Isoform-specific disruption of the TP73 gene reveals a critical role for TAp73γ in tumorigenesis via leptin
Figures
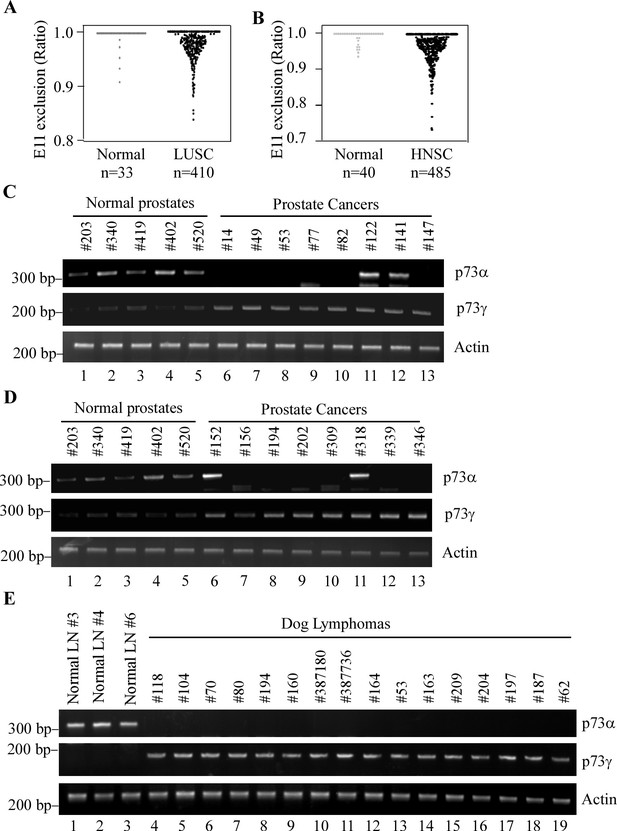
E11 skipping is detected in a subset of human cancers and dog lymphomas.
(A, B) Ratio of TP73 exon 11 exclusion was examined in lung squamous cell carcinoma (LUSC) and head-neck squamous cell carcinoma (HNSC) along with their normal tissues by using TCGA SpliceSeq database. Value of 1 indicates 100% of the transcripts contains all exons, for example, p73α. Values less than 1 represents the extent of expression of the isoforms with exon 11 exclusion, for example, p73γ. (C, D) The levels of p73α, p73γ, and actin transcript were examined in 5 normal prostates and 16 prostate cancer samples. (E) The levels of p73α, p73γ, and actin transcript were examined in 3 normal dog lymph nodes and 16 dog lymphomas.
-
Figure 1—source data 1
Labeled gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig1-data1-v2.pdf
-
Figure 1—source data 2
Raw gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig1-data2-v2.tif
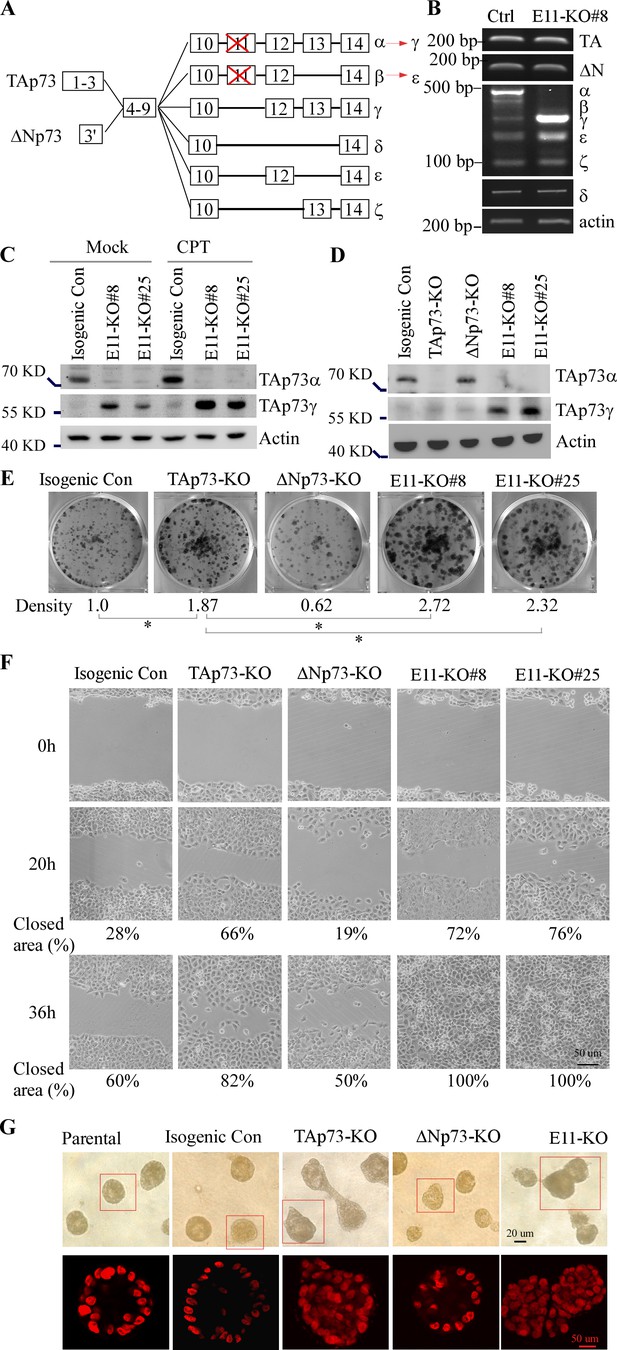
Deletion of exon 11 in the TP73 gene leads to isoform switch from α to γ, resulting in enhanced cell proliferation and migration as well as altered epithelial morphogenesis.
