Hepatic lipid overload triggers biliary epithelial cell activation via E2Fs
Figures
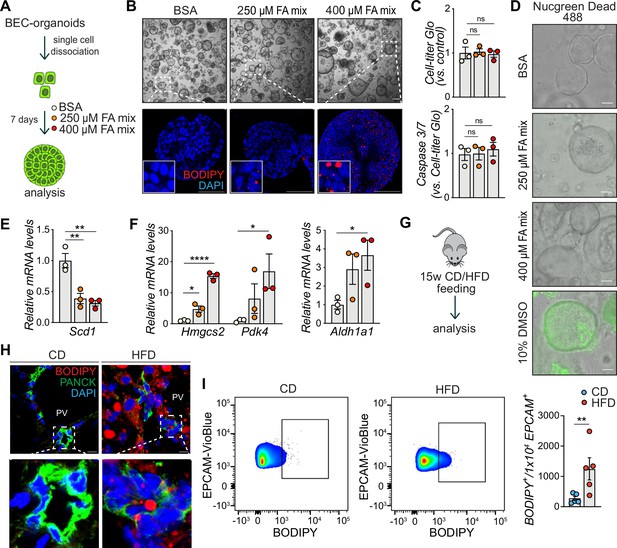
Biliary epithelial cells (BECs) accumulate lipids.
(A) Schematic depicting fatty acid (FA) treatment of BEC-organoids in vitro. (B) Representative bright field and immunofluorescence (IF) images of lipids (BODIPY) in control (BSA) and FA-treated organoids. Close-up IF images were digitally zoomed in four times. n=3. (C) Cell-titer Glo and Caspase 3/7 activity measurement for viability and apoptosis detection relative to panel A. n=3. (D) Representative Nucgreen Dead 488 staining as composite images from bright field and fluorescent microscopy. n=3. (E–F) Quantification of Scd1 (E) and Hmgcs2, Pdk4, and Aldh1a1 (F) mRNA in control (BSA) and FA-treated organoids. n=3. (G) Schematic depicting chow diet (CD) and high-fat diet (HFD) feeding in vivo. (H) Representative images for co-staining of BODIPY and PANCK, relative to panel G. Close-up IF images were digitally zoomed in four times. n=3. (I) Representative quantitative plots of the percentage (left) and quantification (right) of BODIPY staining in EPCAM+ BECs isolated from the liver of C57BL/6 J mice fed CD or HFD for 15 weeks. n=5. Data are shown as mean ± SEM. Absence of stars or ns, not significant (p>0.05); *p<0.05; **p<0.01, ****p<0.0001; one-way ANOVA with Dunnett’s test (C), Fisher’s LSD test (E, F), and unpaired, two-tailed Student’s t-test (I) were used. PV, portal vein. Scale bars, 200 μm (B - bright field), 100 μm (B - IF, D), and 10 μm (H).
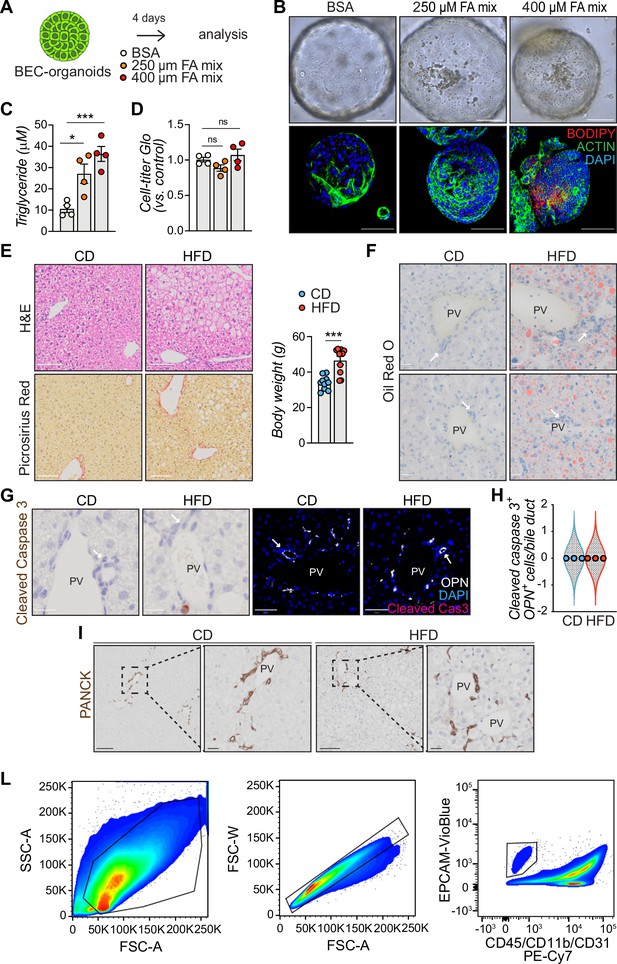
Further characterization of lipid accumulation in biliary epithelial cells (BECs).
