Molecular characterization of the intact mouse muscle spindle using a multi-omics approach
Figures
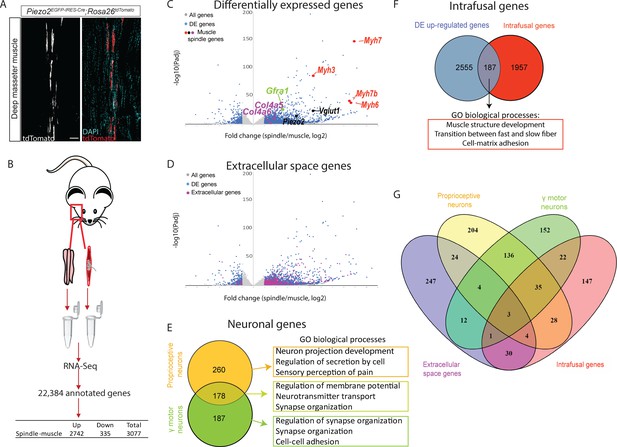
Transcriptomic analysis of intact muscle spindles identified genes expressed in the different spindle tissues.
(A) Confocal images of longitudinal sections of the deep masseter muscle of adult (>P90) Piezo2EGFP-IRES-Cre;Rosa26tdTomato mice. The expression of TdTomato shows the abundance of muscle spindles in this muscle. White (left) and red, tdTomato; cyan, DAPI; scale bar represents 50 μm. (B) Schematic representation of sample isolation and sequencing. Bulk transcriptomic analysis was performed on intact muscle spindles and adjacent extrafusal muscle fibers (muscle). The table contains the number of genes that were differentially expressed between spindle and muscle samples. (C) Volcano plot depicting differentially expressed (DE) genes between spindle and muscle samples. Gray dots represent all detected genes; blue dots represent DE genes. Other colored dots indicate genes known to be expressed in intrafusal fibers (red), proprioceptive neurons (black), γ-motoneurons (green), and muscle spindle capsule (magenta). Y-axis denotes −log10 (p-values), whereas X-axis shows log2 fold change values. (D) Volcano plot depicting DE genes between spindle and muscle samples. Gray dots represent all detected genes; blue dots represent DE genes; magenta represent DE genes that are located at the extracellular. Y-axis denotes −log10 (p-values), whereas X-axis shows log2 fold change values. (E) Left: A Venn diagram showing DE genes potentially expressed by proprioceptive neurons (orange) and γ-motoneurons (green). The overlap between the two datasets is marked by light green. Right: Gene ontology (GO) analysis for enriched biological processes in each dataset using Metascape (see also Supplementary file 4, Supplementary file 5). (F) A Venn diagram showing the overlap between upregulated genes in our analysis (blue) and intrafusal genes previously reported by Kim et al., 2020 (red). Below are the most enriched biological processes in the shared genes, as indicated by GO analysis using Metascape (see also Supplementary file 6; Supplementary file 7). (G) A Venn diagram of the four groups of DE genes displayed in D–F, namely genes associated with the extracellular space (magenta), proprioceptive neurons (yellow), γ-motoneurons (green), and intrafusal fibers (red).
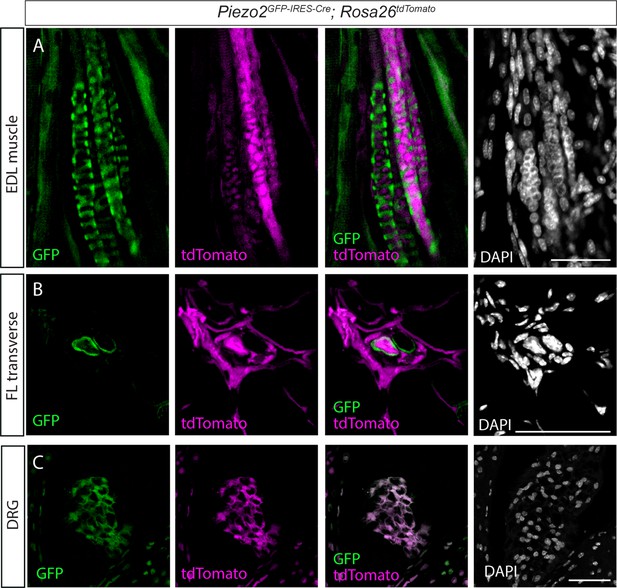
Expression of tdTomato in Piezo2EGFP-IRES-Cre;Rosa26tdTomato mouse.