(A) Schematic representation of p73 isoforms and isoform switch resulting from E11 deletion. (B) The level of p73 splicing variants was examined by RT-PCR in isogenic control and E11-KO H1299 cells. (C) The levels of TAp73α, TAp73γ, and actin protein were measured in isogenic control and E11-KO H1299 cells mock-treated or treated with camptothecin (CPT). (D) The levels of TAp73α, TAp73γ, and actin protein were measured in isogenic control, TAp73-KO, ΔNp73-KO, and E11-KO H1299 cells. (E) Colony formation was performed with isogenic control, TAp73-KO, ΔNp73-KO, and E11-KO H1299 cells. The was quantified as relative density. (F) Scratch assay was performed with isogenic control, TAp73-KO, ΔNp73-KO, and E11-KO H1299 cells. Phase-contrast photomicrographs were taken immediately after scratch (0 hr), 20 hr, and 36 hr later to monitor cell migration. (G) Representative images of parental, isogenic control, TAp73-KO,ΔNp73-KO, and E11-KO MCF10A cells in three-dimensional culture. Top panel: phase-contrast image of acini in 3-D culture. Bottom panel: confocal images of cross-sections through the middle of acini stained with TO-PRO-3.
-
Figure 2—source data 1
Labeled gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig2-data1-v2.pdf
-
Figure 2—source data 2
Raw gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig2-data2-v2.tif
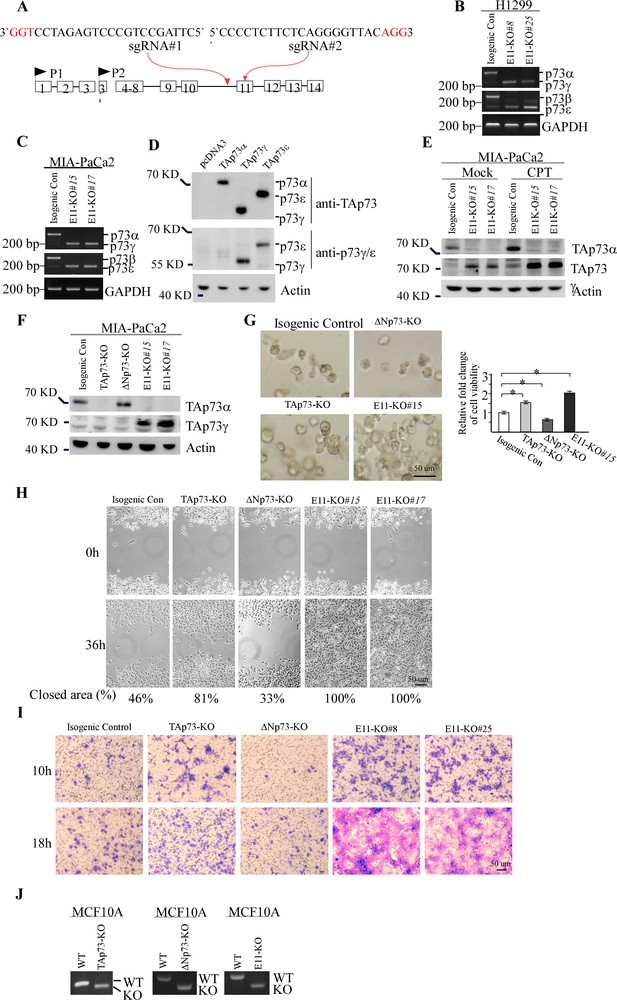
Deletion of E11 in TP73 leads to enhanced TAp73gamma expression and subseuqntly, enhanced cell proliferation and migration.
(A) Schematic representation of the strategy to target TP73 exon 11 using two guide RNAs. (B, C) The levels of p73α/γ, p73β/ε, and GAPDH transcripts were measured in isogenic control and E11-KO H1299 and Mia-PaCa2 cells. (D) H1299 were transiently transfected with an empty pcDNA3 vector or a vector expressing HA-tagged TAp73a, TAp73g, or TAp73e, followed by western blot analysis with antibodies against TAp73, p73g/e, or actin. (E) The levels of TAp73a, TAp73g, and actin protein were measured in isogenic control and E11-KO Mia-PaCa2 cells mock-treated or treated with camptothecin (CPT). (F) The levels of TAp73α, TAp73γ, and actin protein were measured in isogenic control, TAp73-KO, ΔNp73-KO, and E11-KO Mia-PaCa2 cells. (G) 3-D tumor sphere assays were performed with isogenic control, TAp73-KO, DNp73-KO, and E11-KO. Left panel: representative phase-contrast images of tumor spheres; Right panel: the viability of tumor spheres was measured using Cell-Titer Glo assay. (H) Scratch assay was performed with isogenic control, TAp73-KO, DNp73-KO, and E11-KO Mia-PaCa2 cells. Phase-contrast photomicrographs were taken immediately after scratch (0 hr) and 36 hr later to monitor cell migration. (I) Isogenic control, TAp73-KO, ΔNp73-KO, and E11-KO H1299 cells were subjected to transwell migration assays, followed by phase-contrast photomicrographs. (J) Genotyping to verify the TAp73-, ΔNp73-, and E11-KO MCF10A cells.
-
Figure 2—figure supplement 1—source data 1
Labeled gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig2-figsupp1-data1-v2.pdf
-
Figure 2—figure supplement 1—source data 2
Raw gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig2-figsupp1-data2-v2.tif
-
Figure 2—figure supplement 1—source data 3
Numerical data for Figure 2—figure supplement 1G.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig2-figsupp1-data3-v2.xlsx
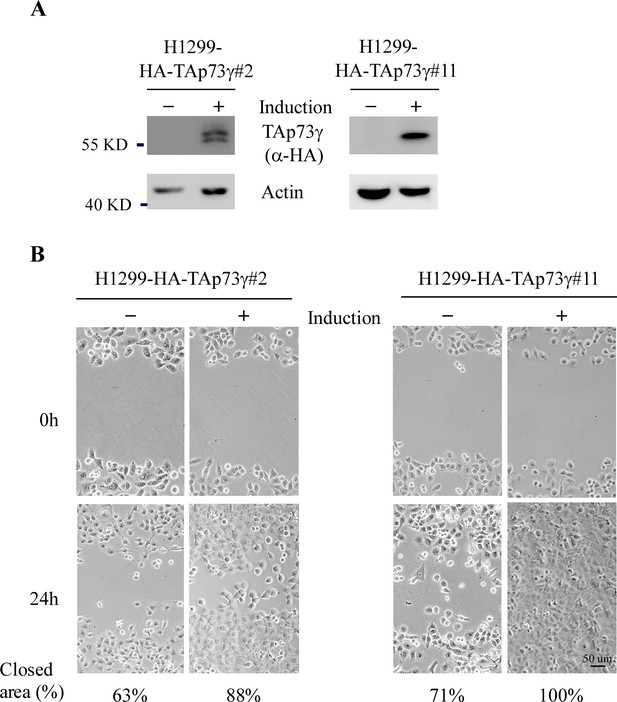
Ectopic TAp73gamma promotes cell migration.
