnikova, 2001). (F) DRP measured from retrogradely labeled MTN-projecting retinal ganglion cells (mean ± SEM). The empirical DRP lacks a full exclusion zone and converges to ~50% of its average density (1.0), indicating that there are likely two ganglion cell types, each forming an independent mosaic. Note that the DRP overshoots 1.0 within the shown domain because ON direction-selective retinal ganglion cells (oDSGCs) are not uniformly distributed across the retina (as shown in [C]).Normalizing to the peak density yields similar results. (G) Polar histogram of preferred directions measured in cell-attached mode across retrogradely labeled retinal ganglion cells identified by epifluorescence targeting. Colored segments of the outer circle indicate preferred direction thresholds for classification of Superior (magenta), Inferior (gray), and other direction-selective retinal ganglion cells (green). Coordinates are in retinal space. Concentric circles indicate the number of cells per bin. Labeled retinal ganglion cells divide into two major physiological types: superior-preferring and inferior-preferring. Only a small fraction of cells prefer horizontal directions (green). (H) Preferred directions of retrogradely labeled cells found in pairs (somas within 30 µm of each other). Paired cells tend to prefer opposite directions of motion (180° apart), further indicating that (1) two separate populations of ganglion cells project to MTN and (2) each population forms an independent mosaic.
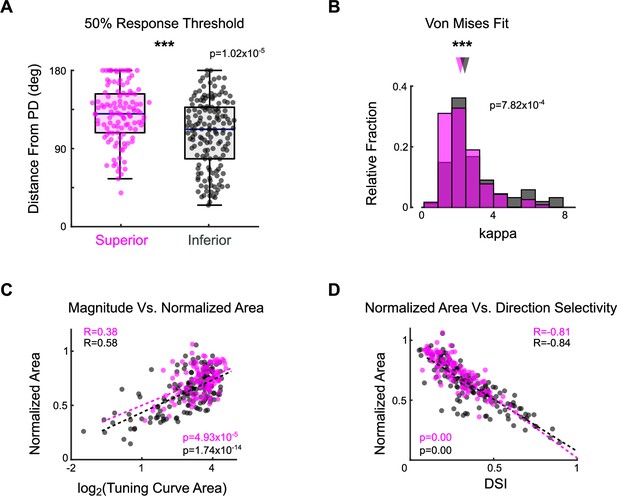
Additional metrics of ON direction-selective retinal ganglion cell (oDSGC) spike tuning curve width.
(A) Distributions of the distance (in degrees) from each cell’s preferred direction to the point at which its response magnitude first drops below 50% of the response in the preferred direction. Larger values indicate a wider tuning curve. Horizontal line represents median, box boundaries are the IQR, and whiskers represent most extreme observation within 1.5× IQR. Points represent individual cells. (B) Histograms of the kappa parameter for the Von Mises fit of the tuning curve of each cell (see ‘Materials and methods’). A smaller kappa value indicates a wider tuning curve. (C) The area of the linear tuning curve and the area of the normalized tuning curve were positively correlated on a cell-by-cell basis. (D) The direction selectivity index and the area of the normalized tuning curve were negatively correlated on a cell-by-cell basis. Dashed lines in (C) and (D) are least-squares linear regressions for Superior (magenta) and Inferior (gray) oDSGCs. R and p values are the Spearman’s rank correlation coefficient and associated two-sided significance, respectively. *p<0.05, **p<0.01, ***p<0.001.
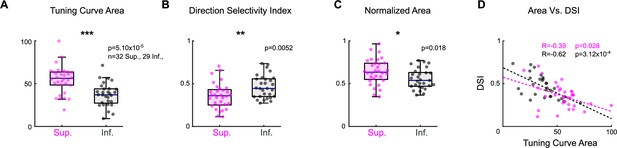
Asymmetries between Superior and Inferior ON direction-selective retinal ganglion cells (oDSGCs) persist under two-photon targeting.
Retrogradely labeled oDSGCs were targeted for cell-attached recordings using a two-photon laser (860 nm). Spikes were measured from Superior and Inferior oDSGCs in response to the drifting bar stimulus. Under two-photon conditions, Superior oDSGCs had (A) greater area of the linear tuning curve, (B) lower direction selectivity indices, and (C) greater area of the normalized tuning curve compared to Inferior oDSGCs. (D) Direction selectivity and tuning curve area were significantly correlated on a cell-by-cell basis for both Superior and Inferior oDSGCs. Dashed lines are least-squares linear regressions for Superior (magenta) and Inferior (gray) oDSGCs. R and p values are the Spearman’s rank correlation coefficient and associated two-sided significance, respectively. These findings confirm the results from experiments in which ganglion cells were targeted by epifluorescence (Figure 2, Figure 2—figure supplement 2). Cells in this two-photon dataset come from tissue that was never exposed to epifluorescence. No cell is in both the epifluorescence and two-photon datasets. *p<0.05, **p<0.01, ***p<0.001.
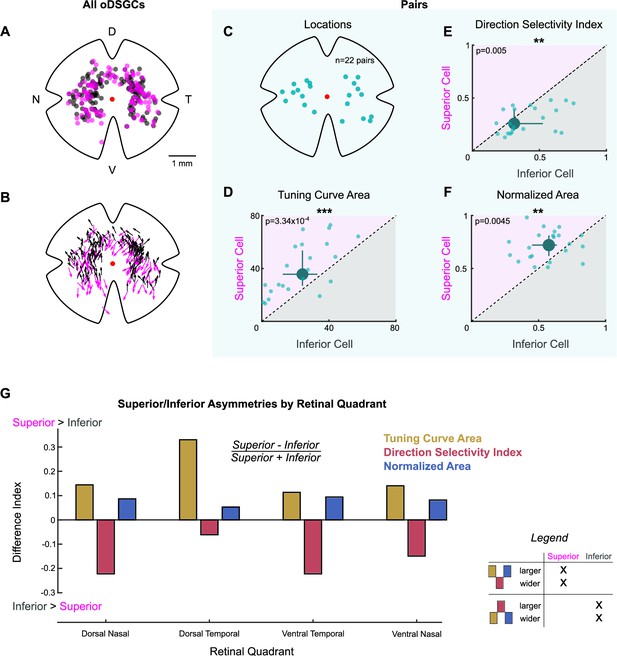
Physiological differences between Superior and Inferior ON direction-selective retinal ganglion cells (oDSGCs) are consistent across retinal topography.
(A) Map of retinal locations of all medial terminal nucleus (MTN)-projecting retinal ganglion cells recorded during cell-attached experiments in which epifluorescence targeting was used. D, T, V, and N denote dorsal, temporal, ventral, and nasal directions, respectively, on the retina and apply to all maps. (B) Map of the preferred direction of each cell in (A). The arrow base marks the location of the cell soma. The arrowhead points in the preferred direction. Preferred directions varied systematically across the retina, as reported previously (Sabbah et al., 2017). (C–F) Data from Superior and Inferior oDSGCs that formed ‘pairs’ (i.e., somas within 30 µm of each other, see Figure 2—figure supplement 1). Pairwise comparisons in these cells remove potential confounds caused by differences in topographic distributions when looking for asymmetries across Superior and Inferior oDSGC populations. (C) Retinal location of each recorded pair. (D–F) Tuning curve metrics for the Superior and Inferior oDSGCs in each pair: (D) linear tuning curve area, (E) direction selectivity index, and (F) area of the normalized tuning curve. Dashed lines indicate unity. Large points represent the univariate medians. Whiskers are 95% confidence intervals for each median, determined via bootstrapping. Significance values indicate pairwise comparisons between Superior and Inferior oDSGCs (two-sided signed-rank). Superior oDSGCs spike more (D), are less direction-selective (E), and have wider tuning curves (F) when compared to the Inferior oDSGCs with which they form pairs. (G) Comparison of tuning curve metrics between Superior and Inferior oDSGCs found in each retinal quadrant. Bars show a difference index [(Superior - Inferior)/(Superior +Inferior)] calculated from the median linear tuning curve area (olive), direction selectivity index (red), or area of the normalized tuning curve (blue) per quadrant for each oDSGC type. Positive values indicate that the metric is greater for Superior cells in that quadrant and negative values indicate that the metric is greater for Inferior cells in that quadrant. Difference indices are bound between –1 and 1. Within each retinal quadrant, Superior oDSGCs have a larger linear tuning curve area, a lower direction selectivity index, and a larger normalized tuning curve area than Inferior oDSGCs. Data match the first possibility illustrated in the legend. *p<0.05, **p<0.01, ***p<0.001.
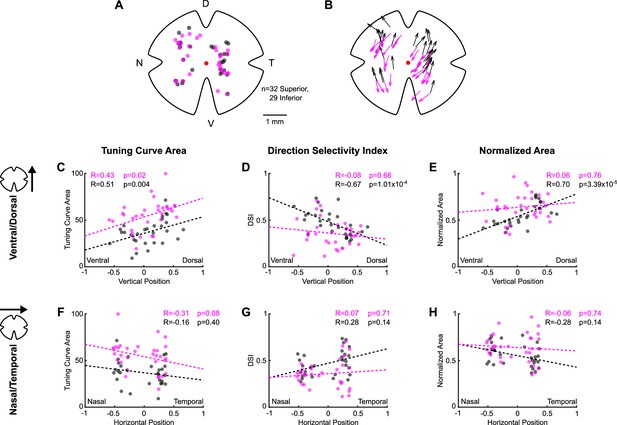
Topographic variation in direction tuning properties across the retina revealed by two-photon targeting.