(A–C) Confocal images of longitudinal sections through the extensor digitorum longus (EDL) muscle (A), transverse section of a muscle spindle in a forelimb (FL) muscle (B), and dorsal root ganglion (DRG) sections (C) from adult (>P90) Piezo2EGFP-IRES-Cre;Rosa26tdTomato mice. tdTomato (magenta) is expressed in DRG neurons (C) as well as in muscle spindle capsule and intrafusal fibers (A,B). Green, GFP; white, DAPI; scale bar represents 50 μm.
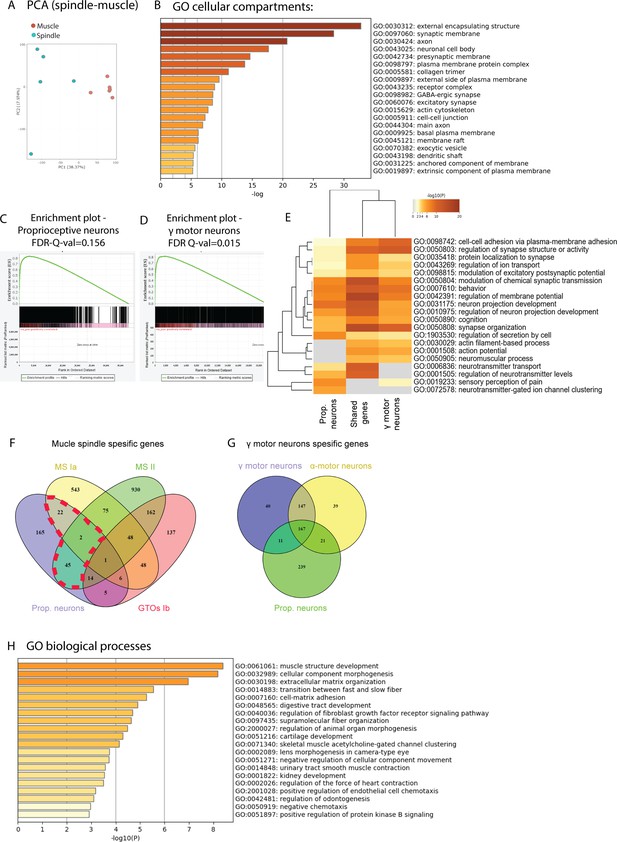
The RNA sequencing (RNA-seq) data contain neuronal transcripts.
(A) Principal component analysis of RNA-seq data from isolated muscle spindles (spindle, blue) and adjacent extrafusal muscle fibers (muscle, red). (B) Results of gene ontology (GO) enrichment analysis for the ‘cellular compartment’ term on differentially expressed (DE) upregulated genes performed by Metascape web tool. The X-axis indicates the −log10 (p-values) of statistically enriched terms. The genes associated with each term are listed in Supplementary file 2. (C,D) Gene set enrichment analysis (GSEA) of proprioceptive neurons (Zheng et al., 2019; C) and γ-motoneurons (Blum et al., 2021; D) pre-ranked by expression values, which were run against spindle DE upregulated genes. Y-axes indicate the enrichment score (ES) and X-axes show the genes (vertical black lines) that are represented in both datasets. All the genes contributing to the enrichment score were used for generating the proprioception and the γ-motoneurons dataset in (E) and in Figure 1E. Significance threshold was set at FDR < 0.05. (E) Heatmap of GO enrichment analysis of biological processes for genes expressed by proprioceptive neurons, by γ-motoneurons and for shared genes (shown in C and D). The −log10 (p-values) of all terms are color-coded (gray is 0 and red is 20). For additional information, see Supplementary file 5. (F) A Venn diagram showing the overlap between our list of spindle afferent genes (prop. neurons, purple) and markers of the three proprioceptive neuron subtypes, muscle spindle afferents Ia (MS-Ia; yellow) and II (MS-II; green) and Golgi tendon organ (GTO) afferents Ib (red), previously reported by Wu et al., 2021. (G) A Venn diagram showing the overlap in gene expression between γ-motoneurons (purple), α-motoneurons (yellow), and proprioceptive neurons (green). (H) Bar graph showing GO enrichment analysis on 187 DE intrafusal genes (shown in Figure 1E). X-axis indicates the −log10 (p-values) of significantly enriched terms. For additional information, see Supplementary file 7.