(A) H1299 cells were uninduced or induced to express HA-tagged TAp73g for 24 hr, followed by western blot analysis with antibodies against HA or actin. (B) H1299 cells were uninduced or induced to express HA-tagged TAp73g for 24 hr, followed by scratch assay. Phase-contrast photomicrographs were taken immediately after scratch (0 hr) and 24 hr later to monitor cell migration.
-
Figure 2—figure supplement 2—source data 1
Labeled gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig2-figsupp2-data1-v2.pdf
-
Figure 2—figure supplement 2—source data 2
Raw gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig2-figsupp2-data2-v2.tif
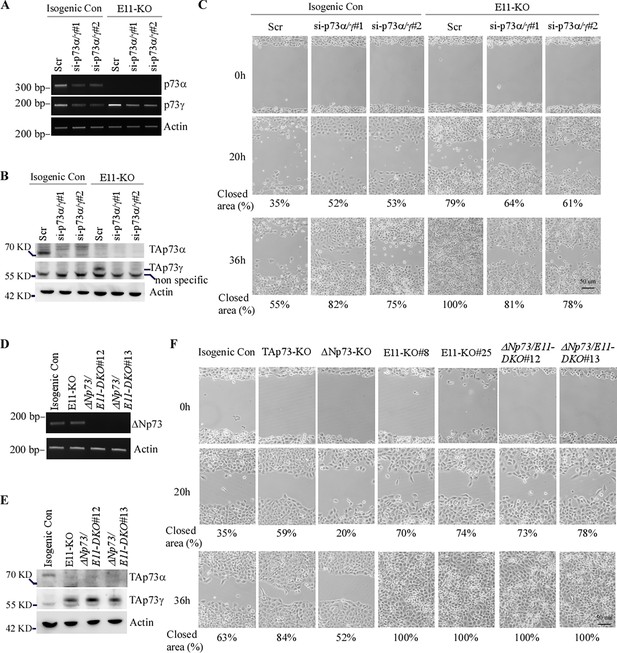
TAp73γ is primarily responsible for the oncogenic effects observed in E11-KO cells.
(A) Isogenic control and E11-KO H1299 cells transfected with a scrambled siRNA or siRNAs against p73α/γ for 3 d, followed by RT-PCR analysis to measure the levels of p73α, p73γ, and actin transcripts. (B) Cell were treated as in (A) and then subjected to western blot analysis to measure the levels of TAp73α, TAp73γ, and actin proteins using antibodies against TAp73, p73α/γ, and actin. (C) Scratch assay was performed with isogenic control and E11-KO H1299 cells transfected with a scrambled siRNA or siRNAs against p73α/γ for 3 d. Phase-contrast photomicrographs were taken immediately after scratch (0 hr), 20 hr, and 36 hr later to monitor cell migration. (D) The levels of ΔNp73 and actin transcripts were measured in isogenic control, E11-KO, ΔNp73/E11-DKO H1299 cells. (E) The levels of TAp73α, TAp73γ, and actin proteins were measured by western blot analysis using isogenic control, E11-KO, ΔNp73/E11-DKO H1299 cells. (F) Scratch assay was performed with isogenic control, TAp73-KO, ΔNp73-KO, E11-KO, and ΔNp73/E11-DKO H1299 cells. Phase-contrast photomicrographs were taken immediately after scratch (0 hr), 20 hr, and 36 hr later to monitor cell migration.
-
Figure 3—source data 1
Labeled gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig3-data1-v2.pdf
-
Figure 3—source data 2
Raw gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig3-data2-v2.tif
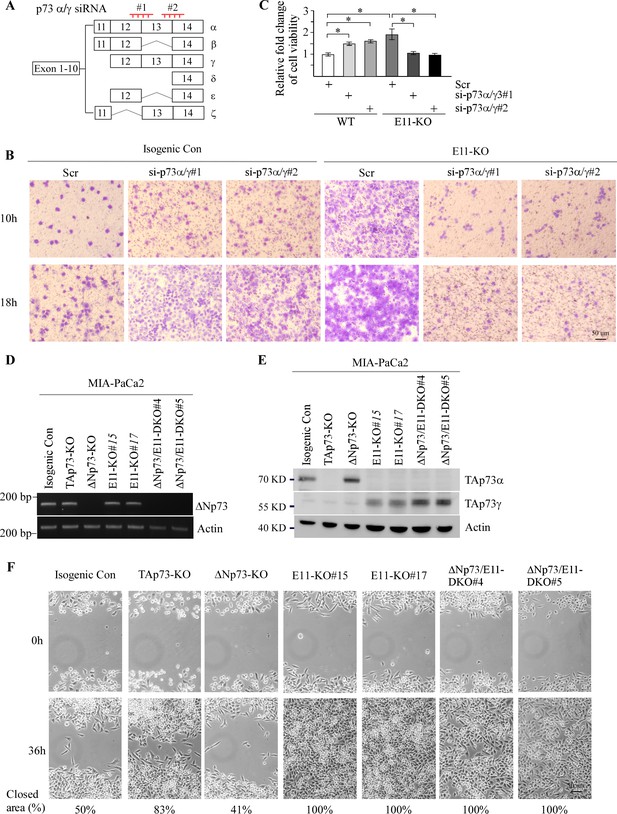
TAp73gamma, but not DNp73 is required for the enhanced cell mgiration and proliferation.