(A) Map of retinal locations of all medial terminal nucleus (MTN)-projecting retinal ganglion cells recorded during cell-attached experiments in which two-photon targeting was used. D, T, V, and N denote dorsal, temporal, ventral, and nasal directions, respectively, on the retina and apply to all maps. (B) Map of the preferred direction of each cell in (A). The arrow base marks the location of the cell soma. The arrowhead points in the preferred direction. Preferred directions varied systematically across the retina, as seen using epifluorescence targeting (Figure 2—figure supplement 4B) and as reported previously (Sabbah et al., 2017). (C–H) Scatter plots of tuning curve metrics as a function of each cell’s position along either the ventral-dorsal (C–E) or nasal-temporal (F–H) axis of the retina. Coordinates are normalized to the size of the retina from which each cell was recorded (normalized coordinates range between –1 and 1, see ‘Materials and methods’). Inferior ON direction-selective retinal ganglion cells (oDSGCs) change tuning curve size (C) and width (D, E) as a function of dorsal-ventral location, whereas only the tuning curve size (C) of Superior oDSGCs is modulated along the same axis. No metric is significantly related to position along the nasal-temporal axis for either cell type (F–H). Further, Superior oDSGCs tend to have larger and wider tuning curves than Inferior oDSGCs across all dimensions (separation between magenta and gray lines). For all scatter plots, dashed lines are least-squares linear regressions for Superior (magenta) and Inferior (gray) oDSGCs. R and p values are the Spearman’s rank correlation coefficient and associated two-tailed significance, respectively. Two-photon targeting was used for all data in this figure so as to avoid confounds associated with epifluorescence exposure and photoreceptor absorption spectra gradients across retinal topography.
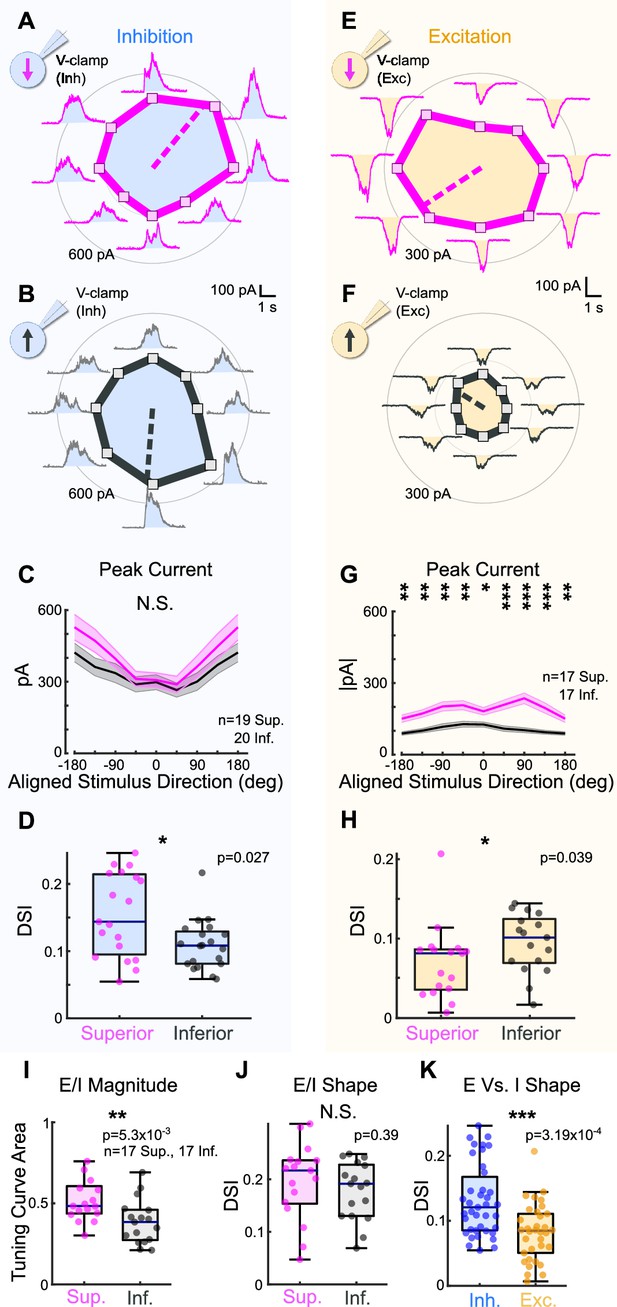
Superior ON direction-selective retinal ganglion cells (oDSGCs) receive similar inhibitory inputs but greater excitatory inputs compared to Inferior oDSGCs.
(A) Inhibitory currents measured from an exemplar Superior oDSGC under voltage-clamp at +10 mV in response to a bar drifting in eight directions. Mean peak inhibitory current is presented as the distance from the origin for each stimulus direction. Dashed line indicates the preferred direction of the peak inhibitory currents. Coordinates are in retinal space. (B) Same as (A) for an exemplar Inferior oDSGC. (C) Population responses for peak inhibitory currents across stimulus directions for Superior (magenta) and Inferior (gray) oDSGCs (mean ± SEM). Stimulus directions are aligned across cell types, where 0° indicates directly superior (for Superior oDSGCs) or inferior (for Inferior oDSGCs) motion. Positive directions are clockwise. (D) Distributions of the direction selectivity index for peak inhibitory currents in individual Superior and Inferior oDSGCs. (E) Excitatory currents measured from an exemplar Superior oDSGC under voltage-clamp at –60 mV in response to a bar drifting in eight directions. Same cell as in (A). (F) Same as (E) for an exemplar Inferior oDSGC. Same cell as in (B). (G) Population responses for peak excitatory currents across stimulus directions for Superior (magenta) and Inferior (gray) oDSGCs (mean ± SEM). (H) Distributions of the direction selectivity index for peak excitatory currents in individual Superior and Inferior oDSGCs. (I, J) The ratio of the peak excitatory current to the peak inhibitory current (E/I) was calculated for each stimulus direction for cells in which both metrics were recorded. (I) Distributions of the linear tuning curve area of E/I. (J) Distributions of the direction selectivity index for E/I. (K) Direction selectivity index for peak inhibitory (blue) and excitatory (yellow) currents collapsed across Superior and Inferior oDSGCs. For box plots, horizontal line represents median, box boundaries are IQR, and whiskers represent the most extreme observation within 1.5× IQR. *p<0.05, **p<0.01, ***p<0.001.
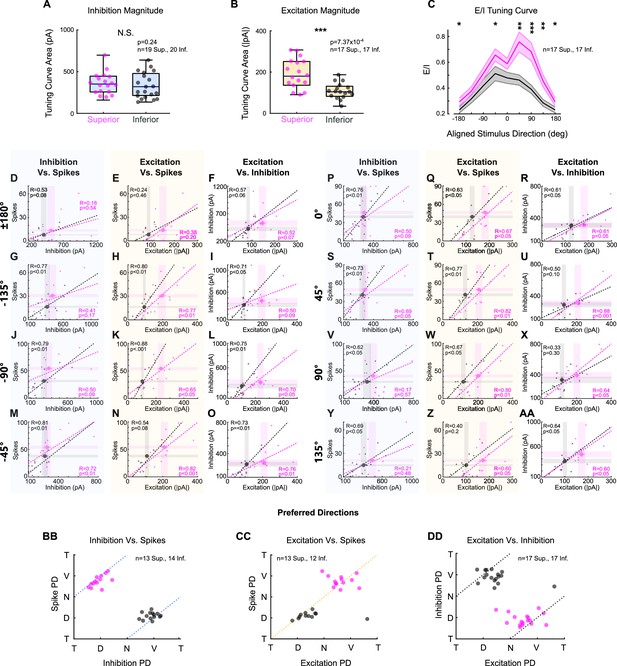
Superior ON direction-selective retinal ganglion cells (oDSGCs) receive more excitatory input, but are less intrinsically excitable, than Inferior oDSGCs.