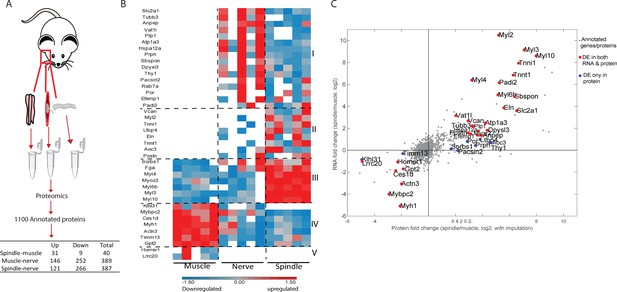
Proteomic analysis of intact muscle spindle identified proteins expressed in its constituent tissues.
(A) Schematic representation of the analyzed samples. Proteomic analysis was performed on intact muscle spindle, extrafusal muscle fibers (muscle), and nerve fibers deprived of their nerve termini and cell bodies (nerve). (B) Heatmap showing clustering of the differentially expressed proteins between muscle spindle and extrafusal fibers. Each horizontal line denotes the relative expression of a single protein (log2-transformed LFQ intensities with row standardization; proteins not detected are in white). Cluster numbers are indicated by Roman letters on the right. (C) Scatter plot showing the correlation between the fold changes of spindle-muscle differentially expressed (DE) proteins and RNA. The X-axis indicates log2 fold change values for proteins, whereas the Y-axis shows the log2 fold change values for transcripts (shown in Figure 1). Gray dots represent all genes and proteins detected, red dots represent DE molecules at both RNA and protein levels, blue dots represent DE proteins only. DE protein symbols are shown on the plot.
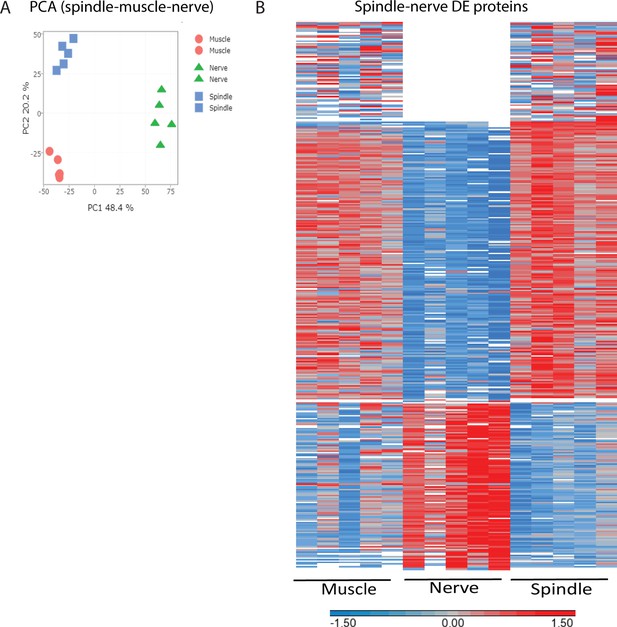
Proteomic analysis of muscle spindles and adjacent muscle and nerve.
(A) Principal component analysis (PCA) of proteomic data from isolated muscle spindles (spindle, blue) extrafusal muscle fibers (muscle, red) and nerve fibers devoid of nerve termini and cell bodies (nerve, blue). (B) Heatmap showing clustering of the differentially expressed proteins between muscle spindle and nerve fibers. Each horizontal line describes the relative expression of a single protein (log2-transformed LFQ intensities with row standardization; proteins not detected are in white).
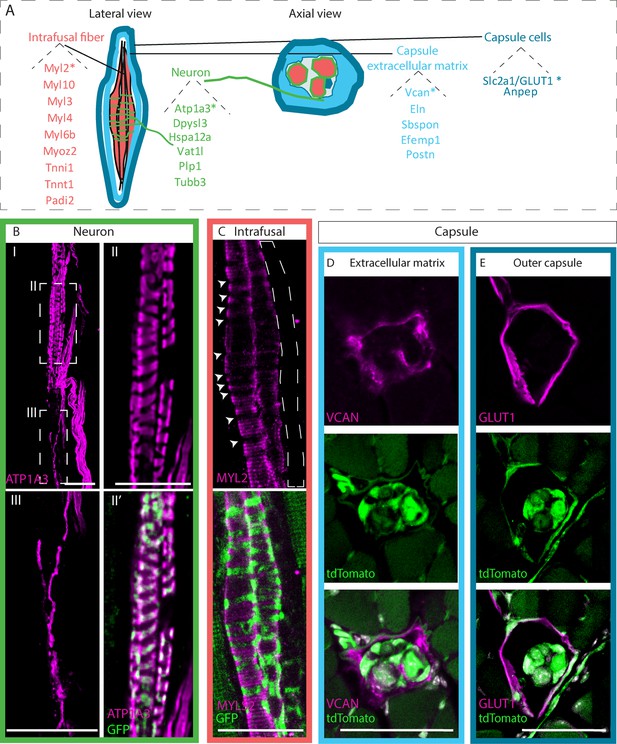
Identifying novel molecular markers for different muscle spindle tissues.