(A) Schematic representation of the locations of p73α/γ siRNAs. (B) Isogenic control and E11-KO H1299 cells were transiently transfected with a scrambled siRNA or siRNA against p73α/γ for 3 d, followed by transwell assay. Representative images were taken by phase-contrast microscope. (C) Isogenic control and E11-KO H1299 cells were transiently transfected with a scrambled siRNA or siRNA against p73α/γ for 3 d, followed by measurement of 3-D tumor spheres viability. Data were presented as Mean ± SEM (n=3). (D) The levels of ΔNp73 and actin transcript were measured in isogenic control, TAp73-KO, ΔNp73-KO, and E11-KO, and DN/E11-DKO Mia-PaCa2 cells. (E) The levels of TAp73α, TAp73γ, and actin protein were measured in isogenic control, TAp73-KO, ΔNp73-KO, E11-KO, and ΔN/E11-DKO Mia-PaCa2 cells. (F) Scratch assay was performed with isogenic control, TAp73-KO, DNp73-KO, E11-KO, and ΔN/E11-DKO Mia-PaCa2 cells. Phase-contrast photomicrographs were taken immediately after scratch (0 hr) and 36 hr later to monitor cell migration.
-
Figure 3—figure supplement 1—source data 1
Labeled gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig3-figsupp1-data1-v2.pdf
-
Figure 3—figure supplement 1—source data 2
Raw gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig3-figsupp1-data2-v2.tif
-
Figure 3—figure supplement 1—source data 3
Numerical data for Figure 3—figure supplement 1C.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig3-figsupp1-data3-v2.xlsx
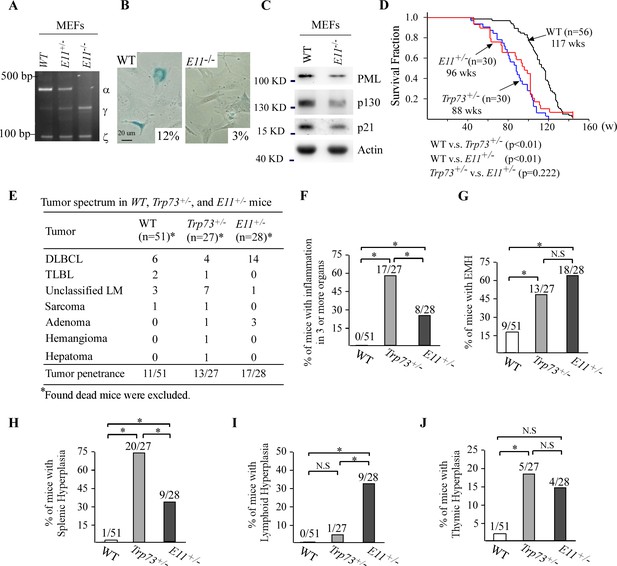
Deletion of E11 in the Trp73 gene leads to shortened lifespan, increased incidence of spontaneous tumor and chronic inflammation in mice.
(A) The levels of Trp73α, Trp73γ, Trp73ζ and actin transcripts were measured in WT, E11-HET, and E11-KO MEFs by RT-PCR analysis. (B) SA-β-gal staining was performed with WT and E11-KO MEFs. The percentage of SA-β-gal-positive cells was shown in each panel (WT vs. E11-KO: p<0.05 by Student’s test).(C) Western blot was performed to measure the levels of PML, p130, p21, and actin proteins in WT and E11-KO MEFs using antibodies against PML, p130, p21, and actin. (D) Kaplan–Meier survival curves of WT (n = 56), Trp73+/- (n = 30), and E11-HET (n = 30) mice. (E) Tumor spectra in WT (n = 56), Trp73+/- (n = 27), and E11-HET (n = 28) mice. (F) Percentage of WT (n=51), Trp73+/- (n=27), and E11-HET (n=28) mice with inflammation in three or more organs.(G) Percentage of WT (n=51), Trp73+/- (n=27), and E11-HET (n=28) mice with extramedullary hematopoiesis. (H) Percentage of WT (n=51), Trp73+/- (n=27), and E11-HET (n=27) mice with splenic hyperplasia. (I) Percentage of WT (n=51), Trp73+/- (n=27), and E11-HET (n=28) mice with lymphoid follicular hyperplasia. (J) Percentage of WT (n=51), Trp73+/- (n=27), and E11-HET (n=28) mice with thymic hyperplasia.
-
Figure 4—source data 1
Labeled gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig4-data1-v2.pdf
-
Figure 4—source data 2
Raw gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig4-data2-v2.tif
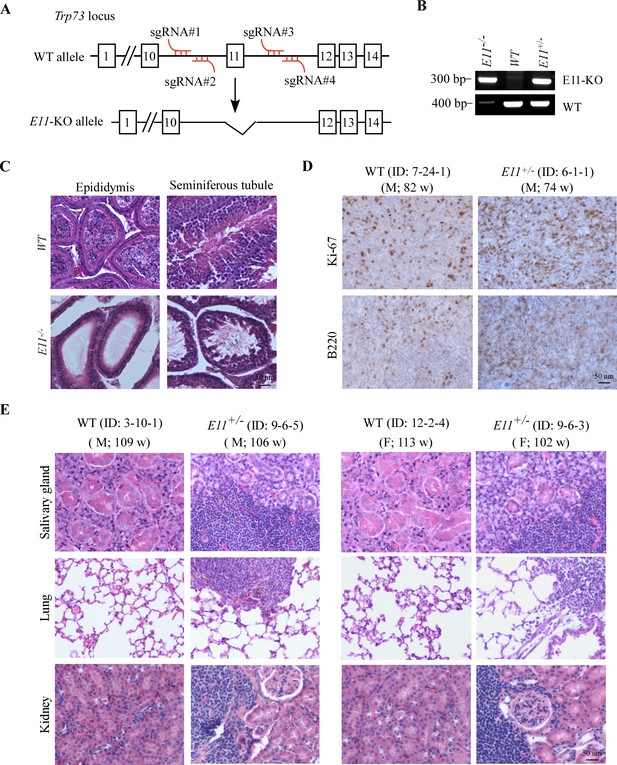
Mice dificient in TP73 E11 were infertile and prone to diffuse large B cell lymphoma and chronic inflammation.