(A, B) Linear tuning curve areas of the peak (A) inhibitory and (B) excitatory current measured in voltage-clamp recordings. Horizontal line represents median, box boundaries are IQR, and whiskers represent most extreme observation within 1.5× IQR. (C) Ratio of peak excitatory to peak inhibitory current (E/I) for each aligned stimulus direction (mean ± SEM), for cells in which both metrics were measured. 0° is directly superior (for Superior oDSGCs) or inferior (for Inferior oDSGCs) motion. Positive directions are clockwise. Statistical significance for each stimulus direction changes depending on how the tuning curves of Superior and Inferior oDSGCs are aligned (e.g., 180° rotation vs. reflection over the x-axis of the polar tuning curve). In general, E/I of Superior oDSGCs is greater than that of Inferior oDSGCs. (D–AA) For each bar direction, inhibition vs. spikes, excitation vs. spikes, and excitation vs. inhibition for cells in which both metrics were recorded (excitation and inhibition are peak values from voltage-clamp recordings, spikes are counts from cell-attached recordings). Dashed lines are least-squares linear regressions for Superior (magenta) and Inferior (gray) oDSGCs. R and p values are the Spearman’s rank correlation coefficient and associated two-sided significance, respectively. Large points represent univariate means ± SEM for each cell type taken from full data sets (i.e., small dots represent only a subset of cells in which both metrics were recorded, but full univariate datasets also consist of cells in which just one metric was recorded). Directions indicate aligned stimulus directions (as in [C]). For excitation vs. spikes, the fit line for Superior oDSGCs tends to fall below the fit line for Inferior oDSGCs, indicating lower intrinsic excitability. However, greater excitatory inputs to Superior oDSGCs outweigh the difference in intrinsic excitability, leading to more total spikes in Superior oDSGCs. Further, inhibition does not intuitively explain spike outputs since there is a positive correlation between inhibitory input and number of spikes across directions. This correlation is likely caused by an additional positive correlation between excitation and inhibition. Therefore, spikes are best explained by excitation. (BB) Preferred direction of inhibition vs. preferred direction of spikes recorded in the same cell. Dashed line represents the prediction for a 180° difference. (CC) Preferred direction of excitation vs. preferred direction of spikes recorded in the same cell. Dashed line represents prediction for 0° difference. (DD) Preferred direction of excitation vs. preferred direction of inhibition recorded in the same cell. Dashed line represents the prediction for 180° difference. Labels of T, D, N, and V correspond to temporal, dorsal, nasal, and ventral directions on the retina, respectively. *p<0.05, **p<0.01, ***p<0.001.
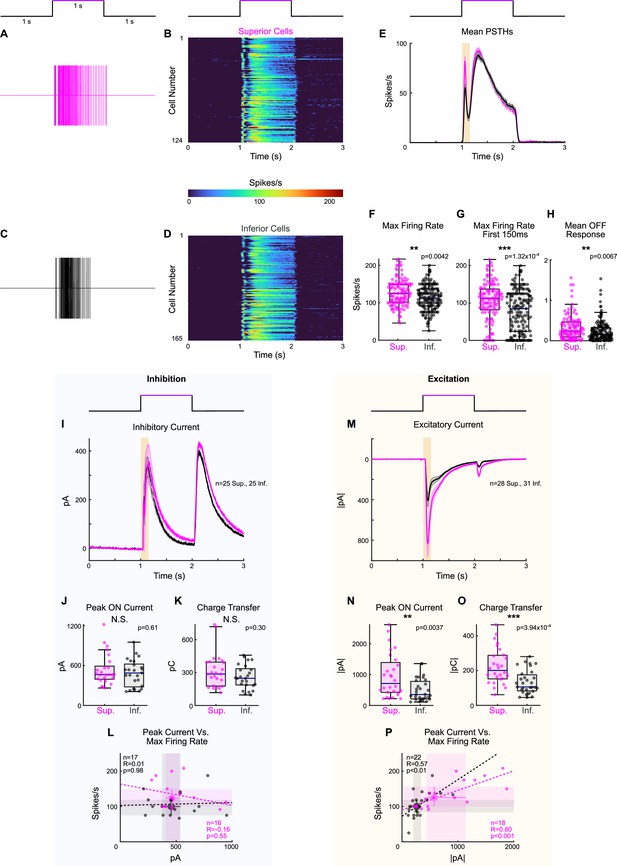
Full-field light increments elicit more spikes and excitation in Superior ON direction-selective retinal ganglion cells (oDSGCs).
(A, C) Example extracellular spike rasters from (A) a Superior and (C) an Inferior oDSGC in response to a 1 s light increment (405 nm). The schematic above shows the timing of the increment relative to the data. (B, D) Peristimulus time histograms (PSTHs) of average cell-attached light increment responses for each of (B) n = 124 Superior and (D) n = 165 Inferior oDSGCs. (E) Average PSTH across all cells shown in (B) and (D) (mean ± SEM). Highlighted region shows the first 150 ms after stimulus onset. (F) Distributions of each cell’s maximum firing rate throughout the entirety of the light increment. (G) Maximum firing rates during the first 150 ms after stimulus onset (i.e., highlighted region in [E]). (H) Mean firing rates ≥50 ms after stimulus offset. (I) Average inhibitory current in response to the 1 s light increment for Superior (magenta) and Inferior (gray) oDSGCs (mean ± SEM; voltage-clamp at +10 mV). (J) Peak inhibitory currents for the duration of the 1 s increment. (K) Total inhibitory charge transfers for the duration of the 3 s stimulus (i.e., including baseline and OFF response). (L) Peak inhibitory currents (as in [J]) versus maximum firing rate (as in [F]) for cells in which both metrics were recorded. (M) Average excitatory current in response to the 1 s light increment for Superior (magenta) and Inferior (gray) oDSGCs (mean ± SEM; voltage-clamp at –60 mV). (N) Peak excitatory currents for the duration of the 1 s increment. (O) Total excitatory charge transfers for the duration of the 3 s stimulus (i.e., including baseline and OFF response). (P) Peak excitatory current (as in [N]) versus maximum firing rate (as in [F]) for cells in which both metrics were recorded. The magenta line falls below the black line, indicating that Superior oDSGCs have a lower ratio of spike output to excitatory input. For scatter plots, large points and whiskers represent univariate medians and 95% confidence intervals (via bootstrapping). Dashed lines are least-squares linear regressions for Superior (magenta) and Inferior (gray) oDSGCs. R and p values are the Spearman’s rank correlation coefficient and associated two-sided significance, respectively. For box plots, the horizontal line represents median, box boundaries are IQR, and whiskers represent the most extreme observation within 1.5× IQR. *p<0.05, **p<0.01, ***P<0.001.
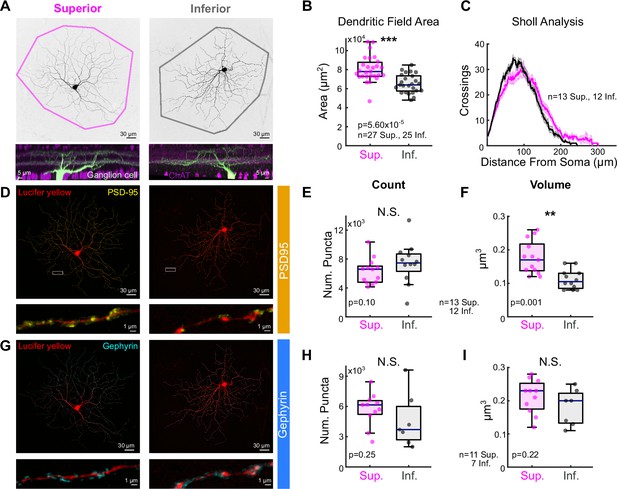
Superior ON direction-selective retinal ganglion cells (oDSGCs) have larger dendritic fields and excitatory postsynaptic sites.
(A) Confocal images of exemplar Superior (left) and Inferior (right) oDSGCs filled with dye. Convex polygons are drawn around the tips of their dendrites. (Bottom) Side views of different Superior and Inferior oDSGCs filled and stained for acetylcholinesterase (ChAT) bands. Both cell types have dendrites that stratify in the ON and OFF ChAT bands, with the majority of dendrites in the ON sublamina. (B) Convex polygon areas across all filled cells. (C) Sholl analysis indicating the number of dendritic crossings as a function of radial distance from the soma (mean ± SEM). (D, G) Ganglion cells with immunostaining for (D) excitatory postsynaptic scaffolding protein PSD-95 or (G) inhibitory postsynaptic scaffolding protein gephyrin. (Bottom) Magnification of a stretch of dendrites with labeled puncta. (E, H) Total number of puncta within each ganglion cell for (E) PSD-95 and (H) gephyrin. (F, I) Quantification of average puncta volume for (F) PSD-95 and (I) gephyrin. For box plots, horizontal line represents median, box boundaries are IQR, and whiskers represent the most extreme observation within 1.5× IQR. *p<0.05, **p<0.01, ***p<0.001.
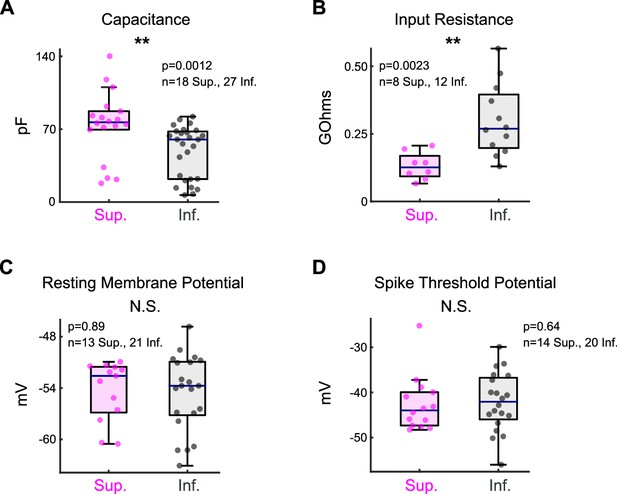
Intrinsic electrophysiological properties of ON direction-selective retinal ganglion cells (oDSGCs).
(A) Membrane capacitance, (B) input resistance, (C) resting membrane potential, and (D) spike threshold potential were measured from Superior (magenta) and Inferior (gray) oDSGCs during whole-cell patch-clamp recordings. Consistent with their larger morphological size (Figure 4B), Superior oDSGCs had larger capacitances and lower input resistances than Inferior oDSGCs. There was no significant difference in either resting membrane potential or spike threshold potential across cell types. For all panels, the blue line represents median, box boundaries are IQR, and whiskers represent the most extreme observation within 1.5× IQR. *p<0.05, **p<0.01, ***p<0.001.