(A) Scheme depicting bovine serum albumin (BSA) or fatty acid (FA) mix treatment for already grown BEC-organoids for 4 days. (B) Representative bright field and immunofluorescence (IF) images of lipids (BODIPY) and ACTIN co-staining, relative to panel A. n=4. (C) Measurement of triglyceride concentration upon FA mix treatment relative to panel A, normalized by protein amount. n=4. (D) Cell-titer Glo measurement for viability detection relative to panel A. n=4. (E) Representative hematoxylin and eosin (H&E) (top) and picrosirius red (bottom) staining of liver sections from C57BL/6 J mice fed chow diet (CD) or high-fat diet (HFD) for 15 weeks. The bar graph represents the body weight of the mice at the end of the experiment. n=10. (F) Representative Oil Red O staining, relative to panel E. n=8. (G–H) Representative cleaved caspase three staining (G) and quantification of apoptotic cells in bile ducts (H), relative to panel E. n=3. (I) Representative PANCK staining (left) and magnification (right), relative to panel E. n=5. (L) Flow cytometry gating strategy for analysis of BECs: individual cells were sequentially gated based on cell size (FSC-A versus SSC-A) and singlets. BECs were then selected based on EPCAM positivity after excluding leukocytes (CD45+), myeloid cells (CD11b+), and endothelial cells (CD31+), yielding a population of single CD45-/CD11b-/ CD31-/EPCAM+ BECs. Data are shown as mean ± SEM. Absence of stars or ns, not significant (p>0.05); *p<0.05; ***p<0.001; one-way ANOVA with Dunnet’s test (C, D) or unpaired, two-tailed Student’s t-test (E) were used. PV, portal vein. Arrowheads mark bile ducts. Scale bars, 100 μm (B, E, I - zoom out), 50 μm (G - IF), and 20 μm (F, G - bright field, I - zoom in).
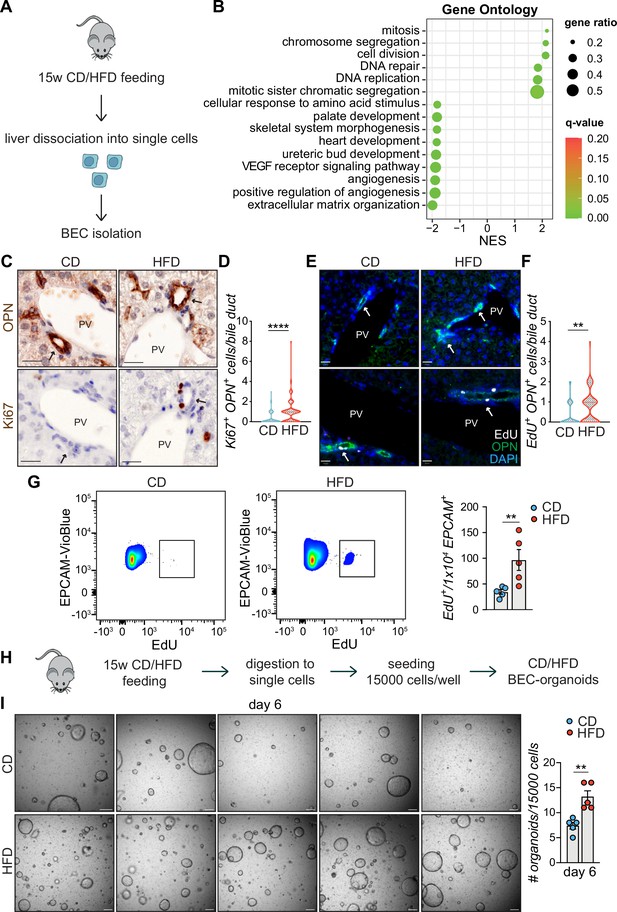
High-fat diet (HFD) feeding induces EPCAM+ biliary epithelial cell (BEC) proliferation.