(A) Schematic representation of the different tissues composing the muscle spindle in a lateral (left) and axial (center) views. Twenty-two potential markers that were found to be upregulated at both RNA and protein levels are listed next to their predicted tissue of expression, namely intrafusal fibers (red), neurons (green), capsule cells (dark blue), and capsule extracellular matrix (light blue). Markers that were further validated are marked with asterisks. (B,C) Confocal images of whole-mount extensor digitorum longus (EDL) muscle from Piezo2EGFP-IRES-Cre mice, in which proprioceptive neurons are fluorescently labeled by GFP (green), which were immunostained for ATP1A3 (B, magenta) or myosin light chain 2 (MYL2) (C, magenta). Anti-ATP1A3 stained proprioceptive neurons (BII) and γ-motoneurons (BIII). (II,III) are high magnifications of the boxed areas in (I). Anti-MYL2 stained intrafusal bag fibers, but not chain fibers (indicated by dashed lines); arrowheads indicate the neuron-muscle interface, where MYL2 staining was absent. Scale bars represent 50 μm. (D,E) Confocal images of transverse sections of forelimb muscles from Piezo2EGFP-IRES-Cre;Rosa26tdTomato mice, in which muscle spindles are fluorescently labeled by tdTomato (green), which were immunostained for versican (VCAN) (D, magenta) or GLUT1 (E, magenta). VCAN was expressed in the extracellular matrix of the capsule, whereas GLUT1 expression was restricted to the outer capsule cells. Scale bars represent 50 μm.
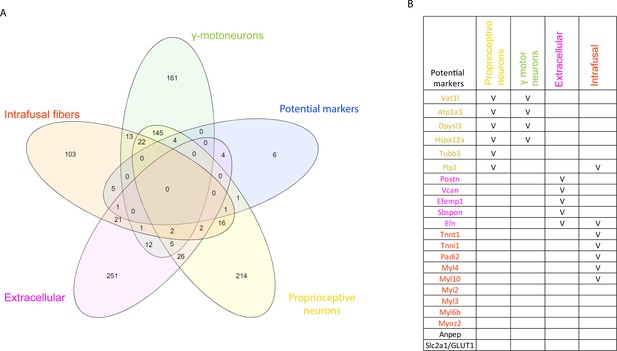
Correlation between protein and RNA upregulation in the spindle.
(A) A Venn diagram of the 22 potential markers that were upregulated at both RNA and protein levels (blue) and the four groups of differentially expressed (DE) upregulated genes shown in Figure 1 (extracellular space, magenta; proprioceptive neurons, yellow; γ-motoneurons, green; intrafusal fibers, red). (B) A Supplementary File of the predicted tissue-specific expression of the 22 potential markers that were upregulated at both RNA and protein levels, color-coded as in (A).