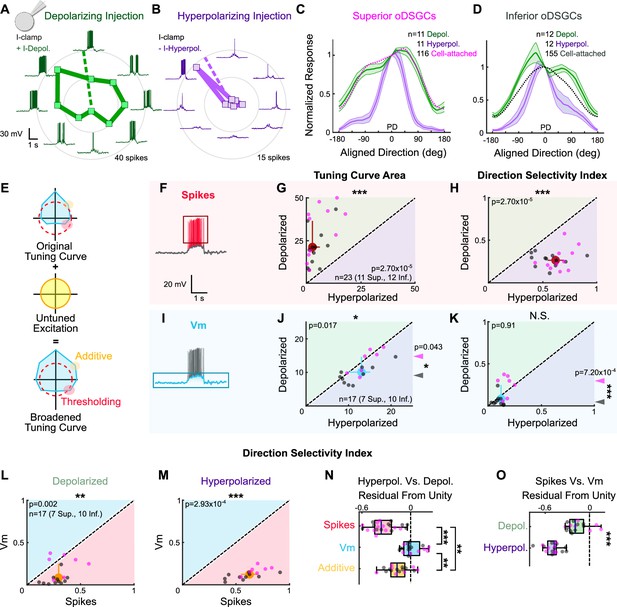
Thresholding differentiates the tuning properties of Superior and Inferior ON direction-selective retinal ganglion cells (oDSGCs).
(A, B) Exemplar Inferior oDSGC in whole-cell current-clamp during (A) depolarizing and (B) hyperpolarizing current injection in response to a bar moving in eight directions. Numbers on concentric circles indicate spike counts. Dashed lines represent preferred directions. Coordinates are in retinal space. (C, D) Mean (± SEM) normalized tuning curves (aligned and normalized to the response of each cell in its preferred direction) for (C) Superior and (D) Inferior oDSGCs under conditions of depolarizing (green) and hyperpolarizing (purple) current injection. Dotted lines indicate the average normalized spike tuning curves of each population from cell-attached recordings (as in Figure 2I). (E) Illustration of the influence of untuned excitation on the tuning curve through additive (yellow) and thresholding (red) effects. The blue area indicates the membrane potential, and the dashed red line indicates the spike threshold. (F, I) Example whole-cell current-clamp recording in which (F) spikes and (I) subthreshold voltages (Vm) have been separated. (G) Linear tuning curve area and (H) direction selectivity index of the spike tuning curve during hyperpolarizing (abscissa) and depolarizing (ordinate) current injections. (J) Linear tuning curve area and (K) direction selectivity index of peak subthreshold membrane potential tuning curves. For (G–H, J–K), regions of green (or purple) indicate that the metric is greater during depolarizing (or hyperpolarizing) injections. Points that fall on the line indicate equivalent metrics under the two conditions. Individual cells are shown as dots (Superior in magenta, Inferior in gray). Large red and blue dots represent univariate medians (collapsed across cell type) and whiskers indicate 95% confidence intervals determined via bootstrapping. Significance values indicate whether the data tend to fall unevenly on one side of the unity line (two-sided signed-rank). Arrowheads in (J, K) represent the median of Superior (magenta) and Inferior (gray) oDSGCs along the unity line, and associated significance values indicate comparison between Superior and Inferior oDSGCs (two-sided rank-sum). (L, M) Direction selectivity index for spikes (abscissa) and simultaneously measured subthreshold voltages (ordinate) under (L) depolarizing and (M) hyperpolarizing conditions. Significance values indicate whether the data tend to fall unevenly on one side of the unity line (two-sided signed-rank). (N) Residuals from the unity line for individual cells from the plots in (H) and (K). Dashed line indicates unity (i.e., no difference across depolarizing and hyperpolarizing conditions). Pairwise comparisons are shown between spikes, Vm, and Vm with additive offset. (O) Residuals from the unity line for individual cells from the plots in (L) and (M). Dashed line indicates unity (i.e., no difference between spikes and subthreshold voltages). Comparisons are made between the depolarizing and hyperpolarizing conditions (two-sided rank-sum). For box plots, the blue line represents median, box boundaries are IQR, and whiskers represent the most extreme observation within 1.5× IQR. *p<0.05, **p<0.01, ***p<0.001.
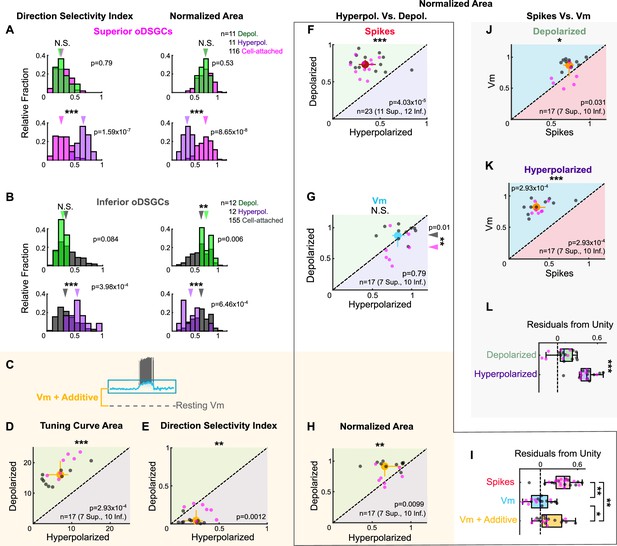
Spike and subthreshold voltage tuning curves with directionally untuned current injections.
(A, B) Comparison of spike tuning curve metrics from cell-attached and current injection recordings. Histograms show the direction selectivity index (left) and area of the normalized tuning curve (right) for cell attached (magenta in [A] or gray in [B]), hyperpolarizing (green, top), and depolarizing (purple, bottom) conditions for (A) Superior and (B) Inferior ON direction-selective retinal ganglion cells (oDSGCs). Arrows indicate medians. (C) Illustration of the voltage offset added via current injection during the depolarizing condition. The same offset was subtracted for the hyperpolarizing condition. Diagram is not shown to scale. (D, E, H) Metrics of the subthreshold membrane potential tuning curve during hyperpolarizing (abscissa) and depolarizing (ordinate) current injections with the additive offset taken into account: (D) linear tuning curve area, (E) direction selectivity index, and (H) area of the normalized tuning curve. Additive effects cause larger and broader membrane potential tuning curves following increases in the amount of untuned excitation. (F–H) Area of the normalized tuning curve during hyperpolarizing (abscissa) and depolarizing (ordinate) current injections for (F) spikes, (G) subthreshold voltages, and (H) subthreshold voltages with the additive offset taken into account. All metrics were measured simultaneously. (I) Residuals from the unity line for individual cells from the plots in (F–H). Dashed line indicates unity (i.e., no difference across depolarizing and hyperpolarizing conditions). Pairwise comparisons are shown between spikes, Vm, and Vm with additive offset. Current injections influence spike tuning curves more than either version of the Vm tuning curve. (J, K) Area of the normalized tuning curve for spikes (abscissa) and simultaneously measured subthreshold voltages (ordinate) under (J) depolarizing and (K) hyperpolarizing conditions. (L) Residuals from the unity line for individual cells from the plots in (J, K). Dashed line indicates unity (i.e., no difference between spikes and subthreshold voltages). Comparisons are made between depolarizing and hyperpolarizing conditions (two-sided rank-sum). For all scatter plots, individual cells are shown as small dots (Superior in magenta, Inferior in gray). Colored regions indicate that the metric is greater for that condition (D–H) or for spikes or Vm (J, K). Points on the line indicate equivalent values under the two conditions. Large dots represent univariate medians collapsed across cell types and whiskers are 95% confidence intervals determined via bootstrapping. Significance values indicate whether the data tend to fall unevenly on one side of the unity line (two-sided signed-rank). For all box plots, the blue line represents median, box boundaries are IQR, and whiskers represent the most extreme observation within 1.5× IQR. *p<0.05, **p<0.01, ***p<0.001.
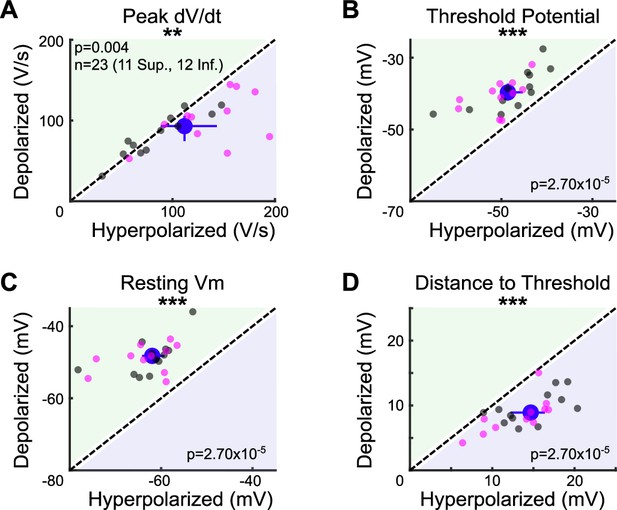
Effects of current injection on intrinsic properties of ON direction-selective retinal ganglion cells (oDSGCs).