(A) Scheme depicting the isolation of EPCAM+ BECs from CD- and HFD-fed mice by fluorescence-activated cell sorting (FACS). (B) Gene set enrichment analysis (GSEA) of Gene Ontology (GO) terms. Top 15 upregulated biological processes (BP), ordered by normalized enrichment score (NES). q-value: false discovery rate adjusted p-values. (C–F) Representative co-staining images (C, E) and quantification (D, F) of BECs stained for OPN and Ki67 (C–D), or OPN and EdU (E–F) in livers of CD/HFD-fed mice. n=10 for Ki67 and n=5 for EdU. (G) Representative quantitative plots of the percentage (left) and quantification (right) of EdU+ EPCAM+ BECs, relative to panel A. n=5. (H) Schematic depicting BEC-organoid formation in vitro from CD/HFD-fed mouse livers. (I) Images of organoid colonies formed 6 days after seeding and quantification of organoids per well. n=5. Violin graphs depict the distribution of data points i.e the width of the shaded area represents the proportion of data located there. Other data are shown as mean ± SEM. **p<0.01; ****p<0.0001; unpaired, two-tailed Student’s t-test was used. PV, portal vein. Arrowheads mark bile ducts. Scale bars, 20 μm (C, E), 200 μm (I).

RNA-seq analysis of EPCAM+ biliary epithelial cells (BECs) upon high-fat diet (HFD).
(A) Fluorescence-activated cell sorting (FACS) gating strategy for isolation of CD45−/CD11b−/CD31−/EPCAM+ BECs: individual cells were sequentially gated based on cell size (FSC-A versus SSC-A) and singlets. BECs were then selected based on EPCAM positivity after excluding leukocytes (CD45+), myeloid cells (CD11b+), and endothelial cells (CD31+), yielding a population of single CD45-/CD11b-/ CD31-/EPCAM+ BECs. (B) Principal component analysis (PCA) of mRNAs measured in EPCAM+ BECs from livers of mice fed chow diet (CD) or high-fat diet (HFD) by RNA-seq. n=5 for CD, n=7 for HFD. (C) Volcano plot of HFD vs. CD differential analysis. Top 20 differentially expressed genes were labeled. Blue dots represent downregulated genes (log2(FC) < –1 & adj. p-value <0.05). Red dots represent upregulated genes (log2(FC) >1 & adj. p-value <0.05). Gray dots represent genes not changing significantly. (D–E) Box plots representing the differential gene expression of Ncam1 (D) and well-established markers of the DR signature (E). n=5 for CD, n=7 for HFD. The Y-axis depicts log2(cpm +1) values. (F) Gene set enrichment analysis (GSEA) of KEGG terms. Top 15 enriched pathways (sorted by q-value). q-value: false discovery rate adjusted p-values. NES: normalized enrichment score. Data are summarized in boxplots. Absence of stars or ns, not significant (p>0.05); **p<0.01; unpaired, two-tailed Student’s t-test (D, E) was used.
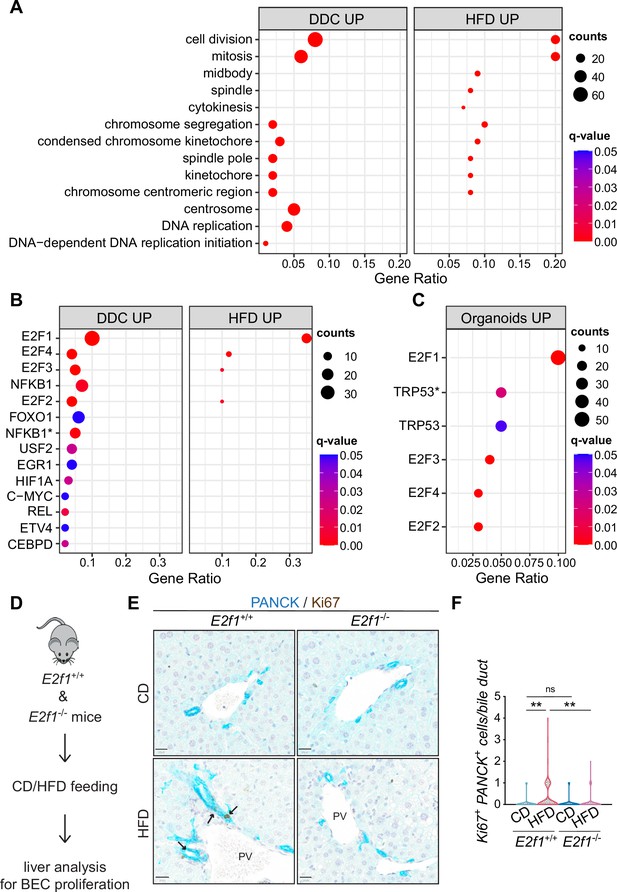
E2Fs are enriched in 3,5-diethoxycarbonyl-1,4-dihydrocollidine (DDC) and high-fat diet (HFD) datasets and mediate biliary epithelial cell (BEC) activation in vivo.