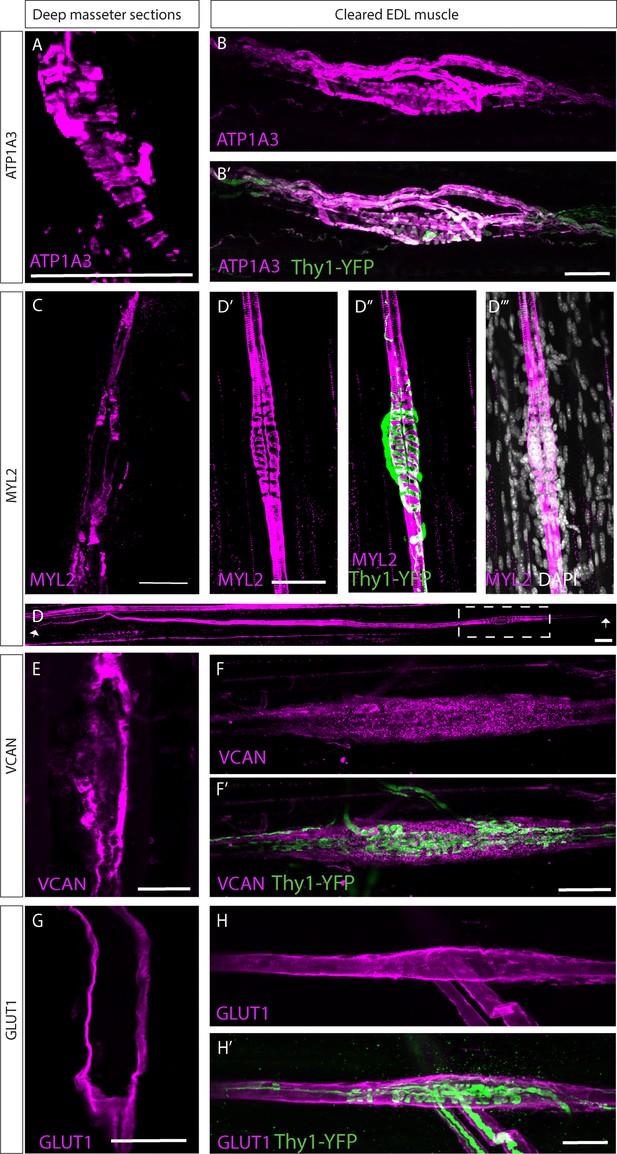
Validation of predicted muscle spindle markers.
Confocal images of longitudinal sections of the deep masseter muscle (A,C,E,G) and of whole-mount extensor digitorum longus (EDL) muscle (B,D,F,H) taken from adult (p>45) mice stained with the indicated antibodies. Muscles in B,D,F,G were taken from mice expressing Thy1-YFP reporter (green) (Feng et al., 2000). ATP1A3 (A,B), myosin light chain 2 (MYL2) (C,D), versican (VCAN) (E,F), GLUT1 (G,H) are in magenta; white is DAPI (D). The boxed area in D is enlarged in D’–D’’’; arrows in D indicate the ends of the bag fibers. Scale bars represent 50 μm.
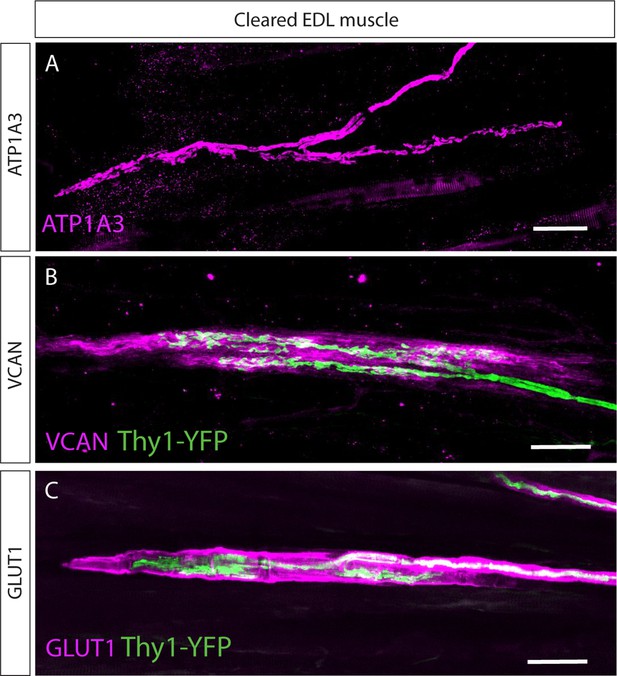
Examination of the expression of muscle spindle markers in the Golgi tendon organ (GTO).
Confocal images of whole-mount extensor digitorum longus (EDL) muscles from adult (p>45) mice stained with antibodies against ATP1A3 (A), versican (VCAN) (B), and GLUT1 (C; all in magenta). Muscles in (B,C) were taken from mice expressing Thy1-YFP reporter (green). Scale bars represent 50 μm.
