Oligodendrocyte-mediated myelin plasticity and its role in neural synchronization
Figures
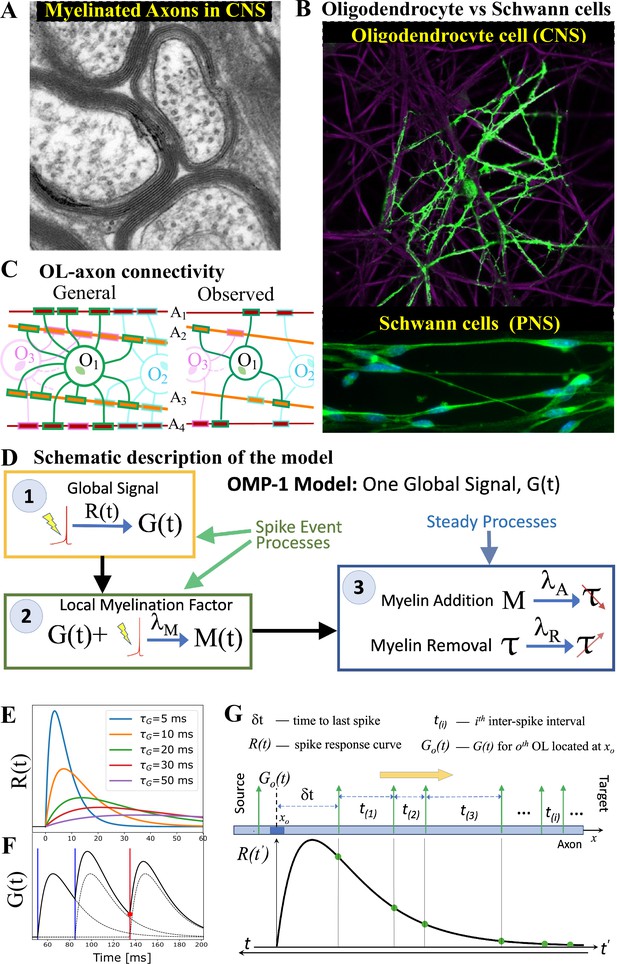
OMP motivation and model description.
(A) Cross-section of myelinated axon bundle in CNS illustrates that myelin thickness on adjacent axons differs. (B) Single OL (green) myelinates many different axons (purple), which is in stark contrast to Schwann cells in the PNS, which myelinate only one axon. (C) OL-axon connectivity: OLs tend to avoid placing multiple processes on a single axon, thus maximizing the number of axons it myelinates. (D) Schematic depiction of the OMP-1 model which contains only the three basic steps required for this type of OMP to work. (E) Spike-response curves for increasing values of . (F) The equation governing the release of is linear and the response to multiple spikes (vertical lines), is the linear sum of individual responses (dashed lines). The release of after each spike at any given OL process/axon will be proportional to the amplitude of at the time of spike (red dot for red spike). (G) The sum over responses can also be viewed as sampling of in reverse time. Panels A and B have been adapted from Chapter 45 Figure 1 in Fields, 2013, used with permission.

OMP-1 model vs OMP-n model.
A more complex OMP models can be constructed; however their structure differs from the simplest OMP-1 model only in step 2, where multiple global factors can participate, together with a cascade of events, which can be triggered by given threshold events, each potential having their own response functions, . Ultimately, the whole cascade will lead to the production of the myelination-promoting factor , as in the case of OMP-1.

Time progression of OMP fast and slow variables.
Time progression of OMP variables when correlated Poisson spikes are conducted through OC with a single OL, , and for axons. The ordering of the spikes carries statistically the signature of the fixed delays. The OMP parameters for this simulation are ms, ms, ms-1, ms, ms-1, and ms-1. (A) A millisecond time-scale dynamics of and , in response to incoming spikes (yellow vertical lines). For clarity, only five of the total axons are shown. Time reported in milliseconds is just an offset from the beginning of the last second epoch. (B) Time progression of (top row) and OMP model variables, as indicated, over seconds, plotted as the averages over second epochs, and different columns showing different values of , as indicated. The second row from the top shows the 10 delays for each axon, for a single trial, combined with the fixed delays (showing the convergence of the arrival times). The third row shows the time evolution of the myelin removal rate, , guided by the homeostatic equation (Equation 17) with black and red dashed lines indicating the simple homeostatic balance estimate () and twice that value, respectively. Differently colored trajectories are the results from three independent trials, with the same parameters but different random initializations of fixed and adaptive delays ( ms). The bottom row shows the concentrations of for each axon in a single trial. For the relatively large, 30 ms, differences in concentrations are not as drastic as they are for 10 ms (see Figure 2—figure supplement 1).