(A) Schematic representation of the strategy to generate E11-deficient mouse model and the location of four guide RNAs. (B) Genotyping to verify WT, E11-HET, and E11-KO MEFs. (C) Representative images of H&E-stained epididymis and seminiferous tubule from male WT and E11-KO mice. (D) Representative images of B220- and Ki-67-stained spleen from WT and E11-HET mice. (E) Representative images of H&E-stained salivary gland, lung, and kidney from male and female WT and E11-HET mice.
-
Figure 4—figure supplement 1—source data 1
Labeled gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig4-figsupp1-data1-v2.pdf
-
Figure 4—figure supplement 1—source data 2
Raw gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig4-figsupp1-data2-v2.tif
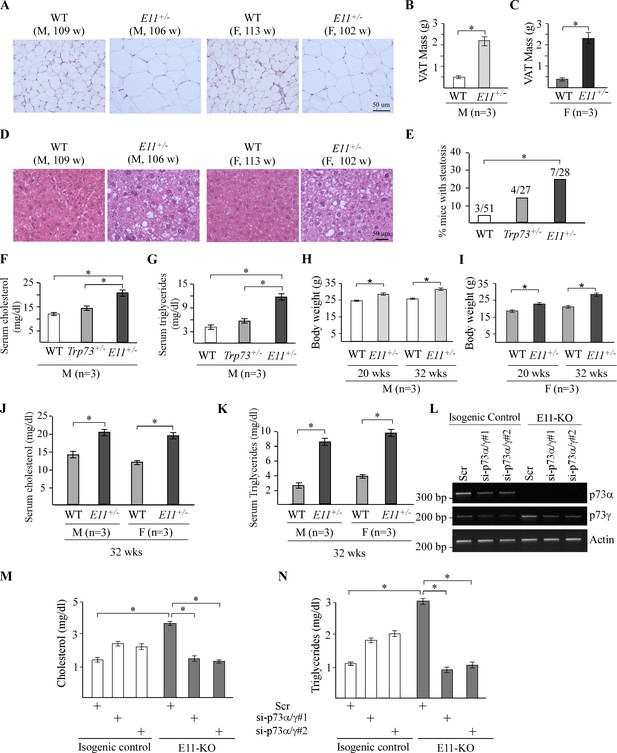
E11-deficient mice are prone to obesity.
(A) Representative images of H&E-stained visceral adipose tissues from male and female WT and E11-HET mice. (B–, C) Visceral adipose tissue (VAT) mass of male (B) and female (C) WT and E11-HET mice at ~100 wk. * indicates p<0.05 by Fisher’s exact test. (D) Representative images of H&E-stained liver tissues from male and female WT and E11-HET mice. (E) Percentage of WT (n=51), Trp73+/- (n=27), and E11-HET (n=28) mice with liver steatosis. * indicates p<0.05 by Fisher’s exact test. (F, G) The levels of serum cholesterol (F) and triglycerides (G) were measured in male WT, Trp73+/-, and E11-HET mice at 100 wk. (H, I) Body weights were measured in male (H) and female (I) WT, Trp73+/-, and E11-HET mice at 20 or 32 wk. (J, K) The levels of serum cholesterol (J) and triglycerides (K) were measured in male and female WT, Trp73+/-, and E11-HET mice at 32 wk. (L) The levels of p73α, p73γ, and actin transcripts were measured in isogenic control and E11-KO H1299 cells transfected with a scrambled siRNA or siRNAs against p73α/γ. (M, N) Cells were treated as in (L), followed by measurement of cholesterol (M) and triglycerides (N). Data were presented as Mean ± SEM (n=3).
-
Figure 5—source data 1
Labeled gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig5-data1-v2.pdf
-
Figure 5—source data 2
Raw gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig5-data2-v2.tif
-
Figure 5—source data 3
Numerical data for Figure 5.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig5-data3-v2.xlsx
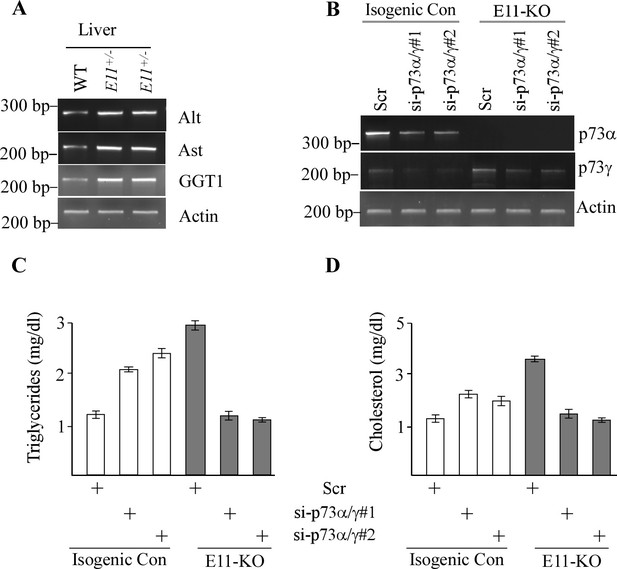
p73gamma maintains the levels of triglycerides and cholesterol in cells.
(A) The levels of alanine aminotransferase (ALT), aspartate aminotransferase (AST), and γ-glutamyltransferase 1 (GGT1) mRNA were measured in the livers from WT and E11+/- mice. (B) The levels of p73a, p73g, and actin transcripts were measured in isogenic control and E11-KO Mia-PaCa2 cells transfected with a scrambled siRAN or a siRNA against p73a/g for 3 d. (C, D) Mia-PaCa2 cells were treated as in (A), followed by ELISA to measure the levels of triglycerides (C) and cholesterol (D). Data were presented as Mean ± SEM (n=3).
