Brown adipocytes local response to thyroid hormone is required for adaptive thermogenesis in adult male mice
Figures
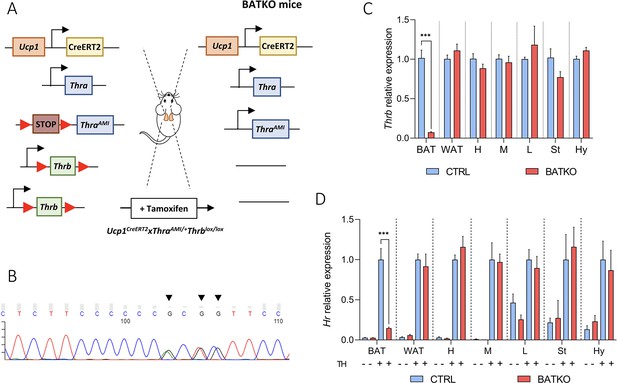
BATKO mice present a brown adipose tissue (BAT)-specific blockade of T3 signaling.
(A) Schematic representation of the BATKO mice. BATKO mice carry the Ucp1CreERT2 transgene, allowing the brown-adipocyte-specific expression of the tamoxifen-sensitive CreERT2 recombinase. BATKO mice are also heterozygous for the ThraAMI allele, which encodes the TRα1L400R dominant-negative receptor after Cre-mediated deletion of a STOP cassette flanked by loxP sequences. BATKO mice are homozygous for the Thrblox allele in which exon 3 is flanked by two tandem-arranged loxP sequences. After tamoxifen injection, Cre-mediated recombination selectively excise the loxP-flanked sequences in brown adipocytes, resulting in the expression of TRα1L400R and elimination of TRβ. Control mice (CTRL, not represented here) had the same genotype except for the absence of the Ucp1CreERT2 transgene and were also tamoxifen treated. (B) Sanger sequencing chromatogram of a fragment of Thra cDNA prepared from BAT RNA of BATKO mice. Arrows indicate the positions of the TRα1L400R mutations. (C) Relative mRNA expression of Thrb in different peripheral and central tissues of CTRL and BATKO mice (H: heart, M: muscle, L: liver, St: striatum, Hy: hypothalamus) (n = 5–6/group). (D) Evaluation of T3 response in propylthiouracil (PTU)-fed CTRL and BATKO mice through the induction of Hr, a well characterized TR target gene, after 24 hr of TH (+ or −; n = 5–7/group). Statistical significance is shown for the comparison of CTRL and BATKO mice treated with TH. Error bars represent the standard deviation (SD). ***p < 0.001 for the indicated comparisons.
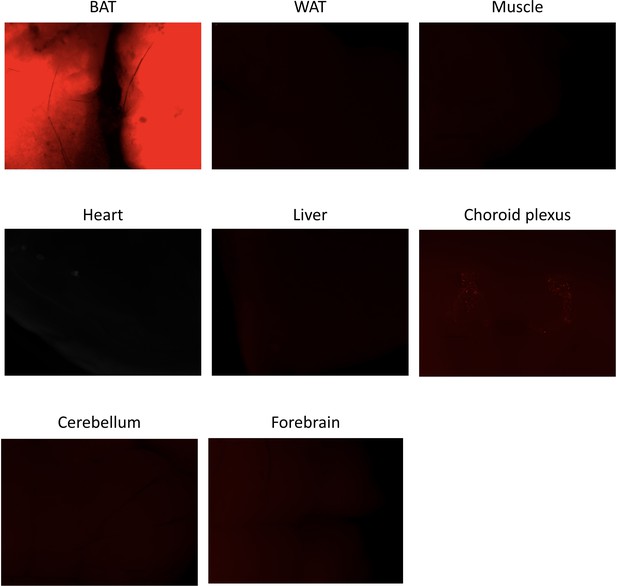
CRE activity in peripheral and central tissues.