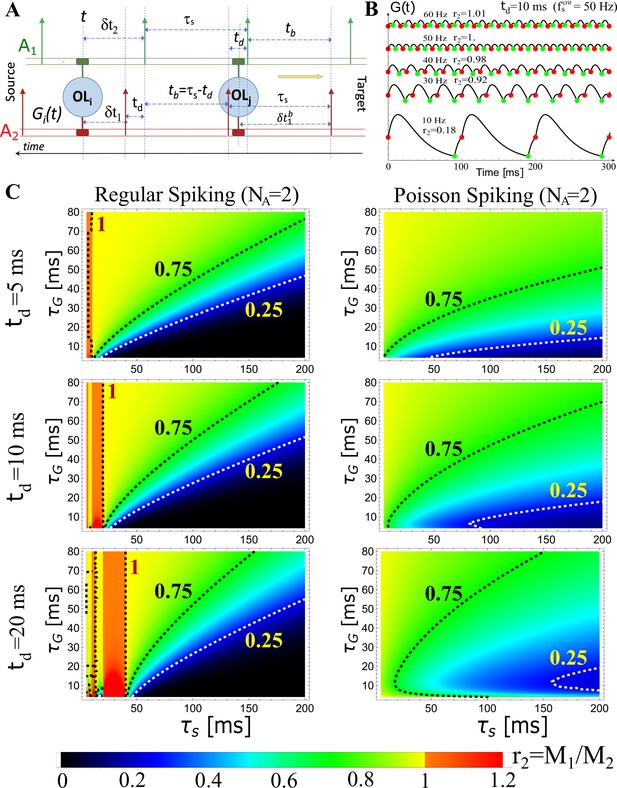
Theoretical predictions of OMP model.
Theoretical predictions for for . (A) Schematic depiction of the temporal relationship between spikes in a pair of axons (; ) in the case of regular spiking, illustrating different scenarios when calculating . The ‘leading’ edge spikes (green) are offset by a fixed delay, td, over the ‘lagging’ spikes (red). (B) Examples of for ms and for five different spiking frequencies Hz, as indicated, showing also the corresponding ratios, . Occurrences of ‘red’ and ‘green’ spikes are plotted together with , indicating also the level of released, as it is proportional to . We see that up to the critical frequency, Hz, the lagging axon will have more myelination factor, , as desired, but this reverses above the critical frequency. (C) Theoretical predictions for the ratio, , as a function of and for three different fixed time-delays, 5, 10, and 20 ms (rows, as indicated), in the case of regular (left column) and Poisson (right column) spiking. The regions with red/orange hues are the regions of instability, where, which will lead to further desynchronization, and is only happening for regular spiking. For Poisson spiking, the ratio is always lt1, as is evident from Equation 3, hence, the ‘lagging’ axon will always have enhanced myelination relative to the ‘leading’ axon, due to a higher concentration of, which is desired for stable synchronization in all regimes.

Theoretical predictions vs simulations.
Comparing theoretical predictions with simulations (). The mean values obtained in simulations (y-axis) are compared to the theoretical prediction (x-axis) given by Equation 4, which ignores jitter. (A) The mean values are from a simulation sets specified in Appendix 1—table 4, for which correlated Poisson spiking was used. The results are shown as a grid plot for different values of jitter and characteristic spike response time, . Individual points from the same parameter set are colored the same, with different parameter sets/runs being colored by cycling through 15 different colors with no particular meaning attached to the coloration. Each set had 2, 5, or 10 points (one per axon). We see that the predictions made for case hold very well for small jitter (), but fail for larger jitters, particularly for ms, making the differences in across axons smaller. Nevertheless, even for large jitter, when is large enough, the prediction given by Equation 4 is in reasonable agreement with the observed/simulated values. (B) Same as in (A), but this time spikes were simulated using a refractory Poisson process with and ms. Because now it violates the assumption of a memoryless process, under which the derivation of Equation 4 is made, the predictions fail for any amount of jitter or . We note that theoretical predictions for a general renewal process with any amount of jitter are outside the scope of the current manuscript.