-
Figure 5—figure supplement 1—source data 1
Labeled gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig5-figsupp1-data1-v2.pdf
-
Figure 5—figure supplement 1—source data 2
Raw gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig5-figsupp1-data2-v2.tif
-
Figure 5—figure supplement 1—source data 3
Numerical data for Figure 5–figure supplement 1C–D.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig5-figsupp1-data3-v2.xlsx
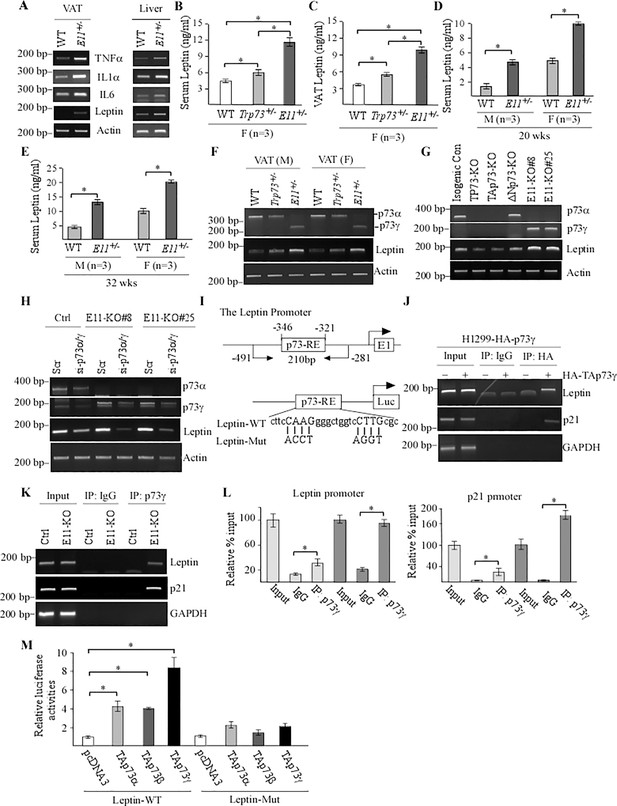
E11 deficiency leads to elevated production of Leptin, a novel target of TAp73g.
(A) The levels of TNFα, IL6, IL-1α, Leptin, and actin transcripts were measured in visceral adipose tissue (VAT) and liver from WT and E11-HET mice. (B, C) The levels of serum (B) and VAT (C) Leptin were measured in female WT, Trp73+/-, and E11-HET mice at 65 wk. (D, E) The levels of serum leptin were measured in male and female WT and E11-HET mice at 20 (D) or 32 (E) wk. (F) The levels of p73, Leptin, and actin transcripts were measured in VATs from male and female WT, Trp73+/-, and E11-HET mice. (G) The levels of p73α, p73γ, Leptin, and actin transcripts were measured in isogenic control, total p73-KO, TAp73-KO, DNp73-KO, and E11-KO H1299 cells. (H) The levels of p73α, p73γ, Leptin, and actin transcripts were measured in isogenic control and E11-KO H1299 cells transfected with a scrambled siRNA or siRNA against for p73α/γ for 3 d. (I) Schematic representation of the Leptin promoter, the location of p73-RE as well as luciferase reporters with WT or mutant p73RE. (J) H1299 cells were uninduced or induced to express TAp73γ for 24 hr, followed by ChIP assay to measure the binding of TAp73γ to the Leptin, p21, and GAPDH promoter. (K, L) Isogenic control and E11-KO cells were used for ChIP assay to examine the binding of p73γ to Leptin and p21 promoter by regular PCR (K) or qPCR (L). Data were presented as Mean ± SEM (n=3).* indicates p<0.05 by Student‘s t-test. (M) Luciferase assay was performed with H1299 cells transfected with a control vector or vector expressing TAp73α, TAp73β, and TAp73g. The relative fold-change of luciferase activity was calculated as a ratio of luciferase activity of each construct versus an empty vector. Data were presented as Mean ± SEM (n=3). * indicates p<0.05 by Student’s t-test.
-
Figure 6—source data 1
Labeled gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig6-data1-v2.pdf
-
Figure 6—source data 2
Raw gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig6-data2-v2.tif
-
Figure 6—source data 3
Numerical data for Figure 6.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig6-data3-v2.xlsx
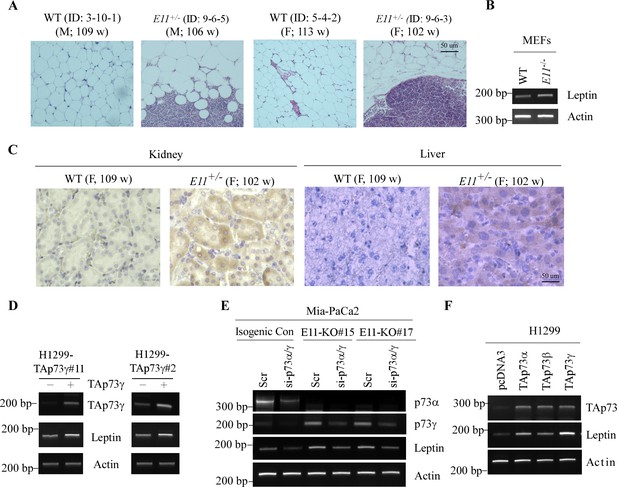
E11 dificiency alters lipid level via increased leptin levels.
(A) Representative images of H&E-stained vesical adipose tissues from male and female WT and E11-HET mice. (B) The levels of Leptin and actin transcripts were measured in WT and E11-KO MEFs. (C) Representative images of Leptin-stained livers and kidneys from WT and E11-HET mice. (D) H1299 cells were uninduced or induced to express HA-tagged TAp73g for 24 hr, followed by RT-PCR analysis to measure the levels of TAp73γ, Leptin, and actin transcripts. (E) The levels of p73α, p73γ, Leptin, and actin transcripts were measured in isogenic control and E11-KO Mia-PaCa2 cells transfected with a scrambled siRNA or siRNA against for p73α/γ for 3 d. (F) The levels of TAp73, Leptin, and actin transcripts were measured in H1299 cells transiently transfected with an empty pcDNA3 vector or a pcDNA3 vector expressing TAp73α, TAp73β, or TAp73γ.