(A) Over-representation analysis results. Top 13 enriched biological processes (BP) upon HFD (own data) and DDC (GSE125688) treatment. q-value: false discovery rate adjusted p-values, counts: number of found genes within a given gene set. (B–C) Enriched transcription factors (TFs) of upregulated genes identified by over-representation analysis in HFD (own data) and DDC (GSE125688) treatment (B), and during the process of organoid formation from single BECs (Organoids vs. T0) (GSE123133) (C). Asterisk (*) marks TFs of the ‘TF_ZHAO’ gene set. (D) Schematic depicting in vivo E2F1 analysis. (E–F) Representative images of PANCK/Ki67 co-staining in livers of E2f1+/+ and E2f1-/- mice fed with chow diet (CD) or HFD (E) and quantification of proliferative BECs in the livers of the indicated mice (F). For CD, n=5 for E2f1+/+ and E2f1-/-. For HFD, n=7 for E2f1+/+, and n=8 for E2f1-/-. Violin graphs depict the distribution of data points i.e., the width of the shaded area represents the proportion of data located there. ns, not significant; **p<0.01; two-way ANOVA with Tukey’s test was used. PV, portal vein. Arrowheads mark bile ducts. Scale bars, 20 μm (E).
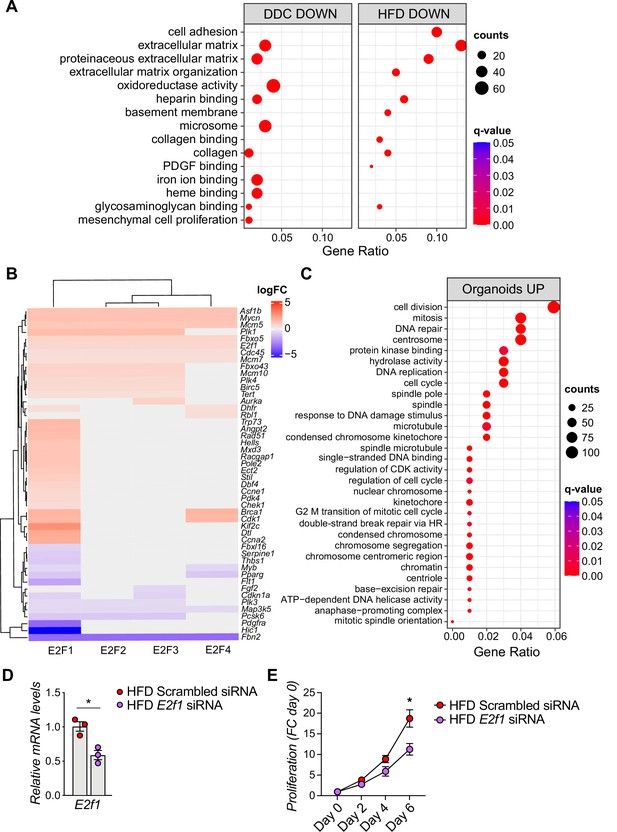
Extended analysis of biliary epithelial cells (BECs) upon high-fat diet (HFD), 3,5-diethoxycarbonyl-1,4-dihydrocollidine (DDC), during BEC-organoid formation and E2F1 silencing.