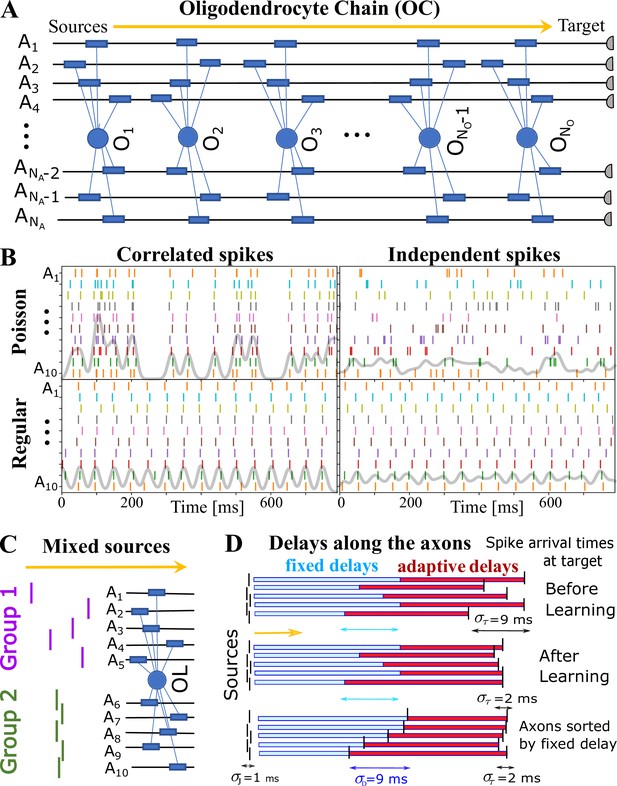
OMP simulations with action potentials/spikes conducted along ‘oligodendrocyte’ chain (OC).
(A) OC with ‘effective oligodendrocytes’ (segments), myelinating axons ( is an matrix of ones). (B) examples of ‘pure’ spikes used in our simulations: correlated versus independent spikes, and Poisson versus regular spiking. Gray lines are moving averages of all spikes using a sliding Gaussian window with 10 ms RMS width. (C) example of ‘mixed’ signals in which axons in Group 1 are conducting independent spikes and those in Group 2 carry correlated spikes. The two groups can potentially interfere with the expected synchronization behavior of each ‘pure’ group. Different groups could also contain spikes time-locked within each group, but independent between the groups. (D) schematic depiction of time delays between the signal source and the target. Fixed delays are not modifiable and essentially represent the spread in spike times as they enter the axonal bundle of myelinated axons, whose delays are adaptive. Note, that the horizontal bars represent the magnitude of the fixed and adaptive delays, not the axons.
Time progression of OMP variables for correlated spikes.
Time progression of the slow variables in the OMP model, similar to one shown in Figure 1—figure supplement 2B, but now with faster spike responses in the OL, ms, and for different (values shown in columns), using only one myelin production rate, , and ms. The independent trials, each have with their own set of randomly chosen fixed delays ( ms). The top row shows the time progression of for each trial (dotted red line is the mean over all trials). The second row are the 10 delays, , for each axon, and the third row are the same delays when combined with the fixed delays (showing the convergence of the arrival times). The fourth row shows the time evolution of , with black and red dashed lines indicating values (the balancing estimate for independent spikes) and twice that value, respectively. The bottom row shows the concentrations of for each axon.

Time progression of OMP variables for independent spikes.
Time progression of the slow variables (averages over s epochs) in the OMP model when the spikes are independent with Poisson ISI. Here, the columns show three independent trials with the same set of OMP parameters but each with its own set of randomly chosen fixed delays ( ms). The OMP parameters for this simulation are ms, ms, , ms, and axons. The top row shows the time progression of for each trial (dotted red line is the mean over all trials). The second row are the 10 axonal delays, and the third row are the same delays when combined with the fixed delays (showing the delay profile that will be fed into the next OL segment in the chain). The fourth row shows the time evolution of , with black and red dashed lines indicating values (the balancing estimate for independent spikes) and twice that value, respectively. The bottom row displays the concentrations of for each axon.
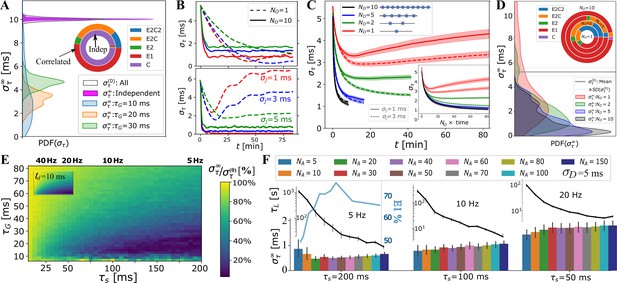
OMP model behavior as a function of OMP and signal parameters.