Ucp1CreERT2:Rosa26TdTomato reporter mice were dissected and their tissues observed under a Leica M205FA fluorescent stereomicroscope. Results are shown for one mouse.
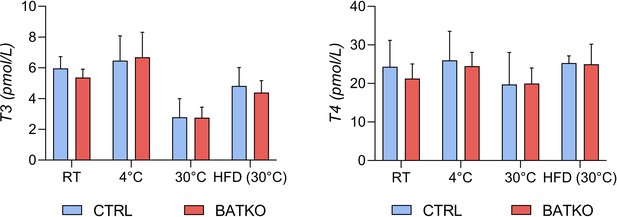
Serum concentration of TH in CTRL and BATKO mice.
Serum concentration (in pmol/l) of free T3 (left panel) and free T4 (right panel) in CTRL and BATKO mice in different experimental conditions (RT: room temperature) (n = 4–5/group).
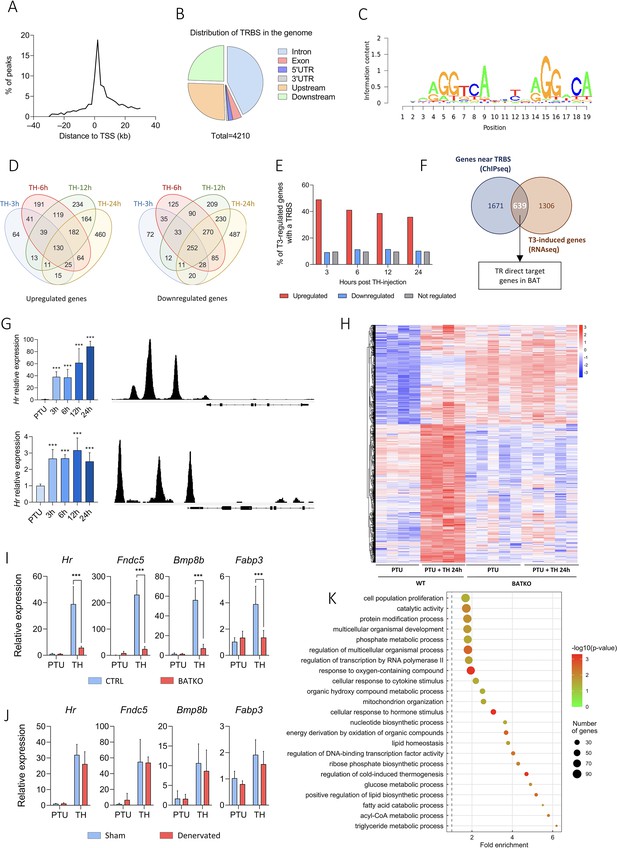
Identification of T3/TR target genes in brown adipocytes.
(A) Consensus sequence found in brown adipose tissue (BAT) TRBS (thyroid hormone receptor-binding site), as identified by de novo motif search. (B) Frequency of TRBS distribution around transcription start sites (TSSs). (C) Pie chart of the 4210 TRBS distribution in the genome (UTR: untranslated region). (D) Venn diagrams of upregulated (left panel) and downregulated (right panel) genes after TH intraperitoneal injection in wild-type propylthiouracil (PTU)-treated mice for different periods. (E) Percentage of genes which possess a TRBS within 30 kb of their TSS among genes whose expression in the BAT is regulated or not by T3 (upregulated in red, downregulated in blue, not regulated in gray). (F) Venn diagram of genes whose expression is induced by T3 in at least one of the time points (in brown, RNAseq data) and genes with a TRBS within 30 kb of their TSS (in blue, chromatin immunoprecipitation sequencing [ChIPseq] data), that is, TR direct targer genes. (G) Left: Time-course analysis of Hr (top) and Ucp1 (bottom) expression in BAT after 24 hr of TH treatment of wild-type hypothyroid mice (RT-qPCR). Statistical significance is shown for the different time points versus untreated PTU-fed mice (n = 4–6/group). Right: Extract of the TRBS in the Mus musculus genome browser around Hr (top) and Ucp1 (bottom). (H) Heatmap representing in both CTRL and BATKO mice the expression of TR direct target genes upregulated after 24 hr of TH injection in CTRL hypothyroid mice. Colors represent the z-scores, see scale besides the heatmap (n = 4–5/group). Relative expression of several TR direct target genes 24 hr after TH treatment in (I) CTRL/BATKO and (J) Sham/denervated PTU-fed C57BL6/J mice. Statistical significance is shown for the comparisons between CTRL-TH and BATKO-TH, or SHAM-TH and BATKO-TH mice (n = 5–7/group). (K) Gene ontology dot plot of the 639 TR direct target genes. Only the ‘biological processes’ terms with a fold-enrichment >1.5 were kept. Some of the terms were shortened to increase readability without affecting the meaning. Error bars represent the standard deviation (SD). ***p < 0.001 for the indicated comparisons.