(A) Over-representation analysis results. Top 15 enriched biological processes (BP) upon HFD (own data) and DDC (GSE125688) treatment in BECs. q-value: false discovery rate adjusted p-values, counts: number of found genes within a given gene set. (B) Heatmap of E2F1-4 target genes from transcription factor (TF) gene sets. Genes with absolute fold change lower than 0.5 were not shown. FC = fold change. (C) GO over-representation analysis of upregulated biological processes (BP) during the process of organoid formation from single BECs (Organoids vs. T0) (GSE123133). (D) Quantification of E2f1 mRNA in EPCAM+ BECs from livers of C57BL/6 J HFD-fed mice. BECs were isolated and treated with a pool of siRNAs specific for E2f1 or with Scrambled siRNA (control). n=3. (E) Proliferation kinetics over 6 days (expressed as FC over day 0), relative to D. n=15 for Scrambled, n=12 for E2f1. Data are shown as mean ± SEM. *p<0.05; unpaired, two-tailed Student’s t-test (D), and mixed-effect analysis with Sidak’s (E), and were used.
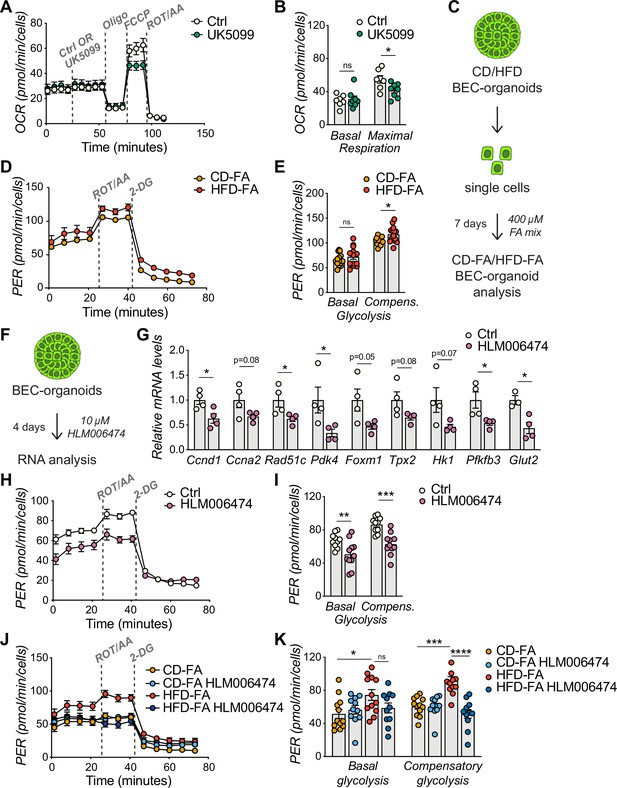
E2Fs promote glycolysis in biliary epithelial cell (BEC)-organoids.
(A–B) Seahorse Substrate Oxidation Assay using UK5099, a mitochondrial pyruvate carrier inhibitor (A), and assessment of the glucose dependency (B) in CD-derived BEC-organoids. n=7 for control (Ctrl), n=8 for UK5099. (C) Scheme depicting the treatment of chow diet (CD)/high-fat diet (HFD)-derived BEC-organoids with fatty acid (FA) mix. (D–E) Proton efflux rate (PER) (D), and basal and compensatory (Compens.) glycolysis (E) were measured using Seahorse XF Glycolytic Rate Assay. Relative to panel C. n=14. (F) Scheme depicting the treatment of CD-derived BEC-organoids with E2F inhibitor, HLM006474. (G) RT-qPCR of selected cell cycle and glycolytic genes, relative to panel F. n=4. (H–I) PER during the Seahorse XF Glycolytic Rate Assay (H), and basal and compensatory (Compens.) glycolysis (I), relative to panel F. n=10 for control (Ctrl), n=11 for HLM006474. (J–K) PER during the Seahorse XF Glycolytic Rate Assay (J), and basal and compensatory glycolysis (K), relative to panel C and treatment with HLM006474. n=12 for CD-FA and HFD-FA, n=11 for HLM006474. Data are shown as mean ± SEM. Absence of stars or ns, not significant (p>0.05); *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001; unpaired, two-tailed Student’s t-test (G), and two-way ANOVA with Sidak’s test (B, E, I, K) were used.

E2F activation correlates with increased glycolysis.