(A) When spikes are independent, consistently show no change in overall synchronization (purple; innermost circle in the model selection pie-chart); for correlated signals significant synchronization occurs, largely depending on and . (B) individual OMP synchronization profiles, , simulations for , , , with ms (top panel) vs. ms (bottom). Dashed lines are for and solid lines for . Colors indicate the jitter level: ms (red), ms (blue), ms (green). (C) dependence of on the number of OLs, , in the OC. We show both the comparison based on the raw time, as well as when matched in terms of the total number of neural spikes encountered by the OLs (inset). (D) density of estimates dependence of on the number of OL in OC. (E) Percent reduction in vs and for ms; (inset) ratio of the concentrations for two axon case, , which matches well the overall pattern of synchronization. (F) dependence of (bar plot) and the synchronization time constant, , (semi-log plot, above) on the number of axons, values are only for runs declared as E1 blue curve inset indicates percent of E1, for different (for all spike rates).
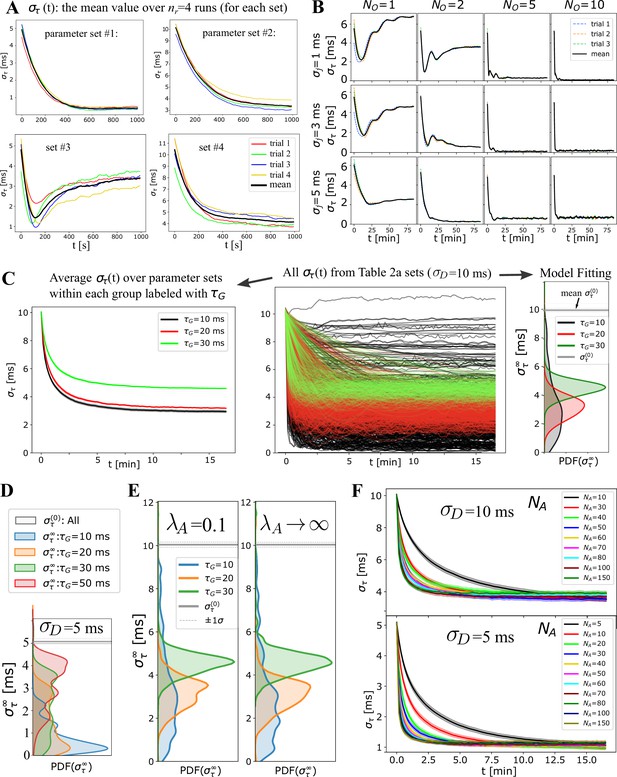
Exploring OMP properties and parameters.
Exploring OMP with a large number of parameter settings. (A) For any given setting of OMP parameters, simulations are repeated nr times each with a random choice of fixed delays (, parameterized with ) and initial adaptive delays (parameterized with and ). These individual temporal profiles/trials (colored lines) are averaged and the mean is reported as the synchronization profile, (that we refer to as a set or run). (B) A grid plot of for correlated spikes using 12 parameter runs (3 trials and the mean), revealing its dependence on and (4 and 3 values, for = 10, ms, , ). Some of these profiles are shown in Figure 3B to illustrate OMP instabilities with small and fixed delays, which are overcome with increasing and . (C) The center panel shows all runs specified in the left panel of Appendix 1—table 1. We summarize such large number of runs in two ways: (1) we average all within a subset of runs grouped by some parameter, , as shown in the left panel; (2) we also fit all to 5 models, as described in Materials and Methods, and plot the distribution of as shown in the right panel. In some cases we also plot the distribution of parameters. (D) Figure 3A results with ms are shown here for ms and also (E) compared to the equivalent runs using the instantaneous myelination OMP model. We used a smaller bandwidth for kernel density estimation to highlight the differences, which are not apparent at smoother KDE values. (F) Summary of average profiles grouped by , shown in Figure 3F for ms, and additionally for ms.
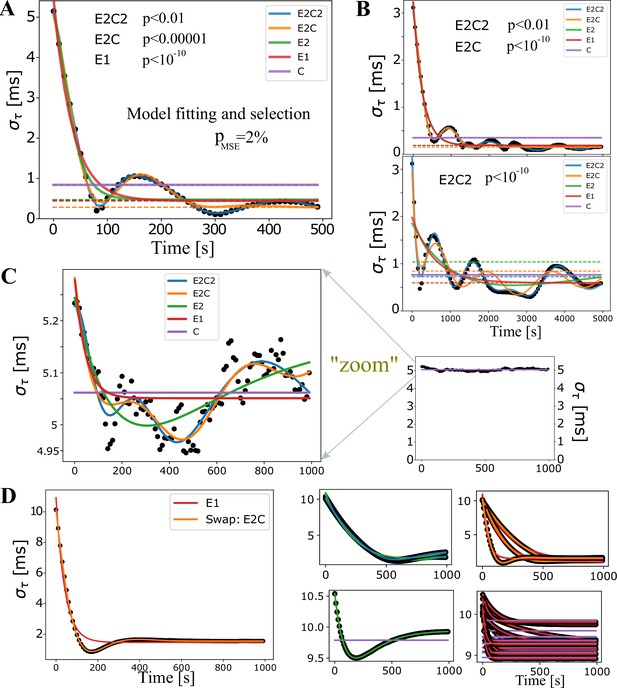
Model fitting for synchronization profiles.