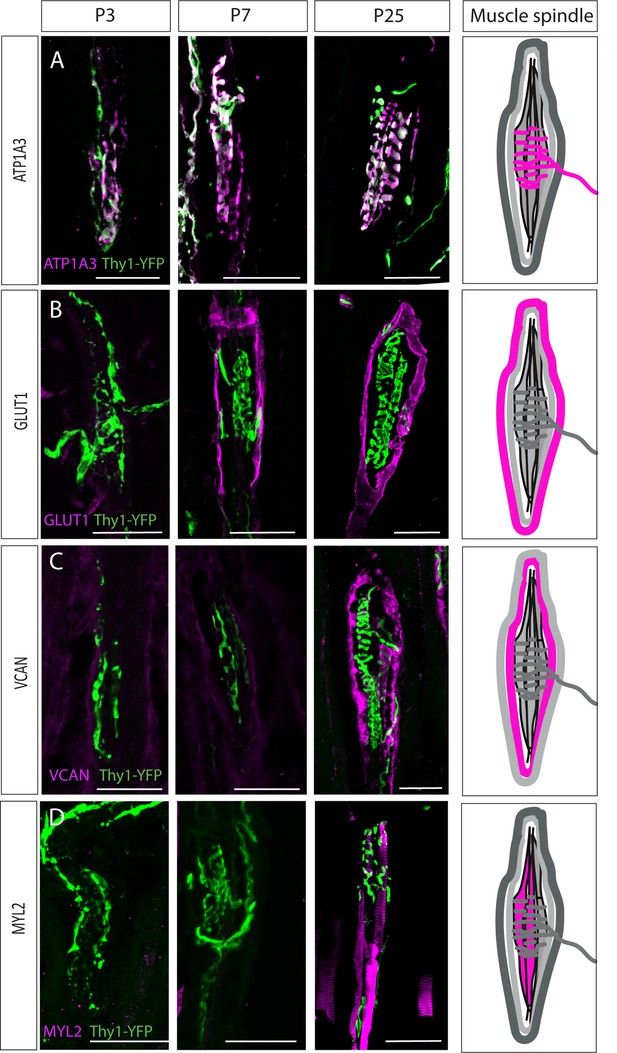
Postnatal muscle spindle development.
(A–D) Confocal images of longitudinal sections of the deep masseter muscle from Thy1-YFP mice (YFP, green) stained with the indicated antibodies at P3, P7, and P25. ATP1A3 (A) was expressed by proprioceptive neurons at all examined time points. GLUT1 expression (B) was detected in the outer capsule cells at postnatal day 7 (P7). Versican (VCAN) expression (C) was detected in the extracellular space at P25. Myosin light chain 2 (MYL2) expression (D) was detected in intrafusal fibers at P25. ATP1A3, GLUT1, VCAN, and MYL2 are in magenta. Scale bars represent 50 μm. On the right, schematic representations of adult muscle spindle with the analyzed tissue in magenta.
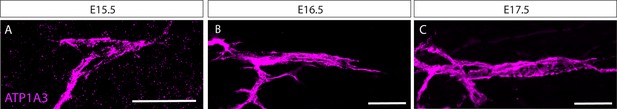
Embryonic expression of ATP1a3.
Confocal images of whole-mount gluteus (A) and extensor digitorum longus (EDL) (B,C) muscles from embryos at the indicated developmental stages stained with anti-ATP1a3 antibody. Scale bars represent 50 μm.
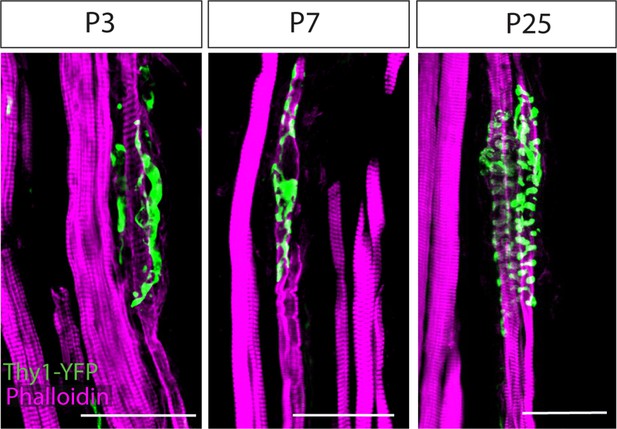
Sarcomeric organization of the muscle spindle.
Confocal images of longitudinal sections of the deep masseter muscle from Thy1-YFP mice (green) stained with phalloidin (magenta) at postnatal days (P) P3, P7, and P25. Scale bars represent 50 μm.