(A) Over-representation analysis results. Top 22 enriched KEGG and EHMN pathways during the process of organoid formation from single biliary epithelial cells (BECs) (Organoids vs. T0) (GSE123133). q-value: false discovery rate adjusted p-values, counts: number of found genes within a given gene set. (B–C) Seahorse Substrate Oxidation Assay using BPTES, n=8 (B), or Etomoxir (C) in CD-derived BEC-organoids. n=7 for control (Ctrl) and n=8 for inhibitor conditions (D–E) Scheme depicting Seahorse Glycolytic Rate Assay (D), and Mito Stress Test (E). (F–G) Oxygen consumption rate (OCR) (F), and basal and maximal respiration (G) were measured using Seahorse XF Mito Stress Test. Relative to Figure 4C. n=14. Data are shown as mean ± SEM. Absence of stars or ns, non-significant (p-value >0.05); ****p<0.0001; two-way ANOVA with Sidak’s test (B, C, G) was used.
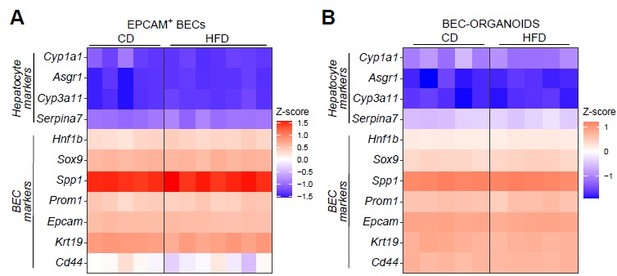
Heatmaps representing the expression of hepatocyte and BEC markers in EPCAM+ BECs isolated from livers of C57BL/6J mice fed with CD or HFD for 15 weeks (A) and BEC-organoids derived from the cells described in A (B).
Heatmaps represent the Z-score calculated as “-1 * Z-score” from the expression data from RNAseq (A) and the δ CT values from RT-qPCR (B), respectively. 36b4 was used as the housekeeping gene..
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-mouse CD45–PE/Cy7 (Rat monoclonal) | BD Biosciences | 552848 | FACS, Flow cytometry (1:100) |
| Antibody | Anti-mouse CD11b–PE/Cy7 (Rat monoclonal) | BD Biosciences | 552850 | FACS, Flow cytometry (1:100) |
| Antibody | Anti-mouse/human CD31–PE/Cy7 (Rat monoclonal) | Abcam | ab46733 | FACS (1:100) |
| Antibody | Anti-mouse CD31–PE/Cy7 (Rat monoclonal) | BD Biosciences | 561410 | FACS, Flow cytometry (1:100) |
| Antibody | Anti-mouse EPCAM–APC (Rat monoclonal) | eBioscience | 17-5791-82 | FACS (1:100) |
| Antibody | Anti-mouse EPCAM–VioBlue (Rat monoclonal) | Miltenyi Biotec | 130-123-871 | Flow cytometry (1:100) |
| Antibody | Anti-Ki67 (Rabbit monoclonal) | ThermoFisher | MA5-14520 | IF (1:100) |
| Antibody | Anti-PANCK (Rabbit polyclonal) | Novusbio | NBP600-579 | IF (1:100) |
| Antibody | Anti-OPN (Goat polyclonal) | R&D Systems | AF808 | IF (1:100) |
| Antibody | Anti-Cleaved caspase 3 (Rabbit polyclonal) | Cell Signaling | 9661 | IF (1:100) |
| Chemical compound, drug | Alexa Fluor Phalloidin 488 | Invitrogen | A12379 | IF (1:1000) |
| Chemical compound, drug | BODIPY 558/568 | Invitrogen | D38D35 | IF (5 µM) FACS (40 nM) |
| Chemical compound, drug | N-acetylcysteine | Sigma-Aldrich | A9165 | (1 mM) |
| Chemical compound, drug | Nicotinamide | Sigma-Aldrich | N0636 | (10 mM) |
| Chemical compound, drug | Y-27632 | Sigma-Aldrich | Y0503 | (10 μM) |
| Chemical compound, drug | HLM006474 | Merck | 324461 | (10 μM) |
| Chemical compound, drug | Oligomycin | Millipore | 495455 | (5 μM) |
| Chemical compound, drug | FCCP | Sigma-Aldrich | C2920 | (2.5 μM) |
| Chemical compound, drug | Rotenone | Sigma-Aldrich | R8875 | (1 μM) |
| Chemical compound, drug | Antimycin A | Sigma-Aldrich | A8674 | (1 μM) |
| Chemical compound, drug | 2-DG | Sigma-Aldrich | D8375 | (50 mM) |
| Chemical compound, drug | UK5099 | Sigma-Aldrich | PZ0160 | (2 μM) |