-
Figure 6—figure supplement 1—source data 1
Labeled gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig6-figsupp1-data1-v2.pdf
-
Figure 6—figure supplement 1—source data 2
Raw gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig6-figsupp1-data2-v2.tif
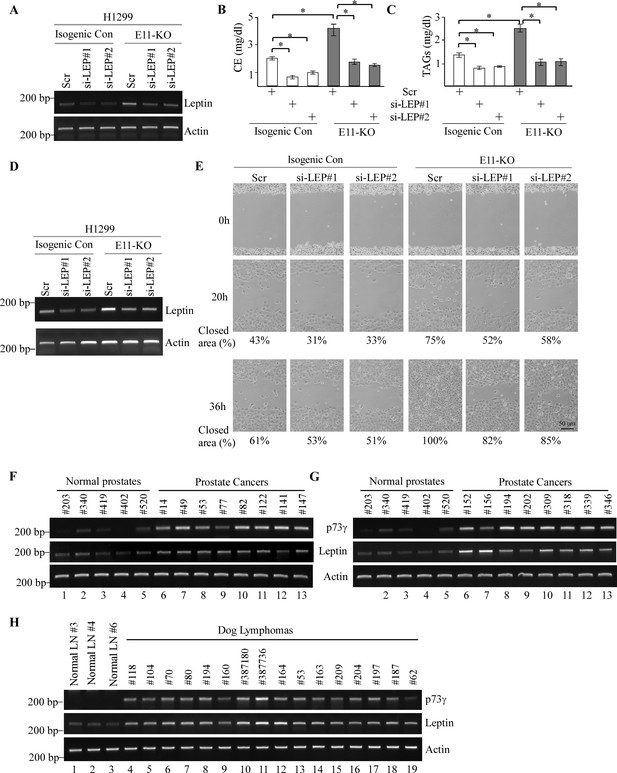
Leptin is a critical mediator of TAp73γ-mediated oncogenesis and altered lipid storage.
(A, D) The levels of Leptin and actin transcripts were measured in isogenic control and E11-KO H1299 cells transfected with a scrambled siRNA or siRNAs against Leptin. (B, C) The levels of cholesterol (B) and triglycerides (C) were measured in cells treated as in (A). Data were presented as Mean ± SEM (n=3). * indicates p<0.05 by Student’s t-test. (E) Scratch assay was performed with H1299 cells treated as in (D). Phase-contrast photomicrographs were taken immediately after scratch (0 hr), 20 hr, and 36 hr later to monitor cell migration. (F, G) The levels of p73γ, Leptin, and actin transcripts were examined in 5 normal prostates and 16 human prostate carcinomas. (H) The levels of p73γ, Leptin, and actin were examined in 3 normal dog lymph nodes and 16 dog lymphomas.
-
Figure 7—source data 1
Labeled gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig7-data1-v2.pdf
-
Figure 7—source data 2
Raw gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig7-data2-v2.zip
-
Figure 7—source data 3
Numerical data for Figure 7.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig7-data3-v2.xlsx
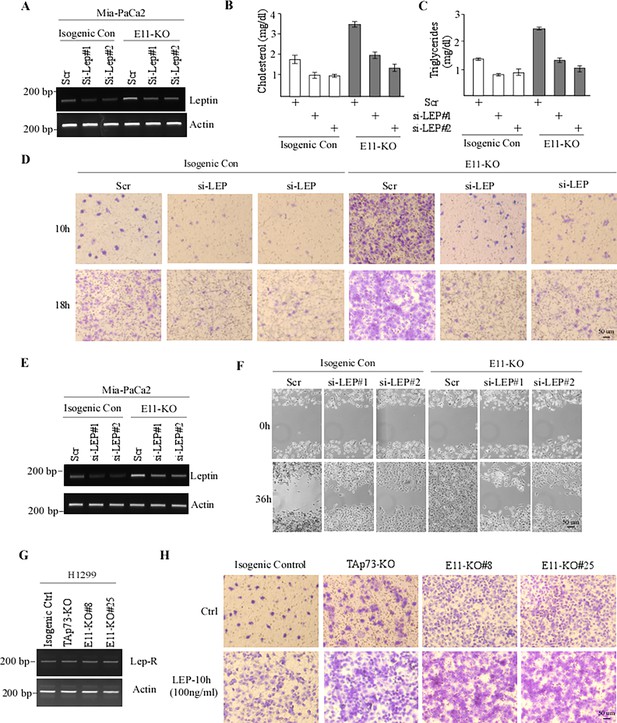
Leptin is required for p73gamma to enhance cell migration.
(A) The levels of Leptin and actin transcripts were measured in isogenic control and E11-KO Mia-PaCa2 cells transiently transfected with a scrambled siRNA or a siRNA against Leptin for 3 d. (B, C) The cells were treated as in (A), followed by ELISA to measure the level of cholesterol (B) and triglycerides (C). Data were presented as Mean ± SEM (n=3). (D) Isogenic control and E11-KO H1299 cells were transiently transfected with a scrambled siRNA or siRNA against Leptin for 3 d, followed by transwell assay. Representative images were taken by phase-contrast microscope. (E) The levels of Leptin and actin transcripts were measured in isogenic control and E11-KO Mia-PaCa2 cells transiently transfected with a scrambled siRNA or a siRNA against Leptin for 3 d. (F) The cells were treated as in (D), followed by scratch assay. Phase-contrast photomicrographs were taken immediately after scratch (0 hr) and 36 hr later to monitor cell migration. (G) The levels of Leptin receptor and actin mRNA were examined in isogenic control, TAp73-KO, and E11-KO H1299 cells. (H) Transwell assay was performed with isogenic control, TAp73-KO, and E11-KO H1299 cells mocked treated or treated with 100 ng/ml Leptin. Representative images were taken by phase-contrast microscope.