-
Figure 2—source data 1
Raw data and compilation of T3/TR target genes in brown adipocytes.
- https://cdn.elifesciences.org/articles/81996/elife-81996-fig2-data1-v2.xlsx
-
Figure 2—source data 2
Raw western blots for brown adipose tissue (BAT) denervation confirmation.
Each lane represents a different sample, from sham mice (+) or denervated mice (−).
- https://cdn.elifesciences.org/articles/81996/elife-81996-fig2-data2-v2.zip
-
Figure 2—source data 3
List of T3/TR target genes that also include PGC1α-binding sites.
- https://cdn.elifesciences.org/articles/81996/elife-81996-fig2-data3-v2.xlsx
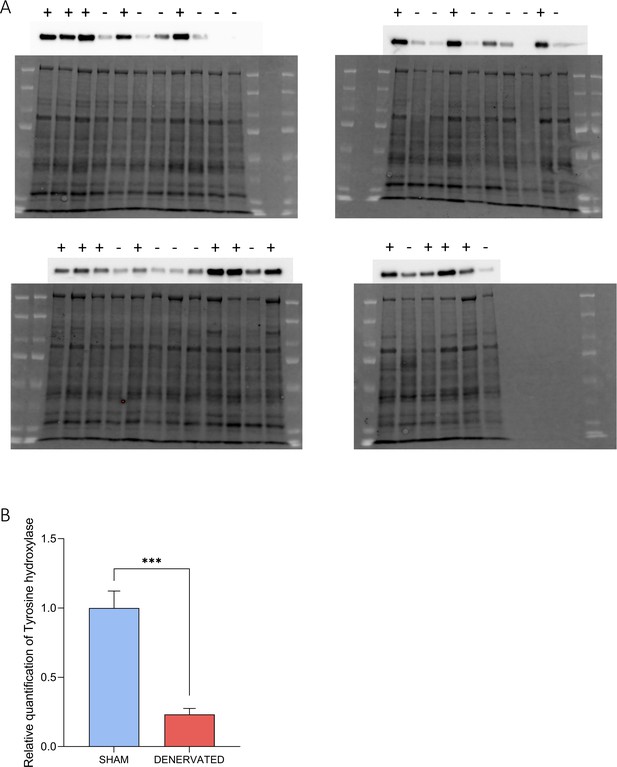
Confirmation of brown adipose tissue (BAT) denervation by quantification of tyrosine hydroxylase.
(A) Western blots with total protein (bottom of the pictures) and tyrosine hydroxylase (top of the pictures) extracted from BAT. Each lane represents a different sample, from sham mice (+) or denervated mice (−). (B) Relative quantification of tyrosine hydroxylase protein in the BAT of sham and denervated mice. Quantity of tyrosine hydroxylase is relative to the quantity of total proteins. Error bars represent the standard error of the mean (SEM). ***p < 0.001 versus SHAM mice.
