Establishment of transgenic fluorescent mice for labeling synapses and screening synaptogenic adhesion molecules
Figures
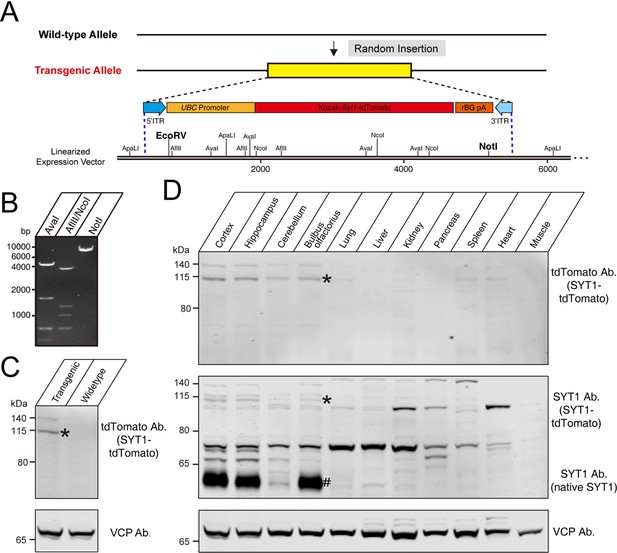
Construction of tdTomato-fused synaptotagmin-1 (Syt1-tdTomato) expression vector and transgenic fluorescent mice.
(A) Schematic diagram of expression vector for generating Syt1-tdTomato transgenic mice. (B) The expression vector was digested by restriction enzymes for confirmation purposes. The size of enzyme digestion band (kb): (left) AvaI: 4.7/1.7/0.7/0.5; (middle) AflII/NcoI: 4.0/1.3/1.1/0.7/0.4/0.2; (right) NotI: 7.6. (C) Immunoblot analysis of protein in transgenic and wild-type mice brain. Equivalent amounts of protein were probed with antibodies to tdTomato and vasolin-containing protein (VCP, used as a loading control). (D) Immunoblot analysis of the expression of the transgenic protein in different tissues of Syt1-tdTomato transgenic mice. Equivalent amounts of protein were examined by immunoblotting with antibodies to tdTomato, SYT1, and VCP. Asterisk shows the band of SYT1-tdTomato and the hash shows the band of native SYT1.
-
Figure 1—source data 1
The original gel of panels B, C and D.
- https://cdn.elifesciences.org/articles/81884/elife-81884-fig1-data1-v2.zip
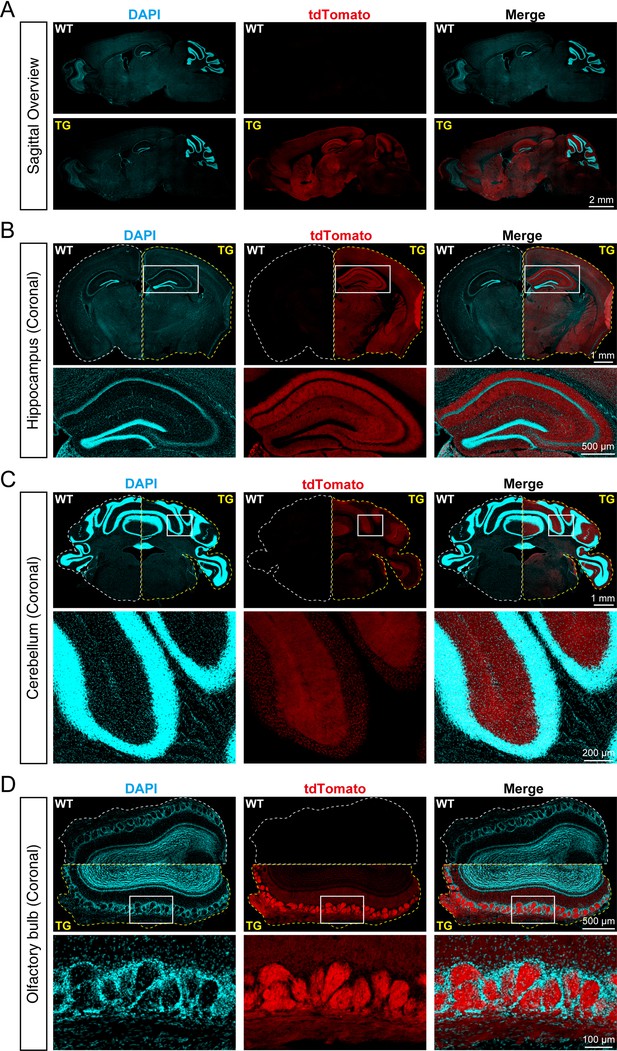
tdTomato-fused synaptotagmin-1 (SYT1-tdTomato) is expressed in different brain regions of transgenic mice.