To measure the effects of depolarizing and hyperpolarizing current injections on the intrinsic properties of oDSGCs, the (A) peak rate of voltage change and (B) spike threshold potential were calculated for every cell under both conditions. Peak dV/dt was on average 10.9 V/s greater under hyperpolarizing relative to depolarizing conditions. The spike threshold potential was on average 9.0 mV more negative under hyperpolarizing conditions. Both of these observations are consistent with an increase in voltage-gated sodium channel availability in the hyperpolarizing condition. This intrinsic change increases the probability of spikes during hyperpolarizing relative to the depolarizing injections, and therefore cannot explain our experimental results of greater spikes and broader tuning curves under depolarizing conditions (Figure 5). The likely reason why our empirical data show increased spiking and broader tuning curves during depolarizing injections is that the relative distance between cells’ resting membrane potentials and spike thresholds was greater in the hyperpolarizing condition. (C) The resting membrane potential was, on average, 12.6 mV more negative during hyperpolarizing injections than during depolarizing injections, whereas the spike threshold only changed by 9.0 mV (B). Thus, the difference between resting membrane potential and spike threshold was greater under hyperpolarizing conditions. (D) On average, cells’ resting membrane potentials were 5.1 mV further away from the spike threshold potential under hyperpolarizing relative to depolarizing conditions. Current injections may also influence conductances of other voltage-gated ion channels, including calcium channels, though evidence that oDSGCs express such channels is limited and these effects are difficult to quantify. For all panels, regions of green (or purple) indicate that the metric is greater for depolarizing (or hyperpolarizing) current injections. Points on the line indicate equivalent values under the two conditions. Individual cells are shown as dots (Superior in magenta, Inferior in gray). Large blue dots represent univariate medians collapsed across cell types and whiskers indicate 95% confidence intervals determined via bootstrapping. Significance values indicate whether the data tend to fall unevenly on one side of the unity line (two-sided signed-rank). *p<0.05, **p<0.01, ***p<0.001.

A parallel conductance model demonstrates how untuned excitation contributes to direction tuning.
An exemplar ON direction-selective retinal ganglion cell (oDSGC) was modeled using parameters recorded directly from oDSGCs, including directionally tuned inhibitory conductances for each of eight drifting bar directions, and a single, untuned excitatory conductance (see ‘Materials and methods’). (A) Inhibitory (pastel colors) and excitatory (yellow) conductances of the model oDSGC in response to bars moving in eight directions. (B) The parallel conductance model uses the empirically measured parameters to model the membrane potential across bar directions, shown here for the case in which the excitatory gain (i.e., a multiplication factor applied to the excitatory conductance) is set to 1.0. The red dotted line indicates the spike threshold. (C) Preferred (solid lines) and null (dotted lines) direction responses of peak subthreshold voltages (blue) and spikes (red) across a range of excitatory gains. Values are normalized to the maximum preferred direction response for each metric. The yellow column indicates the regime in which a null direction stimulus evokes zero spikes but a preferred direction stimulus evokes increasingly more spikes, an example of nonlinear behavior caused by the spike threshold. (D, E) Directional tuning properties as a function of excitatory gain for subthreshold voltages (blue) and spikes (red): (D) area of the linear tuning curve (normalized to the maximum area for each metric), and (E) direction selectivity index. Orange dot (and error bars) in (E) correspond to the empirically measured median (and 95% confidence interval determined via bootstrapping) direction selectivity index from cell-attached recordings, collapsed across cell types.
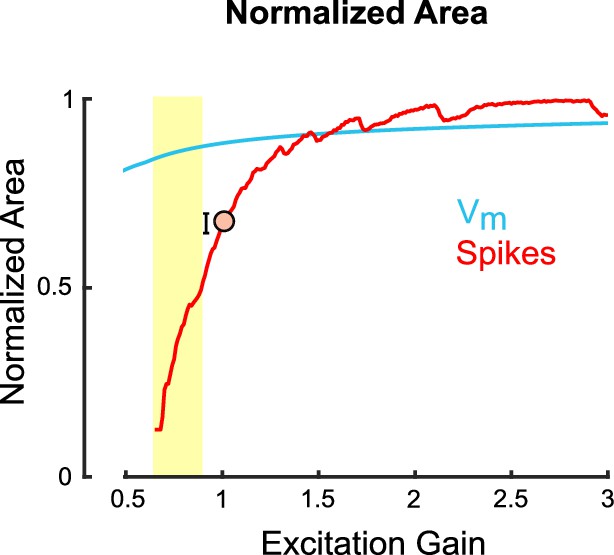
The normalized area of model spike tuning curves, but not subthreshold membrane potential tuning curves, is steeply influenced by excitation gain.
Area of the normalized tuning curve for spikes (red) and underlying subthreshold membrane potentials (blue) as a function of the gain of an untuned excitatory input to a model ON direction-selective retinal ganglion cell (oDSGC). Greater values along the vertical axis indicate a broader tuning curve. Additive excitation contributes to the slow rise of the membrane potential line, whereas thresholding effects cause the steep change in spike tuning curve width. The orange dot and error bars correspond to the empirically measured median and 95% confidence intervals (via bootstrapping), respectively, for this metric collapsed across cell types from cell-attached recordings.
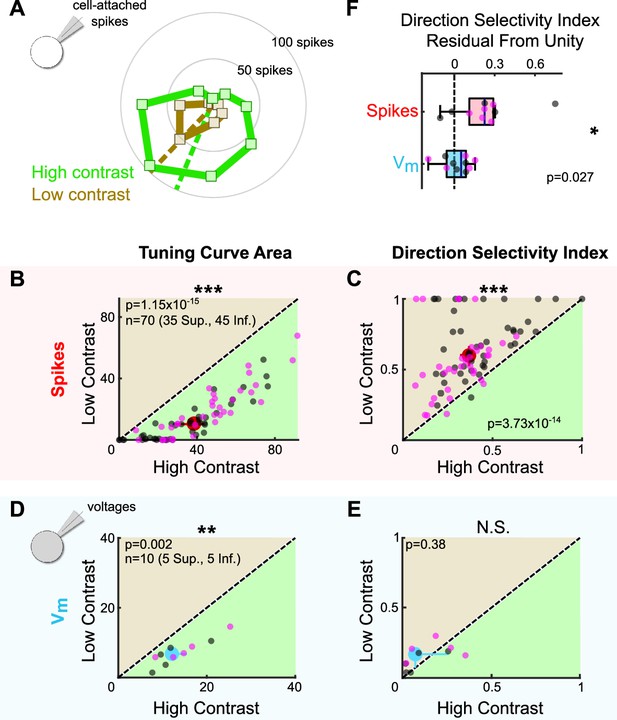
Stimulus contrast modulates the spike tuning curves of ON direction-selective retinal ganglion cells (oDSGCs).
(A) Cell-attached tuning curves from an exemplar Superior oDSGC at high (green) and low (tan, 20% relative) contrasts. Numbers on concentric circles indicate spike counts. Dashed lines represent preferred directions. Coordinates are in retinal space. (B) Linear tuning curve area and (C) direction selectivity index from spike responses to high-contrast (abscissa) and low-contrast (ordinate) bars drifting in eight directions. Differences between Superior (magenta) and Inferior (gray) oDSGCs persist under low contrast (see Figure 2). (D) Linear tuning curve area and (E) direction selectivity index from peak subthreshold voltage responses to high-contrast (abscissa) and low-contrast (ordinate) bars drifting in eight directions. (F) Residuals from the unity line of the direction selectivity index under high- and low-contrast conditions for simultaneously measured spikes and subthreshold voltages. Comparison is made between spikes and subthreshold voltages. For all scatter plots, the region of green (or tan) indicates the metric is greater under high-contrast (or low-contrast) conditions. Points on the line indicate equivalent metrics under the two conditions. Individual cells are represented by small dots. Large dots represent univariate medians (collapsed across cell type). Whiskers indicate 95% confidence intervals determined via bootstrapping. Significance values indicate whether the data tend to fall unevenly on one side of the unity line (two-sided signed-rank). *p<0.05, **p<0.01, ***p<0.001.
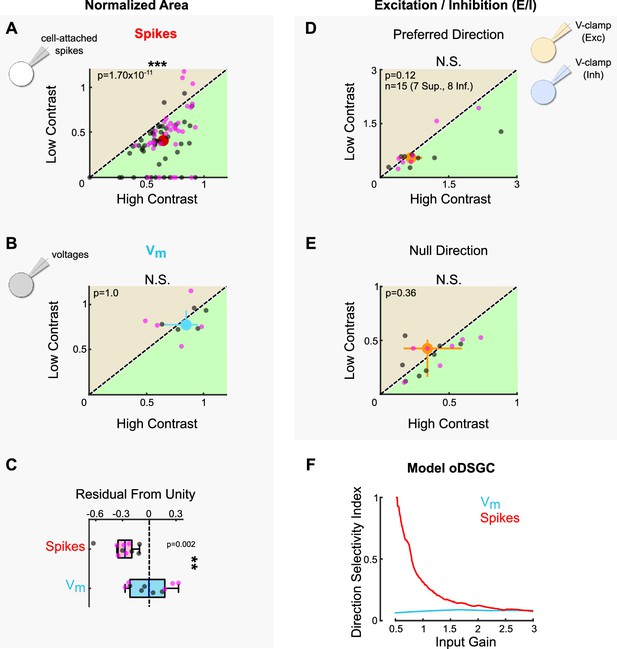
Stimulus contrast modulates spike tuning curve width but not the ratio of excitation to inhibition.