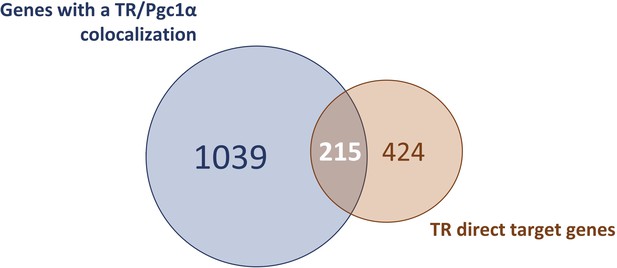
Comparison of TR direct target genes with the genes presenting a PGC1⍺-binding site.
Venn diagram designed to highlight TR direct target genes that include a PGC1⍺-binding site.
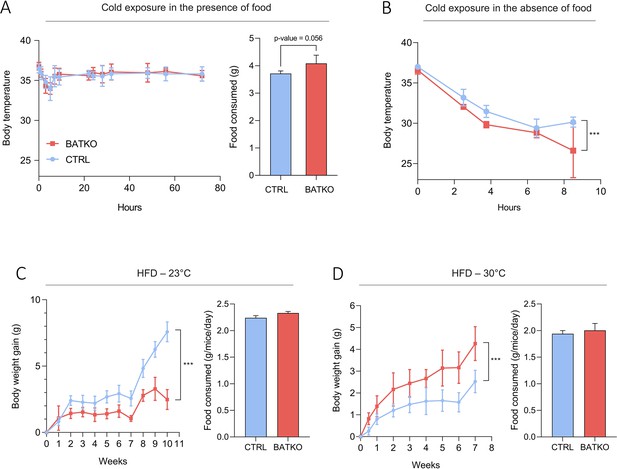
BATKO mice are cold sensitive.
(A) Core body temperature (left) and food consumption (right, over 48 hr) of CTRL and BATKO mice exposed to 4°C in the presence of food (n = 5–7/group). BATKO mice tended to eat more than CTRL mice. (B) Core body temperature in CTRL and BATKO mice exposed to 4°C in the absence of food. After 8 hr of cold exposure, BATKO mice reached severe hypothermia and the experiment was stopped (n = 4–5/group). Body weight gain and food consumption of CTRL and BATKO mice at room temperature (C, n = 4–5/group) or 30°C (D, n = 5–8/group). Error bars represent the standard error of the mean. ***p < 0.001.

Body composition and indirect calorimetry in CTRL and BATKO mice.
(A) Body composition of BATKO and CTRL mice measured by nuclear magnetic resonance (n = 7–9/group). (B) VCO2 (ml/hr/kg), (C) VO2 (ml/hr/kg), (D) metabolic rate, (E) respiratory quotient (ratio of VO2/VCO2), (F) cumulative food consumed by CTRL and BATKO mice, as monitored during 48 hr with 12 hr light/dark cyles (dark phases are represented by gray areas). Error bars represent the standard error of the mean (SEM). ***p<0.001.
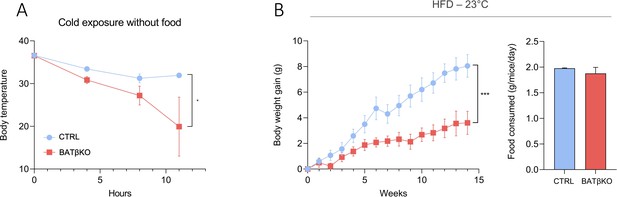
BATβKO mice are also cold sensitive.
(A) Core body temperature in CTRL and BATβKO mice exposed to 4°C in the absence of food. After 11 hr of cold exposure, BATβKO mice reached severe hypothermia and the experiment was stopped (n = 6/group). (B) Body weight gain and food consumption of CTRL and BATβKO mice at room temperature (n = 4–6/group). Error bars represent the standard error of the mean. *p<0.05 and ***p < 0.001.