(A) Representative fluorescence images of the sagittal plane of the brains in Syt1-tdTomato transgenic mice and wild-type (WT) mice (8 weeks). The SYT1-tdTomato signals were shown in channel tdTomato (red). All nuclei were stained with DAPI (cyan). (B–D) Representative fluorescent images in different brain areas were obtained from the Syt1-tdTomato transgenic mice and WT mice (8 weeks). (B) Hippocampus and cortex; (C) cerebellum; (D) olfactory bulb.
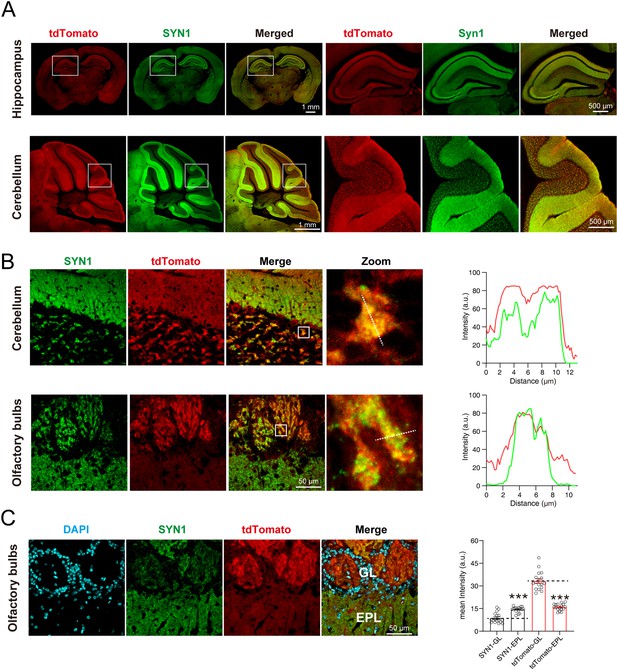
tdTomato-fused synaptotagmin-1 (SYT1-tdTomato) is colocalized with synaptic markers in situ.
(A) The images of SYT1-tdTomato (tdTomato) signals and synapsin1 (SYN1) signals in hippocampus and cerebellum. (B) Magnified images of SYT1-tdTomato and SYN1 in granular layer of cerebellum and the glomerular layer of olfactory bulb. The fluorescence intensity profiles of dash lines in zoomed images are plotted on the right. (C) The represent images (left) and mean intensity (right) of SYT1-tdTomato and SYN1 in glomerular layer (GL) and external plexiform layer (EPL) of olfactory bulb. All summary graphs show the mean ± standard error of the mean (SEM); statistical analysis was made by t-test (n=17 images in each group, ***p<0.001).
-
Figure 3—source data 1
Numerical data corresponding to the graph in panels B and C.
- https://cdn.elifesciences.org/articles/81884/elife-81884-fig3-data1-v2.zip
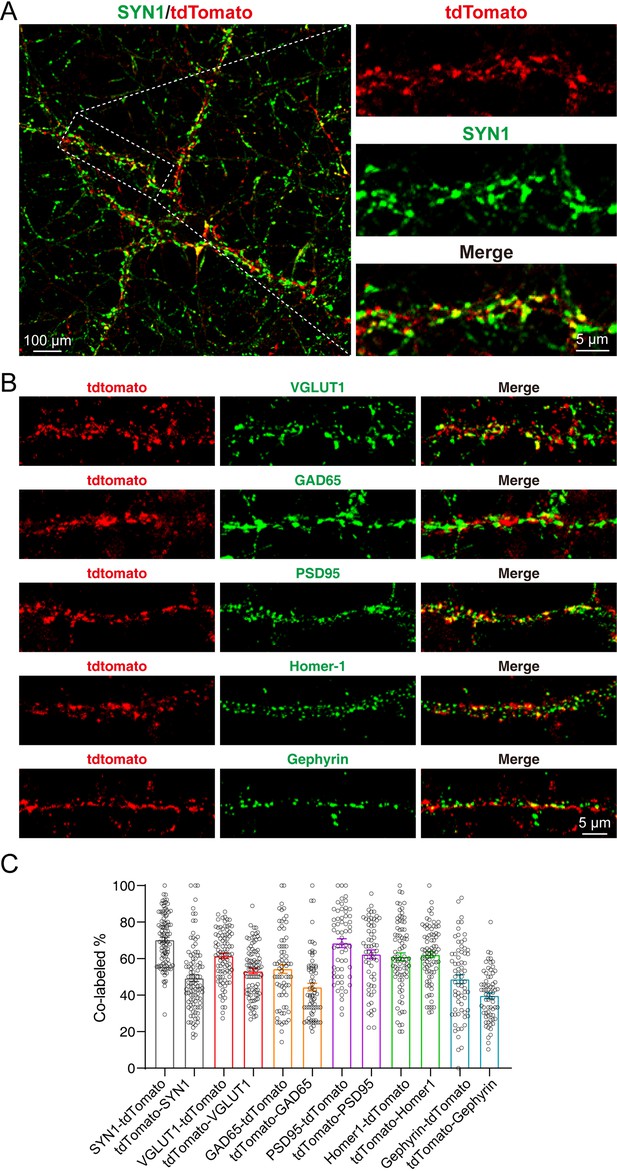
tdTomato-fused synaptotagmin-1 (SYT1-tdTomato) signals localize to the synaptic site in cultured neurons.