-
Figure 7—figure supplement 1—source data 1
Labeled gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig7-figsupp1-data1-v2.pdf
-
Figure 7—figure supplement 1—source data 2
Raw gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig7-figsupp1-data2-v2.tif
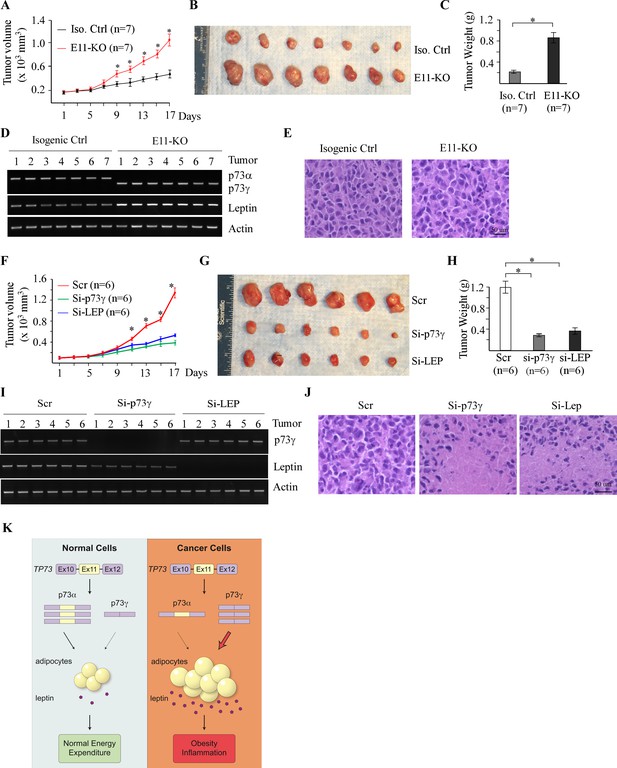
Targeting p73γ or Leptin inhibits tumor growth in vivo.
(A) Xenograft tumor growth in nude mice from isogenic control or E11-KO H1299 cells (error bars represent SEM, * indicates p<0.05 by Student’s t-test). (B) Images of tumors excised from isogenic control and E11-KO groups. (C) Tumor weight distribution between isogenic control and E11-KO groups upon termination of tumor growth experiments at day 17 (*p<0.05 by Student’s t-test). (D) The levels of p73α, p73γ, Leptin, and actin transcripts were measured in the tumors from isogenic control and E11-KO groups. (E) Representative images of H&E-stained tumors from isogenic control and E11-KO groups. (F) Xenograft tumor growth in nude mice from E11-KO H1299 cells transfected with scrambled siRNA or a siRNA against Leptin or p73γ (error bars represent SEM, * indicates p<0.05 by Student’s t-test). (G) Images of tumors excised from E11-KO groups with or without knockdown of Leptin or p73γ. (H) Tumor weight of E11-KO tumors with or without knockdown of Leptin or p73γ (*p<0.05 by Student’s t-test). (I) The levels of p73α, p73γ, Leptin, and actin transcripts were measured in E11-KO tumors with or without knockdown of Leptin or p73γ. (J) Representative images of H&E-stained E11-KO tumors with or without knockdown of Leptin or p73γ. (K) A proposed model to elucidate the role of p73α/γ switch and the p73γ-Leptin axis in tumorigenesis.
-
Figure 8—source data 1
Labeled gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig8-data1-v2.pdf
-
Figure 8—source data 2
Raw gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig8-data2-v2.tif
-
Figure 8—source data 3
Numerical data for Figure 8.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig8-data3-v2.xlsx
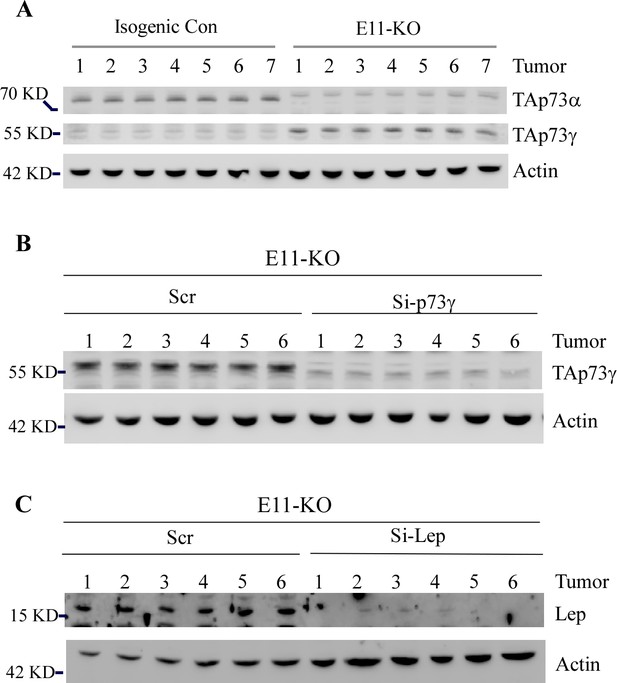
The levels of p73gamma and Leptin are measured in multiple sets of tumors.
(A) TAp73α, TAp73γ, and actin proteins were measured in tumors from isogenic control and E11-KO groups. (B) TAp73γ and actin proteins were measured in E11-KO tumors with or without p73γ knockdown. (C) TAp73γ and Leptin proteins were measured in E11-KO tumors with or without Leptin knockdown.
-
Figure 8—figure supplement 1—source data 1
Labeled gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig8-figsupp1-data1-v2.pdf
-
Figure 8—figure supplement 1—source data 2
Raw gel images.
- https://cdn.elifesciences.org/articles/82115/elife-82115-fig8-figsupp1-data2-v2.tif
Additional files
-
Supplementary file 1
Survival time, tumor spectrum, steatosis, inflammation, and other abnormalities in WT, Trp73+/- and E11+/- mice.
(a) Wild-type (WT) mice (n = 56) – survival time, tumor spectrum, steatosis, inflammation, and other abnormalities. (b) Trp73+/- mice (n = 30) – survival time, tumor spectrum, steatosis, inflammation, and other abnormalities. (c) E11-HET mice (n = 30) – survival time, tumor spectrum, steatosis, inflammation, and other abnormalities.
- https://cdn.elifesciences.org/articles/82115/elife-82115-supp1-v2.docx
-
Supplementary file 2
Primers for generating vectors.
(a) The primers used to generate plasmids’ expression vectors. (b) The primers used for genotyping. (c) The primers used for RT-PCR and ChIP.
- https://cdn.elifesciences.org/articles/82115/elife-82115-supp2-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/82115/elife-82115-mdarchecklist1-v2.docx