Model fits for quantifying synchronization performance across different parameter runs.
(A) An example of fits for all five nested models. Dashed lines represent estimates of the long-time baseline, . (B) In most cases, comparing the dashed lines, the model selection does not affect the estimate of the baseline (top panel), but can sometimes have a significant impact. For example, in the bottom panel, the baselines differ by nearly 50%. For such a complex synchronization profile it is actually hard to determine the true long-time baseline and the only way to resolve those situations is to run really long simulations, which we could not afford in this early and extensive exploratory phase. (C) In the majority of runs, the less restricted models were deemed the best with the standard F-test. When magnified, it is clear that E2C2 offers a statistically better description than the constant model, which we label (C). On the other hand, when considered at the full scale (small graph on the right), we desire to classify such a profile simply as the constant model, (C). This is achieved by introducing a fudge factor, %, which under our modified F-test selects the model C. (D) We also introduced a ‘swapping’ rule. This rule reverts the fit back to the less restricted model when the true MSE in the restricted model is 500 times larger than the one for the restricted model. This did not seem to be consequential, as in most simulations very few swaps occurred, and when they occurred seemed to be justified, as shown in the large panel on the left, where the E1 model suggestion, given by the modified F-test, was reverted to a double exponential fit. The four panels on the right show all swaps obtained with runs specified in Oligodendrocyte-mediated Myelin Plasticity and its Role in Neural Synchronization Appendix 1—table 3, which was the case that had most of the swaps, and most of those were swaps from C to E1 (lower right panel) which had a very small range. Hence, the swap did not change significantly but had a large effect on the value of , which is hence less reliable in our summary statistics.
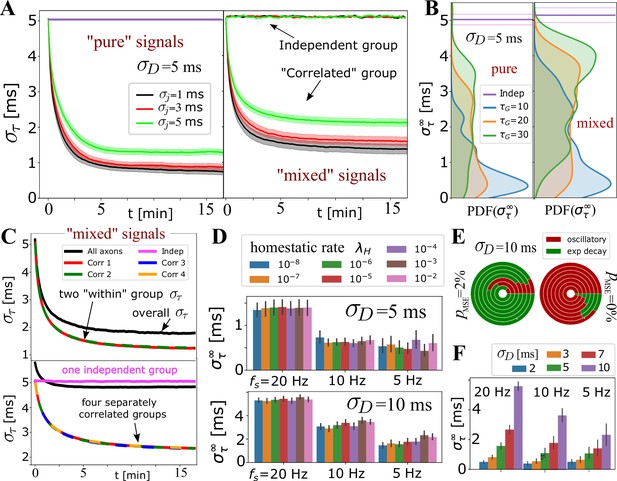
Selective synchronization with mixed signals; effects of and parameters.
(A) Average (grouped by ; shaded regions indicate SE over the parameter sets for and ; see Appendix 1—table 2). In the left panel is the average for the set of axons conducting only ‘pure’ correlated signals, and on the right are the averages within two groups of 10 axons, out of total that OL myelinates, carrying ‘mixed’ signals – one group conducting correlated signals and the other independent signals. The behavior of the correlated groups is clearly distinguishable from the independent ones (indicated by arrows). (B) Estimated density for comparing pure (left) vs mixed signals (right panel) for a wider range of parameters, including and , but with the number of axons carrying correlated signals matched in all cases (, ). (C) Top panel shows for two groups of correlated signals (colored lines) with Poisson spiking (correlated within group, but mutually independent); bottom panel shows results for four separately correlated groups + 1 independent group (cyan). Black lines represent the spread over all axons. (D) Investigating the influence of homeostatic regulation and homeostatic rate, , grouped by different firing rates and different . (E) Model selection chart depicting the relative proportions of oscillatory (E2C2, E2C) vs stable, single exponential (E1) , for two different values of the F-test fudge-factor ; the innermost circle is for and the outermost for . (F) OMP synchronization performance for different magnitudes of fixed delays, .

Supplemental results for Figure 4.