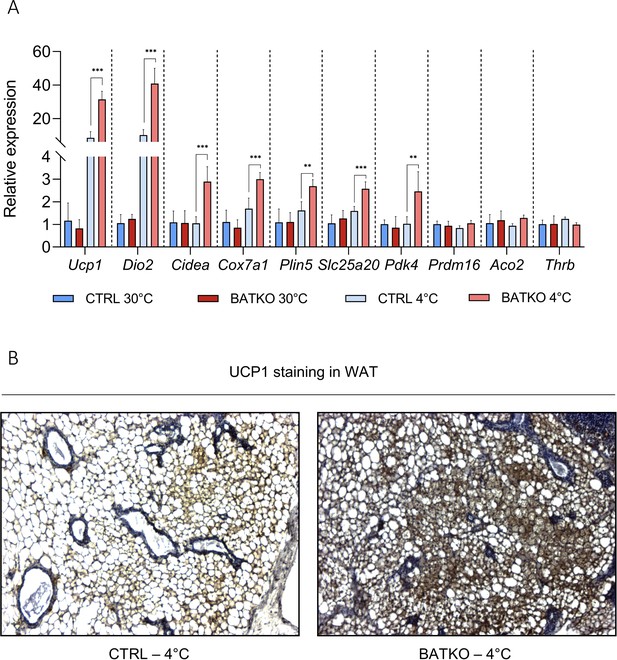
White adipose tissue (WAT) browning is exacerbated in BATKO mice.
(A) Relative expression of genes in the WAT (n = 4–7/group) and (B) representative images of UCP1 staining in the WAT, in CTRL and BATKO mice after a 72 hr 4°C exposure (with ad libitum access to food). Browning was exacerbated in BATKO mice. Thrb was not affected in WAT. Statistical significance is shown for the CTRL 4°C versus BATKO 4°C group comparison. **p<0.01 and ***p<0.001 for the indicated comparisons.
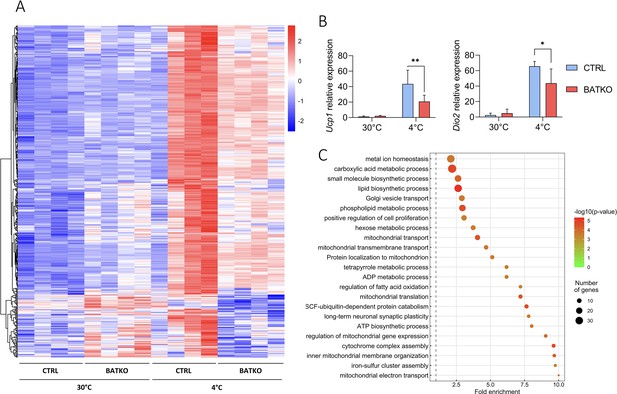
A subset of T3-regulated genes is activated during cold exposure and is necessary for an efficient brown adipose tissue (BAT) thermogenic response.
(A) Heatmap representation of cold-responsive genes altered in BATKO mice, after 24 hr at 4°C. Colors represent the z-scores, see scale in heatmap (n = 3–5/group). (B) Relative mRNA expression of Ucp1 (left panel) and Dio2 (right panel) in CTRL and BATKO mice at 30°C or after 24 hr at 4°C. Statistical significance is shown for the comparison between CTRL and BATKO mice at 4°C (n = 4–6/group). (C) Gene ontology dot plot representation of biological processes enriched in the 491 genes inefficiently induced in BATKO mice at cold. Some of the terms were shortened to increase readability without affecting the meaning. Error bars represent the standard deviation (SD). *p < 0.05, **p < 0.01 for the indicated comparisons.
-
Figure 4—source data 1
Transcriptome analysis of cold response in the brown adipose tissue (BAT) of both CTRL and BATKO mice.
- https://cdn.elifesciences.org/articles/81996/elife-81996-fig4-data1-v2.xlsx
-
Figure 4—source data 2
List of T3/TR target genes which expression is affected in the brown adipose tissue (BAT) of BATKO mice during cold exposure.