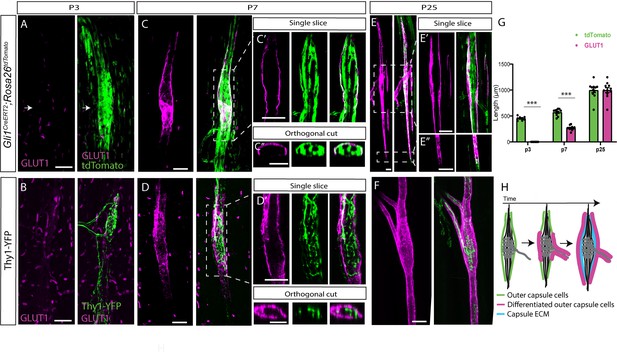
Postnatal development of muscle spindle capsule cells.
(A,C,E) Confocal images of whole-mount extensor digitorum longus (EDL) muscle taken from Gli1CreERT2;Rosa26tdTomato mice stained for GLUT1 (magenta). Gli1+ were labeled by a single tamoxifen administration at postnatal day 1 (P1) and analyzed at P3 (A), P7, (C) and P25 (E). Arrows in (A) show the location of the capsule cells, as marked by TdTomato (green). The center of the spindle (boxed area in C) is shown in a single slice (C’) and in an orthogonal view (C’’). The center of the spindle (top boxed area in E) and its edges (bottom boxed area in E) are shown in single slices (E’ and E’’, respectively). (B,D,F) Confocal images of whole-mount EDL muscle from Thy1-YFP mice stained with anti-GLUT1 antibody at P3 (B), P7 (D), and P25 (F). The center of the spindle (boxed area in D) is shown in a single slice (D’) and in an orthogonal view (D’’). (G) Quantification of capsule length, as measured in (A,C,E) based on GLUT1 and tdTomato labeling (nP3 = 7, p = 9.07E-08; nP7 = 10, p = 1.09E-08; nP25 = 10, p = 0.989; two-tailed t-test; data are presented as mean ± SEM; each dot represents one spindle). (H) Schematic representation of the maturation process of the outer capsule throughout postnatal development. Scale bars represent 50 μm in (A–F) and 20 μm in (B’,C’).
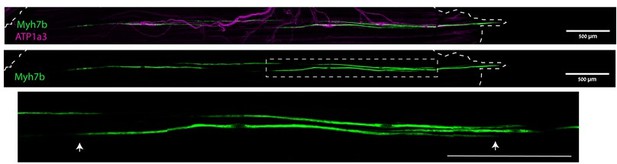
The anchoring of muscle spindles.
Confocal images of whole-mount EDL muscle from adult mice stained for Myh7b (green), to mark bag 2 intrafusal fiber, and ATP1a3 (magenta). The boxed area, which is enlarged below, demarcates an intrafusal fiber ending within the muscle belly. Arrows indicate the ends of the bag 2 fiber, dashed line indicates the end of the muscle. Scale bars represent 50 μm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Mus musculus) | B6(SJL)-Piezo2tm1.1(cre)Apat/J | Jackson Laboratory | Stock #027719 RRID:IMSR_JAX:027719 | |
| Gene (Mus musculus) | Gli1tm3(cre/ERT2)Alj/J | Jackson Laboratory | Strain #:007913 RRID:IMSR_JAX:007913 | |
| Gene (Mus musculus) | B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J | Jackson Laboratory | Strain #:007909 RRID:IMSR_JAX:007909 | |
| Gene (Mus musculus) | B6.Cg-Tg(Thy1-YFP)16Jrs/J | Jackson Laboratory | Strain #:003709 RRID:IMSR_JAX:003709 | |
| Antibody | Anti-ATP1a3 (rabbit polyclonal) | Millipore | Cat# 06-172I, RRID:AB_310066 | Section 1:100 Whole mount 1:100 |
| Antibody | Anti-VERSICAN (rabbit polyclonal) | Abcam | Cat# ab19345, RRID:AB_444865 | Section 1:300 Whole mount 1:50 |
| Antibody | Anti-MYL2 (rabbit polyclonal) | Abcam | Cat# ab79935, RRID:AB_1952220 | Section 1:100 Whole mount 1:100 |
| Antibody | Anti-GLUT1 (rabbit monoclonal) | Abcam | Cat# ab195020, RRID:AB_2783877 | Section 1:400 Whole mount 1:200 |
| Antibody | Anti-GFP (biotin goat polyclonal) | Abcam | Cat# ab6658 RRID:AB_305631 | Section 1:100 Whole mount 1:100 |
| Antibody | Cy5 conjugated donkey anti-rabbit (polyclonal) | Jackson ImmunoResearch Laboratories | Cat# 711-175-152 RRID:AB_2340607 | Section 1:100 Whole mount 1:200 |
| Peptide, recombinant protein | Phalloidin, synthetic peptide (TRITC) | Sigma-Aldrich | Cat# P1951 RRID:AB_2315148 | Section 2 μg/ml |
| Peptide, recombinant protein | Native Streptavidin protein (DyLight 488) | Abcam | Cat# ab134349 | Section 1:100 Whole mount 1:200 |
| Software, algorithm | ImageJ software | ImageJ (http://imagej.nih.gov/ij/) | RRID:SCR_003070 |
Primer sequences and amplicon sizes used for PCR.