(A) Individual runs for the averages reported in Figure 4A for mixed signals, one conducting correlated spikes and the other independent spikes. Individual runs for independent spikes here are manually offset for clarity, not to overlap with profiles obtained with the correlated spikes, but are mainly shown to demonstrate that the actual spread in the performance is much larger than what the shaded standard error (SE) indicates for the independent group, appearing only as lines when averaging over many runs (and drawn as a ‘thick’ purple line for pure independent signals in Figure 4A, where the SE was even smaller). (B) Influence of on synchronization efficiency. (C–D) Averages over all runs with ms, grouped by jitter, comparing (C) pure signals with (D) mixed signals with one independent and one correlated group, as was similarly shown in Figure 4A for ms.
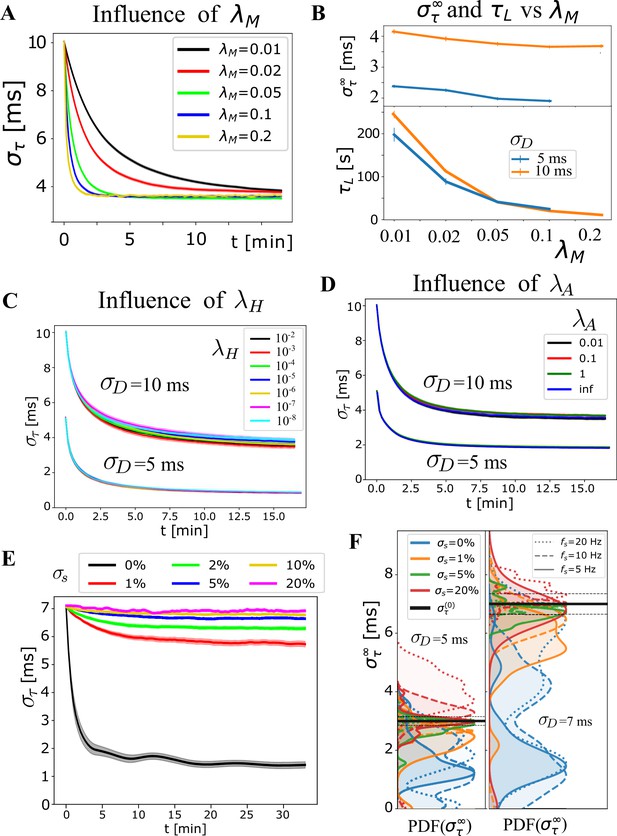
Influence of additional OMP and signal parameters.
Exploring additional OMP and signal parameters. (A) Average , grouped by , which highly influences the learning rate, as it sets the time scale for changes in myelination. (B) Bottom panel shows this explicitly via ‘learning time’, , as a function of for 5 and 10 ms. The dependence of on (top panel) was not strong and it appears to slightly favor faster rates. This ‘slight’ trend does not appear consistently and depends on the range of parameters chosen for averaging. (C) The average for and ms, respectively, show that does not have a strong influence on the behavior of the OMP model (also emphasized in Figure 4E). (D) Average for and ms now grouped by the myelin conversion rate, , including the instantaneous myelination model, Equation 15, and showing that has a very small influence on synchronization. This might change when the saturation limits , become narrower, as well when different values in Equation 17 are used. We use saturation limits that allow highly heterogeneous values for the delays (3 ms to 100 ms), making it harder to achieve synchronization. (E) The sensitivity of OMP to firing rate variability, specified via in the case of regular spiking, having ‘beating’ synchronized periods, but also even longer non-synchronized periods, potentially detrimental to the synchrony. Averages, grouped by , indicate that when the spiking-rate variability is smaller than 5%, synchronization still occurs but is very inefficient. This might not be a problem since myelin plasticity operates on a very slow time scale, measured in days and months, but only if does not get worse during non-synchronized epochs. (F) The sensitivity to quantified via , showing that variations greater than 5% exhibit de-synchronization effects but these might not arise for Poisson spiking.