- https://cdn.elifesciences.org/articles/81996/elife-81996-fig4-data2-v2.xlsx
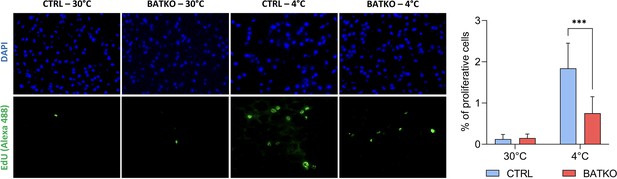
Local control of brown adipocytes proliferation by T3.
Representative images (left) and quantification (right) of EdU-positive proliferative cells in brown adipose tissue (BAT) from both CTRL and BATKO mice exposed 72 hr to 4°C and injected with EdU after 24 and 48 hr of cold. Nuclei are stained in blue with DAPI. Percentage of proliferative cells is the ratio of proliferative cells on the number of nuclei (n = 4–7/group). Statistical significance is shown for the comparison CTRL 4°C versus BATKO 4°C. Error bars represent the standard deviation (SD). ***p < 0.001r for the indicated comparisons.
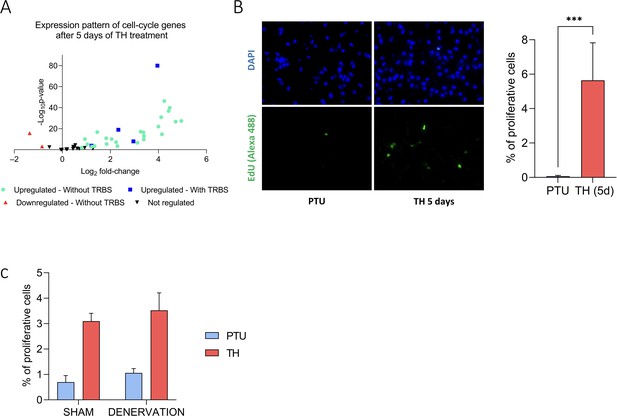
TH trigger brown adipose tissue (BAT) proliferation.
(A) RNAseq expression pattern of cell cycle genes (Whitfield et al., 2002) after 5 days of TH treatment in wild-type propylthiouracil (PTU)-fed mice. Genes are divided into four groups, according to their regulation by TH treatment (n = 4/group) and the presence of a thyroid hormone receptor-binding site (TRBS) within 30 kb from their promoter. (B) Representative images (left) and quantification (right) of EdU-positive proliferative cells (green cells) in BAT from wild-type hypothyroid mice treated or not with TH during 5 days and co-injected with EdU at day 4. Nuclei are stained in blue with DAPI. Percentage of proliferative cells is the ratio of proliferative cells on the number of nuclei (n = 5–6/group). (C) Quantification of EdU-positive cells in BAT from PTU-fed denervated or sham mice treated or not with TH during 5 days and co-injected with EdU at day 4. Percentage of proliferative cells is the ratio of proliferative cells on the number of nuclei (n = 5/group). Error bars represent the standard deviation (SD). ***p<0.001.
Tables
List of T3/TR target genes dysregulated during cold exposure in BATKO mice.