(A) Area of the normalized tuning curve from spike responses to high-contrast (abscissa) and low-contrast (ordinate) bars drifting in eight directions. Differences between Superior (magenta) and Inferior (gray) ON direction-selective retinal ganglion cells (oDSGCs) persist under low contrast (see Figure 2). (B) Area of the normalized tuning curve from subthreshold voltages in response to high-contrast (abscissa) and low-contrast (ordinate) bars drifting in eight directions. (C) Residuals from the unity line for simultaneously measured spikes (as in [A]) and subthreshold voltages (as in [B]). The dashed line indicates unity (i.e., no difference between high and low contrast). Comparison is made between spikes and subthreshold voltages. (D, E) Excitation-to-inhibition (E/I) ratios for high- and low-contrast bars drifting in (D) the preferred and (E) the null direction of each cell. (F) Direction selectivity index generated from a parallel conductance model of an oDSGC under different scale factors applied jointly to excitatory and inhibitory inputs (constant E/I). For all scatter plots, the region of green (or tan) indicates the metric is greater under high-contrast (or low-contrast) conditions. Points on the line indicate equivalent metrics under the two conditions. Individual cells are represented by small dots. Large dots represent univariate medians (collapsed across cell type). Whiskers indicate 95% confidence intervals determined via bootstrapping. Significance values indicate whether the data tend to fall unevenly on one side of the unity line (two-sided signed-rank). All data acquired following epifluorescence targeting. *p<0.05, **p<0.01, ***p<0.001.
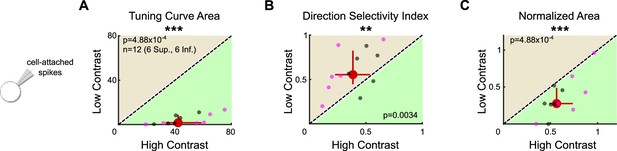
Two-photon targeting confirms that ON direction-selective retinal ganglion cells (oDSGCs) are contrast sensitive.
(A) Tuning curve area, (B) direction selectivity index, and (C) normalized area of the spike tuning curve were measured in the cell-attached configuration in response to high- and low-contrast drifting bars following two-photon targeting of oDSGCs. As occurred following epifluorescence targeting (Figure 7, Figure 7—figure supplement 1), two-photon targeting revealed that the spike tuning curves of oDSGCs were smaller (A) and narrower (B, C) in response to low-contrast stimuli. For all plots, regions of green (or tan) indicate that the metric is greater for high-contrast (or low-contrast) stimuli. Points on the line indicate equivalent metrics under the two conditions. Individual cells are shown as small dots (Superior in magenta, Inferior in gray). Large red dots represent univariate medians collapsed across cell types. Whiskers indicate 95% confidence intervals determined via bootstrapping. Significance values indicate whether the data tend to fall unevenly on one side of the unity line (two-sided signed-rank). *p<0.05, **p<0.01, ***p<0.001.
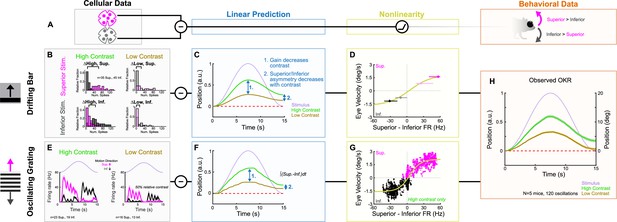
ON direction-selective retinal ganglion cell (oDSGC) responses predict the optokinetic reflex (OKR) across stimulus types, directions, and contrasts.
(A) Schematic of the putative computation between oDSGCs and OKR, consisting of a subtraction between Superior and Inferior oDSGC spikes and a nonlinearity. (B–H) Two separate implementations of the subtraction model described in (A). (B–D) Prediction of OKR behavior from oDSGC spike responses to the drifting bar stimulus. (B) Distributions of Superior (magenta) and Inferior (gray) oDSGC spike responses across high-contrast (left) and low-contrast (right, 20% relative) superior (top) and inferior (bottom) drifting bars. The brackets denote the difference between the medians of the Superior and Inferior oDSGC response distributions in each condition. (C) Linear predictions of OKR are made for the average eye velocity over the course of each half oscillation cycle based on the difference in firing rate between Superior and Inferior oDSGCs (i.e. brackets in [B]) to high- and low-contrast bars drifting in the corresponding stimulus direction. The shape of the curves as sinusoids is inferred from the stimulus position over time (lavender). (D) The empirically computed nonlinearity shows the relationship between linear behavioral predictions (as in [C]) from the drifting bar stimulus and the corresponding average eye velocities measured during behavioral OKR experiments for superior (magenta points) and inferior (gray points) stimuli at high (dark points) and low (light points) contrast. The points indicate univariate medians for each condition and whiskers are 95% confidence intervals computed via bootstrapping (vertical error bars are too small to see). The solid line is a fitted sigmoid function of the form described in Equation 6 and has parameters , , , . (E–G) Prediction of OKR behavior from oDSGC responses to an oscillating sinusoidal grating. (E) Median responses of Superior (magenta) and Inferior (gray) oDSGCs across a single cycle of the oscillating grating for high-contrast (left) and low-contrast (right, 50% relative) stimuli. Lavender traces represent the relative position of the stimulus across time. Directions of superior/inferior motion are in visual space and indicated by arrows. These recordings were made using two-photon targeting. (F) Linear predictions of OKR across time made by subtracting the instantaneous firing rates of Superior and Inferior oDSGCs to generate predictions for instantaneous eye velocity, and integrating to predict eye position. (G) The empirically computed nonlinearity shows the difference between Superior and Inferior oDSGC firing rates plotted against the time-matched average eye velocity of behaving animals. Each small point represents the average firing rate difference and eye velocity at a single time point over the course of one stimulus oscillation cycle. Magenta points represent superior eye velocities (i.e., above 0 on the ordinate), and gray points represent inferior eye velocities (i.e., below 0 on the ordinate). The solid line is a fitted sigmoid curve of the form described in Equation 6 and has parameters , , , and Only data for the high-contrast condition were used. (H) Mean eye position of behaving mice in response to a single oscillation of a high-contrast (green, as in Figure 1I) or low-contrast (tan, 20% relative) gratings. Compare to linear predictions in (C) and (F).
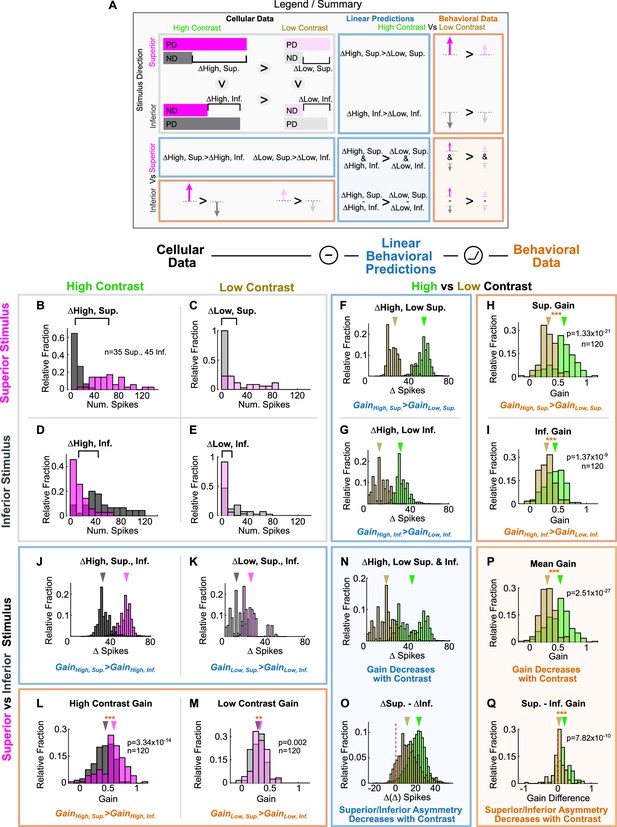
Behavioral prediction for the optokinetic reflex (OKR) from spike responses to the drifting bar stimulus.