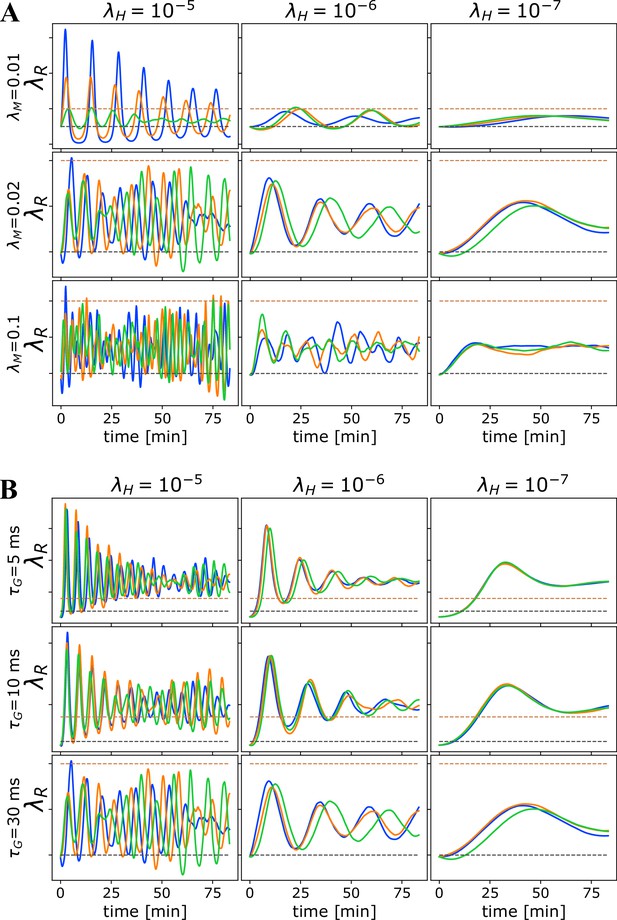
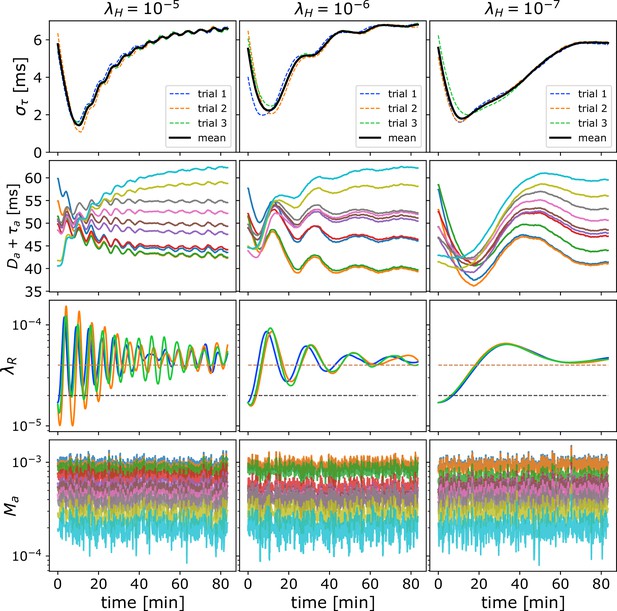
Oscillations due to homeostatic regulation.
Oscillations in due to homeostatic regulation. The theoretical guess for the balance between addition and removal of myelin, that we initialize to at the start of each simulation, should work well for independent spikes, if saturation effects can be ignored. However, in most cases, this prediction does not match the true balancing value for , and hence for fast homeostatic rates, the approach to equilibrium will be too fast and will induce oscillations. (A) Time progression of in the OMP model for three different values of (columns) and three different values of when a set of correlated spikes with exponential ISI (Poisson) are conducted through = 10 axons. For this set of runs , , and . (B) Time progression of in the OMP model for three different values of (columns) and three different values of when a set of correlated spikes with exponential ISI (Poisson) are conducted through = 10 axons. For this set of runs, , and . The spike-response time, , does not influence the oscillation frequency of as strongly as .
Tables
List of symbols used in the manuscript: OMP model parameters, OMP variables, spiking signal parameters, and data analysis and quantification parameters and other symbols.
For each we provide a short description and the range of the parameter values, or its dimensionality, in the case of the variables.
| List of symbols used in this manuscript | ||
|---|---|---|
| Symbol | Description | Explored Values/Dimensionality |
| OMP model parameters | ||
| OL transient response curve | see | |
| characteristic time for () | [2-100] ms | |
| production rate for factor | [0.01, 0.5] ms-1 | |
| myelin conversion/addition rate | [0.01, 0.5] ms-1 | |
| myelin removal rate | Equation 16; variable for | |
| homeostatic rate | [0, 10-2] ms-2 | |
| number of OL in OC | [1-20] | |
| number of axons | [2-150] | |
| myelination matrix | : {full connectivity} | |
| minimal delay attainable on axons | 3 ms | |
| maximal delay attainable on axons | 100 ms | |
| nominal/homeostatic delay | [10-90] ms | |
| initial adaptive delays parameter | ||
| percent spread of initial adaptive delays | 5% | |
| OMP model variables | ||
| global OL signal (Equation 8) | variables | |
| concentration of on axon (Equation 11) | variables | |
| conduction delay(s) for axon (Equation 12) | or variables | |
| for OC segment (Equation 12) | variables | |
| myelin removal rate (Equation 17) | OMP parameter when | |
| Spiking signal parameters | ||
| inter-spike time interval parameter | [10-250] ms | |
| mean firing rate | [4-100] Hz | |
| refractory period in refractory Poisson process | [0–100] ms | |
| amount of jitter given to spikes | [0–10] ms | |
| SD for fixed delays | [0–20 ] ms | |
| percent variability in firing rates | [0–20] % | |
| total duration of simulation | [10 min - 10 hrs ] | |
| number of recorded epochs during simulation | [20–500 ] | |
| duration of each recorded epochs | [10 sec - 5 min] | |
| number of replicated simulations/trials | [3 - 10] | |
| Quantification parameters/other symbols | ||
| OC spread SD () | evaluated after each epoch | |
| synchronization profile during OMP learning | values for ne epochs | |
| initial spread before learning, | determined by , , and | |
| long-time baseline, | estimated via model fitting | |
| characteristic time for synchronization | estimated via model fitting | |
| learning/synchronization rate | = | |
| generic name for fixed delays | NA | |
| pre-specified/normalized fixed delay on axon | random values, | |
| ISI for the interval | NA | |
| probability density function of ISI | NA | |
Parameter sets used in our OMP model simulations.