| Reaction | Amplicon (bp) | Sequences |
|---|---|---|
| Cre | 800 | F: CCTGGAAAATGCTTCTGTCCGTTTGCC R::GAGTTGATAGCTGGCTGGTGGCAGATG |
| Cre-ERT2 | 800 | F: CCTGGAAAATGCTTCTGTCCGTTTGCC R: GAGTTGATAGCTGGCTGGTGGCAGATG |
| tdTomato (wild type) | 297 | F: AAG GGA GCT GCA GTG GAG TA R: CCG AAA ATC TGT GGG AAG TC |
| tdTomato (tdTomato-flox allele) | 196 | F: GGC ATT AAA GCA GCG TAT CC R: CTG TTC CTG TAC GGC ATG G |
| YFP (GFP) | 300 | F: GACGGCAACATCCTGGGGCACAAG R: CGGCGGCGGTCACGAACTCC |
Additional files
-
Supplementary file 1
Differentially expressed genes between spindle and muscle samples.
DE, differentially expressed.
- https://cdn.elifesciences.org/articles/81843/elife-81843-supp1-v2.xlsx
-
Supplementary file 2
Gene ontology (GO) enrichment analysis for upregulated genes in the various cellular compartment using Metascape.
- https://cdn.elifesciences.org/articles/81843/elife-81843-supp2-v2.xlsx
-
Supplementary file 3
Differentially upregulated genes localized to the extracellular space.
Ingenuity pathway analysis revealed 325 differentially upregulated genes localized to the extracellular space.
- https://cdn.elifesciences.org/articles/81843/elife-81843-supp3-v2.xlsx
-
Supplementary file 4
Gene set enrichment analysis (GSEA) results.
Genes of proprioceptive neurons (Zheng et al., 2019) and of γ-motoneurons (Blum et al., 2021) that contributed to the enrichment score are listed, as are 178 common genes between proprioceptive neurons and γ-motoneurons, muscle spindle-specific genes, γ-motoneuron-specific genes, and common genes between proprioceptive neurons, γ-motoneurons, and α-motoneurons.
- https://cdn.elifesciences.org/articles/81843/elife-81843-supp4-v2.xlsx
-
Supplementary file 5
Gene ontology (GO) enrichment analysis of neuronal genes (listed in Supplementary file 4) using Metascape.
- https://cdn.elifesciences.org/articles/81843/elife-81843-supp5-v2.xlsx
-
Supplementary file 6
Intrafusal genes.
Comparison of differentially upregulated genes in our dataset with the expression profile of intrafusal fibers (Kim et al., 2020) revealed 187 overlapping genes.
- https://cdn.elifesciences.org/articles/81843/elife-81843-supp6-v2.xlsx
-
Supplementary file 7
Gene ontology (GO) enrichment analysis of intrafusal genes (listed in Supplementary file 6) using Metascape.
- https://cdn.elifesciences.org/articles/81843/elife-81843-supp7-v2.xlsx
-
Supplementary file 8
Proteomic analysis results – differentially expressed proteins between spindle, muscle, and nerve samples.
The expression intensity of all detected proteins as well as differentially expressed (DE) proteins between spindle and muscle samples and between spindle and nerve samples are shown.
- https://cdn.elifesciences.org/articles/81843/elife-81843-supp8-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/81843/elife-81843-mdarchecklist1-v2.pdf