Spikes of Superior and Inferior ON direction-selective retinal ganglion cells (oDSGCs) in response to the drifting bar stimulus were used to predict the magnitude of OKR gain in behaving animals. (A) Legend and summary for panels (B–Q). Cellular data (gray box): the responses of Superior (magenta) and Inferior (gray) oDSGCs to preferred (PD) and null (ND) direction drifting bars at high (left column) and low (20% relative, right column) contrasts. Linear predictions (blue boxes): the differences between responses in the preferred and null directions across cell types, represented by Δ, provide predictions of the relative OKR gain across stimulus directions and contrasts. Behavioral results (orange boxes): arrows represent the magnitude of OKR gain measured in behaving mice across stimulus directions and contrasts. Dark arrows represent high-contrast stimuli, and light arrows represent low-contrast stimuli. (B–E) Cellular data: distributions of superior (magenta) and Inferior (gray) oDSGC spike responses to superior (top row) and inferior (bottom row) stimuli at high (left column) and low (right column) contrast. Brackets above show the difference between the medians of the two distributions. Same data plotted in Figure 8B. (F–G, J–K, N–O) Linear predictions from data in (B–E) for how behavioral OKR gain changes across stimulus directions and contrasts. The asymmetries in the tuning curves of Superior and Inferior oDSGCs resulted in four first-order linear predictions for how OKR gain changes across stimulus conditions: (1) for superior stimuli, gain is greater in response to high-contrast stimuli than to low-contrast stimuli (F), (2) for inferior stimuli, gain is greater in response to high-contrast stimuli than to low-contrast stimuli (G), (3) at high-contrast, gain is greater in response to superior stimuli than to inferior stimuli (J), and (4) at low-contrast, gain is greater in response to superior stimuli than to inferior stimuli (K). Two additional second-order hypotheses were made that rely on an approximately linear relationship between differences in oDSGC output and gain in the tested regime: (5) gain is greater under high-contrast, regardless of stimulus direction (N), and (6) behavioral asymmetries in response to superior and inferior stimuli diminish with decreasing contrast (O). (H–I, L–M, P–Q) The six linear predictions were tested by measuring OKR in behaving mice in response to superior and inferior oscillating gratings at high- and low-contrast. All six linear predictions accurately matched behavior. Distributions in these panels are made up of the average OKR gain over the course of individual half oscillation cycles, arrows indicate medians (two-sided signed-rank). *p<0.05, **p<0.01, ***p<0.001.
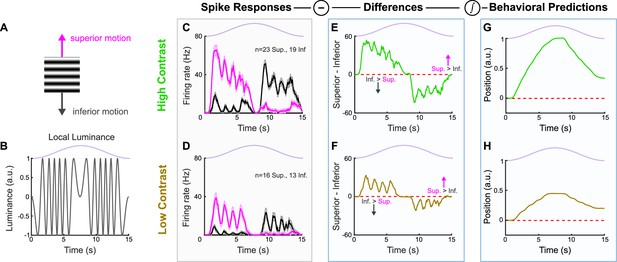
ON direction-selective retinal ganglion cell (oDSGC) responses to oscillating gratings.
(A) Oscillating sinusoidal gratings used in oDSGC electrophysiology were equivalent to those used in behavioral experiments. Motion directions apply to all panels. (B) Luminance of a single point in space as the sinusoidal grating completes one oscillation cycle. A value of 1.0 along the ordinate indicates full luminance and –1.0 indicates minimal luminance. This luminance function is determined by both the spatial frequency of the grating and the velocity of the sinusoidal oscillation. The light available to a single oDSGC follows such luminance fluctuations over the course of a single stimulus oscillation cycle, but the position of the oDSGC relative to the phase of the grating will cause a corresponding phase shift in its luminance function. These luminance oscillations therefore likely account for the high-frequency oscillations seen in (C–F). Stimulus position (in the oscillation cycle) is shown above in lavender. (C) Median spike responses of Superior (magenta) and Inferior (gray) oDSGCs over the course of a single high-contrast oscillation cycle. Two-photon targeting was used during all recordings in this data set. Error bars represent standard error of the mean. (D) Same as (C), but for a low-contrast stimulus, which, for this experiment, was defined as 50% contrast relative to the high-contrast stimulus (see ‘Materials and methods’). (E, F) Moment-by-moment subtraction of the median Inferior oDSGC firing rate from the median Superior oDSGC firing rate for (E) high- and (F) low-contrast gratings. Values fall above unity (dashed red line) when Superior oDSGCs spike more than Inferior oDSGCs, and below unity when Inferior oDSGCs spike more than Superior oDSGCs. This subtraction constitutes a linear prediction of eye velocity for each point in time. (G, H) Linear predictions of eye position across time for a single (G) high- and (H) low-contrast oscillation cycle. These predictions are the cumulative integral of the firing rate differences shown in (E) for high-contrast and (F) for low-contrast. For all plots, the lavender trace indicates the position of the grating stimulus in its oscillation cycle across time. Two-photon targeting was used during all recordings in this data set.
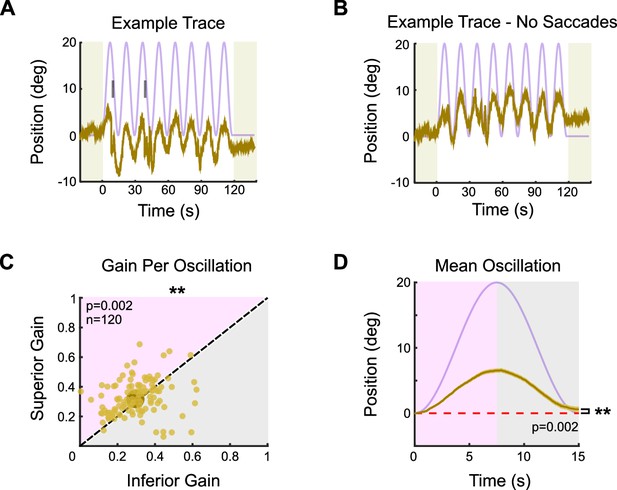
The optokinetic reflex (OKR) at low contrast.
Eye movements were measured from head-fixed mice in response to an oscillating sinusoidal grating. All parameters of the grating were the same as under high-contrast conditions (Figure 1), except for the grating contrast, which was five times lower (i.e., 20% relative contrast). (A) Example of OKR in response to the vertically oscillating low-contrast grating. The eye position is shown in tan, and the stimulus position is shown in lavender. Saccades are indicated by tick marks. (B) Same as (A), but with saccades (‘fast nystagmuses’) removed to reveal the asymmetry between superior and inferior OKR. For each epoch, animals viewed eight oscillation cycles lasting a total of 120 s, flanked by 20 s of a static grating (shaded regions). (C) Average gain of slow nystagmus during the superior versus inferior stage of individual oscillations. Each small dot is a single oscillation. The region of magenta (or gray) indicates that gain was greater for the superior (or inferior) stage of the oscillation. Points that fall on the line indicate equivalent gain for both stimulus directions. Large point and whiskers represent univariate medians and 95% confidence intervals (via bootstrapping), respectively. Significance value indicates whether the points tend to fall unevenly on one side of the unity line (two-sided signed-rank). (D) Eye position (tan) and stimulus position (lavender) averaged across all oscillations and all animals (mean ± SEM). Starting eye position is normalized to 0° at cycle onset. The average ending eye position is displaced in the superior direction (two-sided signed-rank). N = 5 mice for all experiments; n = number of trials.
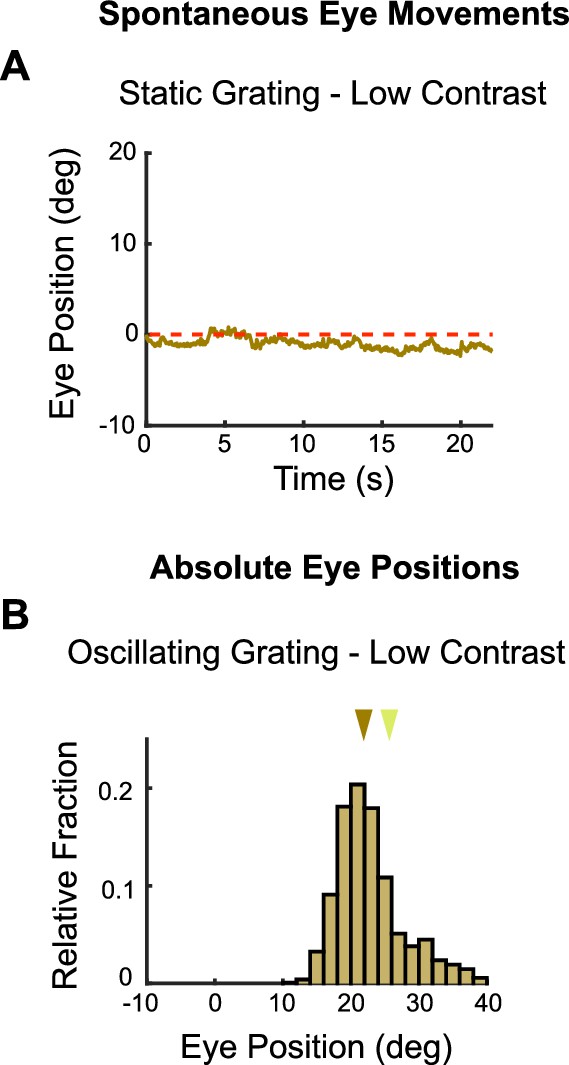
Baseline vertical eye movements to low-contrast stimuli (see also Figure 1—figure supplement 2).
Vertical eye movements were measured in response to static, low-contrast (20% relative) gratings to calculate eye drifts for baseline subtraction. (A) Example raw eye trace over 22 s of a static low-contrast grating. The calculated position of the eye drifts downward over time. This may reflect true eye movements or a calibration error in our recording configuration, but these two possibilities cannot be disambiguated (see ‘Materials and methods’). The magnitude of eye position drift during static gratings is approximately 11-fold less than the magnitude of the eye movements elicited by low-contrast moving gratings (see Figure 1—figure supplement 2A for comparison to high-contrast grating). (B) Absolute vertical position of the eye without drift correction during low-contrast gratings. Brown arrow marks the median of the distribution. Yellow arrow indicates median position of the eye prior to stimulus onset (as in Figure 1—figure supplement 2C). Spontaneous drift velocity was baseline subtracted from eye positions in response to low-contrast oscillating gratings since median eye position was similar to that measured in response to static gratings. For further data on eye drift and spontaneous eye movements to high-contrast gratings, see Figure 1—figure supplement 2.