(A–B) Representative images of hippocampal neurons from Syt1-tdTomato transgenic mice stained with antibodies against SYN1, VGLUT1, GAD65, PSD95, Homer-1, and Gephyrin at DIV 14. (C) Summary graphs of co-labeling rate of SYT1-tdTomato with SYN1, VGLUT1, GAD65, PSD95, Homer-1, and Gephyrin (SYN1: n=98/3, VGLUT1: n=90/3, GAD65: n=71/3, PSD95: n=61/3, Homer-1: n=81/3, Gephyrin: n=69/3). All summary graphs show the mean ± standard error of the mean (SEM).
-
Figure 4—source data 1
Numerical data corresponding to the graph in panel C.
- https://cdn.elifesciences.org/articles/81884/elife-81884-fig4-data1-v2.zip
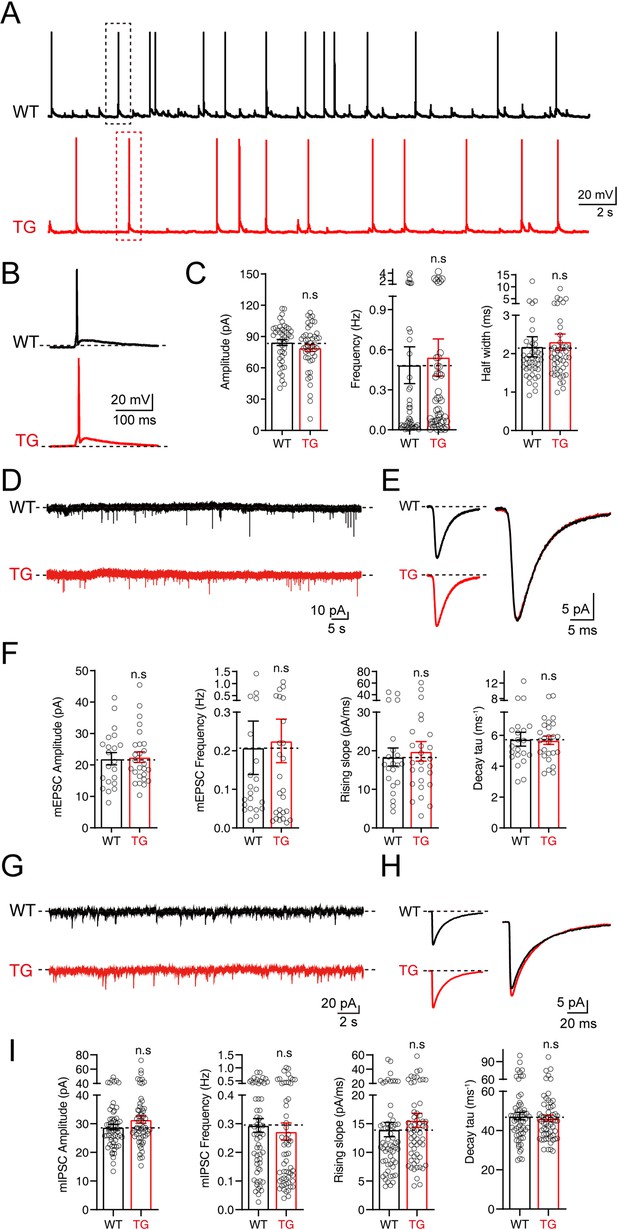
No significant changes in electrophysiological properties were observed in neurons of tdTomato-fused synaptotagmin-1 (Syt1-tdTomato) transgenic mice.