| Chemical compound, drug | Etomoxir | Sigma-Aldrich | E1905 | (4 μM) |
| Chemical compound, drug | BPTES | Sigma-Aldrich | SML0601 | (3 μM) |
| Commercial assay or kit | Anti-rabbit IgG Polymer Detection Kit (ImmpRESS Horse HRP conjugated secondary) | VectorLabs | MP-74-01-15 | IF (100 µl) per tissue section |
| Commercial assay or kit | Anti-mouse IgG Polymer Detection Kit (ImmpRESS Horse HRP conjugated secondary) | VectorLabs | MP-74-02-15 | IF (100 µl) per tissue section |
| Commercial assay or kit | Click-iT EdU Alexa Fluor 647 | ThermoFisher | Cat. # C10340 | (50 μg per g of mouse weight) |
| Commercial assay or kit | EdU Alexa Fluor 488 | ThermoFisher | C10425 | |
| Commercial assay or kit | MT Cell Viability Assay | Promega | G9711 | |
| Commercial assay or kit | RNeasy micro kit | QIAGEN | 74104 | |
| Commercial assay or kit | RNAqueous total RNA isolation kit | Invitrogen | AM1931 | |
| Commercial assay or kit | QuantiTect Reverse Transcription Kit | Qiagen | 205314 | |
| Commercial assay or kit | SYBR Green | Roche | 4887352001 | |
| Commercial assay or kit | Triglyceride kit | Abcam | ab65336 | |
| Commercial assay or kit | Cell-titer Glo | Promega | G7570 | |
| Commercial assay or kit | Caspase 3/7 activity | Promega | G8091 | |
| Commercial assay or kit | Nucgreen Dead 488 | Invitrogen | R37109 | |
| Peptide, recombinant protein | Gastrin | Sigma-Aldrich | G9145 | (10 nM) |
| Peptide, recombinant protein | EGF | Peprotech | AF-100–15 | (50 ng/ml) |
| Peptide, recombinant protein | FGF10 | Peprotech | 100–26 | (100 ng/ml) |
| Peptide, recombinant protein | HGF | Peprotech | 100–39 | (50 ng/ml) |
| Peptide, recombinant protein | Wnt3a | Peprotech | 315–20 | (1 µg/ml) |
| Strain, strain background (Mus musculus) | C57BL/6JRj | Janvier Labs | C57BL/6JRj | Males, 8-week-old |
| Strain, strain background (Mus musculus) | E2f1+/+ and E2f1-/- | The Jackson Laboratory | (B6;129S4-E2f1tm1 Meg/J) | Males, 8-week-old |
| Software, algorithm | FlowJo | FlowJo | v10.8 | |
| Software, algorithm | Prism | GraphPad | Prism 9 | |
| Software, algorithm | STAR | Dobin et al., 2013 | version 2.6.0 a | |
| Software, algorithm | DESeq2 | Love et al., 2014 | version 1.34.0 | |
| Software, algorithm | clusterProfiler | Yu et al., 2012 | ||
| Software, algorithm | ggplot2 | Wickham, 2016 | ||
| Software, algorithm | QuPath | Bankhead et al., 2017 | version 0.2.3 | |
| Software, algorithm | Fiji | Schindelin et al., 2012 | version 2.3.0 | |
| Other | Chow Diet | SAFE | SAFE 150 | Section ‘Mouse studies and ethical approval’ For C57BL/6JRj |
| Other | High Fat Diet | Research Diets Inc | D12492i | Section ‘Mouse studies and ethical approval’ For C57BL/6JRj |
| Other | Chow Diet | Kliba Nafag | 3336 | Section ‘Mouse studies and ethical approval’ For E2f1+/+ and E2f1-/- |
| Other | High Fat Diet | Envigo | TD93075 | Section ‘Mouse studies and ethical approval’ For E2f1+/+ and E2f1-/- |
| Other | BEC-organoids from mouse | This paper | Original protocol: Broutier et al., 2016 | Section ‘Culture of mouse liver BEC-organoids from single bile duct cells of digested livers’ |
| Other | DMEM/Glutamax | ThermoFisher | 31966–021 | Section ‘EPCAM+ BEC isolation, FACS, and flow cytometry analysis’ |