Parameter sets in the table on the left were used for exploring the general OMP model behavior with both, correlated and independent spikes, yielding runs. Simulations with ms were used to create Figure 3A, while other runs from this set are used in Figure 3—figure supplement 1 and Figure 4—figure supplement 2. The parameter sets on the right () were used in Figure 3E, in which we comprehensively examined the two most important parameters, and .
| Parameter | Values | Parameter | Values | |
|---|---|---|---|---|
| 0.01, 0.02, 0.05, 0.1, 0.2 | 0.02, 0.05 | |||
| 0.01, 0.1, 1 | 0.01 | |||
| 10-5, 10-6 | 4, 6, 8, …, 82 | |||
| 10, 25 | 10 | |||
| 5, 10 | 5, 10, 15, … 195, 200 | |||
| 25, 50, 100, 200 | 1 | |||
| nr | 5 | nr | 10 | |
| Te | 60 |
Parameter sets used in our OMP model simulations.
On the left are parameter sets used to create Figure 3B, C and D exploring the influence of the number of OL, , in the OC ( runs). On the right are parameter sets exploring mixed signals shown in Figure 4A, B and C, which were repeated in three scenarios: (1) two equally sized groups of axons, one carrying correlated and the other independent spikes, (2) two equal and separately correlated groups, and (3) one independent and four equal separately correlated groups. Scenario 1 was used in Figure 4A and B, and scenarios 2 and 3 are shown in Figure 4C. Two additional matched sets of runs are performed for "pure" correlated or independent spikes, matched in terms of within a group, yielding a total of runs.
| Parameter | Values | Parameter | Values | |
|---|---|---|---|---|
| 0.01, 0.1, 1 | 20, 50 | |||
| 1, 2, 5, 10 | 5, 10 | |||
| 5 | 25, 50, 100, 200 | |||
| 0, 30 | nr | 5 | ||
| 25, 50, 100, 200 | ||||
| ne | 500/ |
Parameter sets used in our OMP model simulations.
On the left are parameter sets used to explore the influence of homeostatic rate () shown in Figure 4D and E and Figure 4—figure supplement 2. Parameters used in Figure 4—figure supplement 2E, F, exploring the influence of the spiking rate variability on synchronization ().
| Parameter | Values | Parameter | Values | |
|---|---|---|---|---|
| 0.01, 0.02, 0.05, 0.1, 0.2 | 0.02, 0.05, 0.1 | |||
| 10−2, 10−3, … , 10−7, 10−8 | 0.1 | |||
| 10, 20 | 10, 20 | |||
| 10 | 10, 20, 30, 50 | |||
| 1, 3 | 5, 3, 7 | |||
| nr | 5 | 1 | ||
| 0, 1, 2, 5, 10, 20 | ||||
| 20 |
Two parameter sets for simulations used in Figure 1—figure supplement 4A (left) and Figure 1—figure supplement 4B (right), comparing theoretical predictions for the pure Poisson synchronized spikes with to the values obtained in simulations with non-zero .
For the table on the left, with ms there were runs and for the one on the right, with refractory Poisson spikes ( and 80 ms) there were runs.
| Parameter | Values | Parameter | Values | |
|---|---|---|---|---|
| 0.01, 0.1 | 0.01, 0.1 | |||
| 0.1, 0.01, 0.001 | 10-7 | |||
| 10-7 | 5, 10, 30 | |||
| 2, 5, 10 | 5 | |||
| 1, 5, 10 | 30, 80 | |||
| 5, 10, 30 | 100, 200 | |||
| 0.1, 1, 3, 5 | 0.1, 1, 3, 5 | |||
| nr | 3 | nr | 2 | |
| ne | 1000 | ne | 1000 | |
| 20 | 5 |