| Biological function | TR-dependent cold-induced genes |
|---|---|
| Lipid metabolism | Fabp3, Pla2g12a, Acsl5, Aspg, Mcee |
| Glucose metabolism | Slc2a4, Ogdh, Idh3a |
| Cell cycle progression | Cux1, Ccnd1 |
| Mitochondrial activity | Ppargc1α, Coq10a, Uqcc2, Cyb561 |
| Futile cycle | Alpl |
| Secreted peptides | Bmp8b, Fgf1 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (male mice) | C57BL/6J (male) | Charles River | ||
| Genetic reagent (male mice) | C57BL/6J-BATKO (male) | This paper | See ‘Material and methods’, section ‘Genetically modified mouse models’ | |
| Genetic reagent (male mice) | C57BL/6J-BATβKO (male) | This paper | See ‘Material and methods’, section ‘Genetically modified mouse models’ | |
| Antibody | Anti-UCP1 (rabbit polyclonal) | Abcam | Abcam: ab10983 | 1:400 |
| Antibody | Anti-rabbit linked to peroxidase (goat polyclonal) | Promega | Promega: W401B | 1:300 |
| Antibody | Anti-tyrosine hydroxylase (rabbit polyclonal) | Merck | Merck: AB152 | 1:500 |
| Antibody | Anti-rabbit IgG linked to peroxidase (goat polyclonal) | Bio-Rad | Bio-Rad: STAR124P | 1:5000 |
| Sequence-based reagent | See Table 2 | This paper | ||
| Commercial assay or kit | Pierce BCA Protein Assay Kit | Thermo Fisher Scientific | Thermo Fisher Scientific: 23225 | |
| Commercial assay or kit | Clarity Western ECL Substrat | Bio-Rad | Bio-Rad: 1705060 | |
| Commercial assay or kit | Diaminobenzidine staining | Sigma-Aldrich | Sigma-Aldrich: D5905 | |
| Commercial assay or kit | Click-iT EdU Cell Proliferation Kit | Thermo Fisher | Thermo Fisher: C10337 | |
| Commercial assay or kit | RNA SENSE Kit | Lexogen | ||
| Commercial assay or kit | Accel-NGS 2S Plus DNA kits | Swift Biosciences | ||
| Chemical compound, drug | Tamoxifen | Sigma-Aldrich | Sigma-Aldrich: T5648 | |
| Chemical compound, drug | PTU-containing diet | Harlan Teklad | Teklad Custome Diet: TD95125 | |
| Chemical compound, drug | T3 | Sigma-Aldrich | Sigma-Aldrich: T2877 | |
| Chemical compound, drug | T4 | Sigma-Aldrich | Sigma-Aldrich: T2376 | |
| Chemical compound, drug | 6-OHDA | Sigma-Aldrich | Sigma-Aldrich: H4381 | |
| Other | Cobas 600 | Roche | T3/T4 serum quantification | |
| Other | Leica TP1020 | Leica | Tissue section dehydration |
Primer sequences.
| Gene | Primer forward | Primer reverse |
|---|---|---|
| Ucp1 | AAGCTGTGCGATGTCCATGT | AAGCCACAAACCCTTTGAAAA |
| Hr | AGAGGTCCAAGGAGCATCAAGG | TTCCTCTTGTTGCTCTGCCTCC |
| Fndc5 | ATGAAGGAGATGGGGAGGAA | GCGGCAGAAGAGAGCTATAACA |
| Bmp8b | ACCTGTACCGTGCCATGACG | CGGTCGCGTTCCACTATGTTG |
| Idh3a | AAGAGGTTTTGCTGGTGGTGTTC | TTGCGCTCCTCCCACTGAATAG |
| Fabp3 | CATCGAGAAGAACGGGGATA | TCATCTGCTGTCACCTCGTC |
| Dio2 | AACAGCTTCCTCCTAGATGCC | ATTCAGGATTGGAGACGTGC |
| Cidea | GGACAGAAATGGACACCGGGTAG | TGACATTGAGACAGCCGAGGAAG |
| Cox7a1 | AGGCTCTGGTCCGGTCTTTTAG | GGTCATTGTCGGCCTGGAAG |
| Plin5 | GCGCCACACAGCAGAATGTC | GGCAAAGCCACCACTCGATTC |
| Slc25a20 | TCAGGCTTCTTCAGGGGAGAAC | CCACTGGCAGGAACATCTCG |
| Pdk4 | TTTCCAGGCCAACCAATCCAC | GTGGCCCTCATGGCATTCTTG |
| Prdm16 | TAGCTGCTTCTGGGCTCAAGG | ACGTCACCGTCACTTTTGGC |
| Aco2 | TGCCTAAGGTGGCTGTACCATC | CACTTCCTGGTTTATGTCCTTGGC |
| Thrb | CTCTTCTCACGGTTCTCCTC | AACCAGTGCCAGGAATGT |