| Other | Collagenase | Merck/Sigma | C9407 | Section ‘EPCAM+ BEC isolation, FACS, and flow cytometry analysis’ |
| Other | Advanced DMEM/F-12 | GIBCO | 12634010 | Section ‘EPCAM+ BEC isolation, FACS, and flow cytometry analysis’ |
| Other | Glutamax | ThermoFisher | 35050061 | Section ‘EPCAM+ BEC isolation, FACS, and flow cytometry analysis’ |
| Other | HEPES | ThermoFisher | 15630–056 | Section ‘EPCAM+ BEC isolation, FACS, and flow cytometry analysis’ |
| Other | Penicillin/Streptomycin | ThermoFisher | 15140–122 | Section ‘EPCAM+ BEC isolation, FACS, and flow cytometry analysis’ |
| Other | Dispase II | ThermoFisher | 17105–041 | Section ‘EPCAM+ BEC isolation, FACS, and flow cytometry analysis’ |
| Other | DNAase | Merck/Sigma | DN25 | Section ‘EPCAM+ BEC isolation, FACS, and flow cytometry analysis’ |
| Other | E2f1 ON-TARGETplus siRNAs | Horizon | L-044993-00-0005 | Section ‘Silencing of E2F1 in EPCAM+ BECs and organoid formation’ |
| Other | Scrambled ON-TARGETplus siRNAs | Horizon | D-001810-10-05 | Section ‘Silencing of E2F1 in EPCAM+ BECs and organoid formation’ |
| Other | TransIT-X2 | Mirus | MIR6000 | Section ‘Silencing of E2F1 in EPCAM+ BECs and organoid formation’ |
| Other | Matrigel | Corning | 356231 | Section ‘Culture of mouse liver BEC-organoids from single bile duct cells of digested livers’ |
| Other | 1 X B27 | Gibco | 17504044 | Section ‘Culture of mouse liver BEC-organoids from single bile duct cells of digested livers’ |
| Other | TrypLE | GIBCO | 12605028 | Section ‘Culture of mouse liver BEC-organoids from single bile duct cells of digested livers’ |
| Other | Palmitic acid | Sigma | P0500 | Section ‘Culture of mouse liver BEC-organoids from single bile duct cells of digested livers’ |
| Other | Oleic acid | Sigma | O1008 | Section ‘Culture of mouse liver BEC-organoids from single bile duct cells of digested livers’ |
| Other | DAPI | ThermoFisher | 62248 | Section ‘Liver immunohistochemistry (IHC) and immunofluorescence (IF)’ |
| Other | Cell Recovery Solution | Corning | 354253 | Section ‘BEC-organoid whole-mount immunofluorescence’ |
| Other | Glucose | Agilent | 103577–100 | Section ‘Bioenergetics with Seahorse extracellular flux analyzer’ |
| Other | Pyruvate | Gibco | 11360070 | Section ‘Bioenergetics with Seahorse extracellular flux analyzer’ |
| Other | Glutamine | Agilent | 103579–100 | Section ‘Bioenergetics with Seahorse extracellular flux analyzer’ |
| Other | Hoechst | ThermoFisher | 62249 | Section ‘Bioenergetics with Seahorse extracellular flux analyzer’ |
Additional files
-
Supplementary file 1
Differential expression analysis results of EPCAM+ BECs upon HFD.
Related to Figure 2.
- https://cdn.elifesciences.org/articles/81926/elife-81926-supp1-v2.xlsx
-
Supplementary file 2
Over-representation analysis of upregulated and downregulated genes in HFD, DDC, and BEC-organoid formation datasets.
Related to Figure 3.
- https://cdn.elifesciences.org/articles/81926/elife-81926-supp2-v2.xlsx
-
Supplementary file 3
Primers used for qPCR analysis.
- https://cdn.elifesciences.org/articles/81926/elife-81926-supp3-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/81926/elife-81926-mdarchecklist1-v2.docx