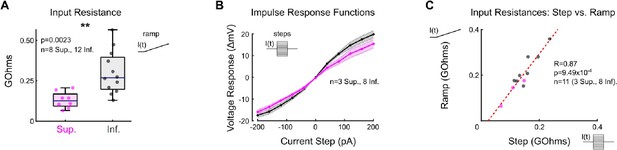
Superior oDSGCs are less excitable than Inferior oDSGCs.
(A) Input resistance in Superior (magenta) and Inferior (gray) oDSGCs calculated from current injection ramps in which current increases linearly from 0 to 50 pA over 0.25 seconds. (B) Steady state current-voltage relationship for Superior (magenta) and Inferior (gray) oDSGCs in response to current steps from -200 to 200 pA lasting 0.5 seconds each. (C) Input resistances calculated from current ramps and steps were positively correlated. For panel (C), R and p values are Spearman’s rank correlation coefficient and associated 2-sided p-value. Dashed line is least squares linear regression.
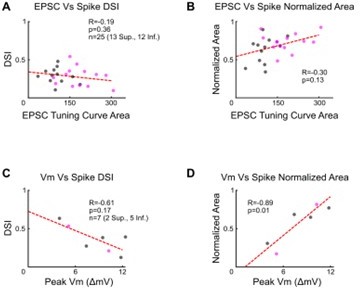
Strength of excitatory input and spike tuning curve properties.
(A-B) Relationships between excitatory postsynaptic current (EPSC) tuning curve area and (A) direction selectivity index or (B) normalized area of the spike tuning curve. Data taken from cells in which both voltage-clamp and cell-attached recordings were made. (C-D) Relationships between the peak subthreshold membrane potential in the cell’s null direction and (C) the direction selectivity or (D) normalized area of the spike tuning curve for cells recorded in the current-clamp configuration with no current injection. For all panels, R and p values are Spearman’s rank correlation coefficient and associated 2-sided p-value. Dashed lines are least squares linear regressions.
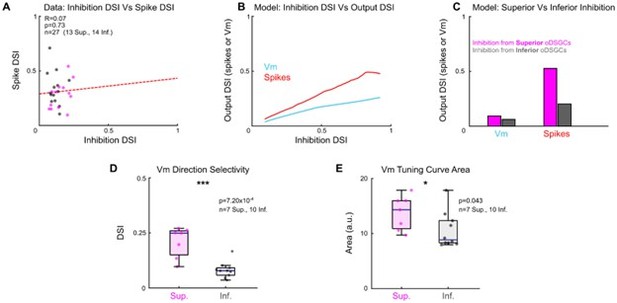
(A) Comparison of the direction selectivity index (DSI) computed for inhibitory inputs (from voltage-clamp recordings) and for spikes (cell-attached recordings) for cells in which both metrics were recorded.
There is no significant relationship, indicating that inhibition is a poor predictor of spike tuning, but this may be caused by noise contributed by other circuit and cell-intrinsic processes. R and p values are Spearman’s rank correlation coefficient and associated 2-sided significance values, respectively. The dashed line is a least squares linear regression. (B) Using our parallel conductance model, the subthreshold membrane potential (Vm) and spike DSI were computed as a function of the DSI of inhibition. For oDSGCs, narrower inhibitory tuning curves predict narrower Vm and spike tuning curves. (C) Output of two additional iterations of the parallel conductance model in which inhibitory conductances were taken only from data recorded from Superior oDSGCs (magenta bars) or only from data recorded from Inferior oDSGCs (gray bars). All other parameters of the model (including excitation, input resistance, threshold potential, and resting membrane potential) were held constant across conditions. All else equal, the model predicts that the inhibitory inputs to Superior oDSGCs predict sharper Vm and spike tuning curves than the inhibitory inputs to Inferior oDSGCs do. (D) Direction selectivity indices of Vm tuning curves, recorded empirically in current-clamp mode. Superior oDSGCs have sharper Vm tuning curves than Inferior oDSGCs. This result matches the prediction of the model from (C), and indicates that sharper inhibitory tuning in Superior oDSGCs does not predict their broader spike tuning curves. These data are also shown in Figure 5K. (E) Area of linear Vm tuning curves recorded empirically in current-clamp mode. Superior oDSGCs have larger Vm tuning curves. This can be accounted for by their larger excitatory inputs (Figure 3G) and explains their broader spike tuning curves. These data are also shown in Figure 5J.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | WT C57BL/6J Mice | The Jackson Laboratory | N/A | |
| Antibody | Anti-Lucifer yellow (rabbit polyclonal) | Invitrogen | Cat #A5750; RRID:AB_1501344 | IF (1:500) |
| Antibody | Anti-gephyrin (mouse monoclonal) | Synaptic System | Cat# 147111; RRID:AB_887719 | IF (1:500) |
| Antibody | PSD-95 MAGUK scaffolding protein (mouse monoclonal) | Neuromab | Cat# 75-028; RRID:AB_2292909 | IF (1:500) |
| Antibody | Anti-choline acetyltransferase (goat polyclonal) | Millipore | Cat# AB144P; RRID:AB_2079751 | IF (1:500) |
| Antibody | Anti-mouse-Dylight 405 (donkey polyclonal) | Jackson ImmunoResearch | Cat# 715-475-150; RRID:AB_2340839 | IF (1:1000) |
| Antibody | Anti-mouse-Alexa 647 (donkey polyclonal) | Jackson ImmunoResearch | Cat# 715-605-151; RRID:AB_2340863 | IF (1:1000) |
| Antibody | Anti-mouse IgG, Fc_Subclass 1 Specific-Dylight 405 (goat polyclonal) | Jackson ImmunoResearch | Cat# 115-475-205; RRID:AB_2338799 | IF (1:1000) |
| Antibody | Anti-mouse moncolonal IgG, Fc_Subclass 2a Specific-Alexa 647 (goat polycolonal) | Jackson ImmunoResearch | Cat# 115-605-206; RRID:AB_2338917 | IF (1:1000) |
| Antibody | Anti-rabbit-Alexa 488 (donkey polyclonal) | Jackson ImmunoResearch | Cat# 711-545-152; RRID:AB_2313584 | IF (1:1000) |
| Antibody | Anti-goat IgG (H+L)-Alexa 647 (donkey polyclonal) | Jackson ImmunoResearch | Cat# 705-605-147; RRID:AB_2340437 | IF (1:1000) |
| Antibody | Streptavidin 488 conjugate antibody | Molecular Probes | Cat# S32354; RRID:AB_2315383 | IF (1:400) |
| Chemical compound, drug | Ames’ Medium | United States Biological | Cat# A1372-25 | |
| Chemical compound, drug | Vectashield | Vector Laboratories | Cat# H-1000; RRID:AB_2336789 | |
| Chemical compound, drug | Red Retrobeads | Lumafluor | https://lumafluor.com/information | |
| Chemical compound, drug | Lucifer yellow CH dilithium salt | Sigma | Cat# L0259 | |
| Chemical compound, drug | Biocytin | Invitrogen | Cat# B1592 | |
| Software, algorithm | Amira | Thermo Fisher Scientific | https://www.fei.com/software/amira-avizo/; RRID:SCR_014305 | |
| Software, algorithm | Bassoon | Harris, 2022 | https://doi.org/10.5281/zenodo.6757605; RRID:SCR_023333 | |
| Software, algorithm | Igor Pro | Igor Pro | RRID:SCR_000325 | |
| Software, algorithm | ImageJ | NIH | https://imagej.nih.gov/ij/; RRID::SCR_003070 | |
| Software, algorithm | Imaris | Bitplane | http://www.bitplane.com/; RRID:SCR_007370 | |
| Software, algorithm | MATLAB | MathWorks | https://www.mathworks.com/products/matlab.html; RRID:SCR_001622 | |
| Software, algorithm | Meshmapper | Paul Bourke | http://paulbourke.net/dome/meshmapper/ | |
| Software, algorithm | ObjectFinder | Della Santina et al., 2013 | https://github.com/lucadellasantina/ObjectFinder; https://zenodo.org/record/4767847; RRID:SCR_023319 | |
| Software, algorithm | Psychopy | Open Science Tools Ltd. Peirce, 2007 | https://psychopy.org/about/index.html | |
| Software, algorithm | ScanImage | MBF Bioscience | https://www.mbfbioscience.com/products/scanimage | |
| Software, algorithm | StreamPix | NorPix | https://www.norpix.com/products/streampix/streampix.php | |
| Software, algorithm | Symphony and Stage | Cafaro, 2019 | https://github.com/Symphony-DAS/symphony-matlab; https://github.com/Stage-VSS/stage-v1 | |
| Software, algorithm | VolumeCut | Della Santina et al., 2021; Della Santina, 2021 | https://github.com/lucadellasantina/VolumeCut; https://doi.org/10.5281/zenodo.5048331 |