(A–C) The spontaneous action potentials were recorded in hippocampal neurons of wild-type (WT) mice and Syt1-tdTomato transgenic mice (TG) at DIV 13–15. (A) Representative traces of action potential; (B) representative traces of single action potential; (C) summary graphs of the amplitude (left), frequency (center), and half widths (right). (WT: n=43/3; TG: n=45/3, amplitude: p=0.235, frequency: p=0.768, half widths: p=0.716). (D–F) The mEPSCs were recorded in hippocampal neurons of WT mice and Syt1-tdTomato transgenic mice in 1 μM tetrodotoxin (TTX) and 0.1 mM picrotoxin (PTX) at DIV 13–15. (D) Representative traces of mEPSCs; (E) representative traces of normalized traces of mEPSCs; (F) summary graphs of the frequency (left), amplitude (center left) of mEPSCs and the rising slope (center right), decay τ (right) of normalized traces of mEPSCs. (WT: n=21/3; TG: n=26/3, amplitude: p=0.837, frequency: p=0.842, rising slope: p=0.681, decay τ: p=0.894). (G–I) The mIPSCs recordings in hippocampal neurons of WT mice and Syt1-tdTomato transgenic mice in 1 μM TTX and 10 μM CNQX at DIV 13–15. (G) Representative traces of mIPSCs; (H) representative traces of normalized traces of mIPSCs; (I) summary graphs of the frequency (left), amplitude (center left) of mIPSCs and the rising slope (center right), decay τ (right) of normalized traces of mIPSCs. (WT: n=59/4; TG: n=59/4, amplitude: p=0.102, frequency: p=0.578, rising slope: p=0.355, decay τ: p=0.654). All summary graphs show the mean ± standard error of the mean (SEM); statistical comparisons were made by a two-tailed unpaired t-test (n.s, not significant).
-
Figure 5—source data 1
Numerical data corresponding to the graph in panel C, F, I.
- https://cdn.elifesciences.org/articles/81884/elife-81884-fig5-data1-v2.zip
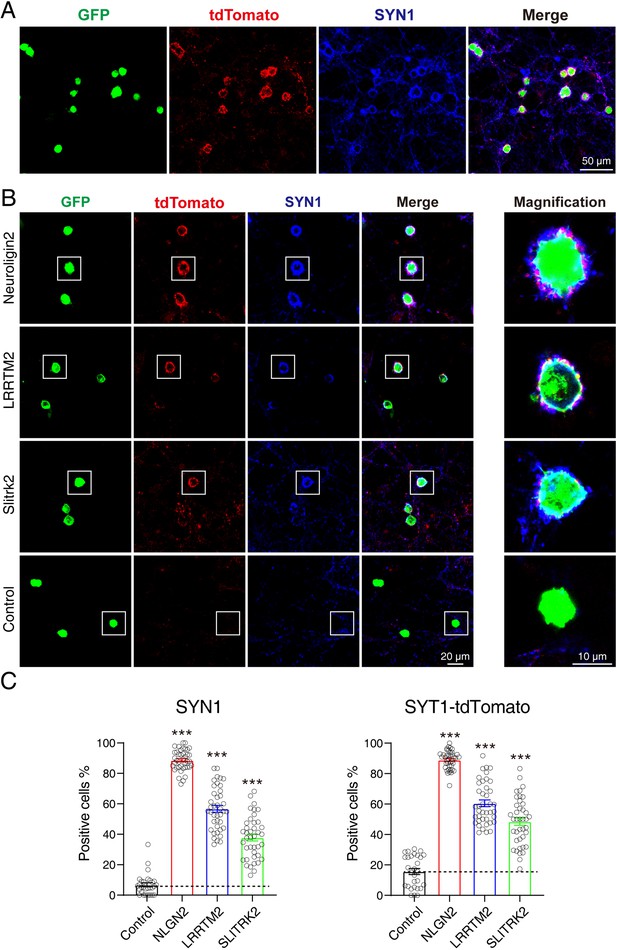
tdTomato-fused synaptotagmin-1 (Syt1-tdTomato) transgenic mice hippocampal neurons can be used in artificial synapse formation experiments to screen for synaptogenic molecules.
(A) Representative fluorescent images of co-cultured neurons from Syt1-tdTomato transgenic mice with HEK293T cells transfected with plasmid encoding NLGN2 and GFP, which are also stained with synapsin antibody. (B) Representative fluorescent images of co-cultured neurons from Syt1-tdTomato transgenic mice with HEK293T cells transfected with plasmid encoding NLGN2, LRRTM2, SLITRK2, and GFP. (C) Summary graphs of the percentage of synapse-positive HEK293T cells from the panel B. (SYN1: Control: n=54 cells out of 854 cells/3 cultures; NLGN2: n=1417 cells out of 1599 cells/3 cultures; LRRTM2: n=692 cells out of 1259 cells/3 cultures; SLITRK2: n=374 cells out of 1032 cells/3 cultures. tdTomato: Control: n=148 cells out of 854 cells/3 cultures; NLGN2: n=1411 cells out of 1599 cells/3 cultures; LRRTM2: n=740 cells out of 1259 cells/3 cultures; SLITRK2: n=486 cells out of 1032 cells/3 cultures). All summary graphs show the mean ± standard error of the mean (SEM); statistical analysis was made by one-way ANOVA (***p<0.001).
-
Figure 6—source data 1
Numerical data corresponding to the graph in panel C.
- https://cdn.elifesciences.org/articles/81884/elife-81884-fig6-data1-v2.zip





