Aggregating in vitro-grown adipocytes to produce macroscale cell-cultured fat tissue with tunable lipid compositions for food applications
Figures
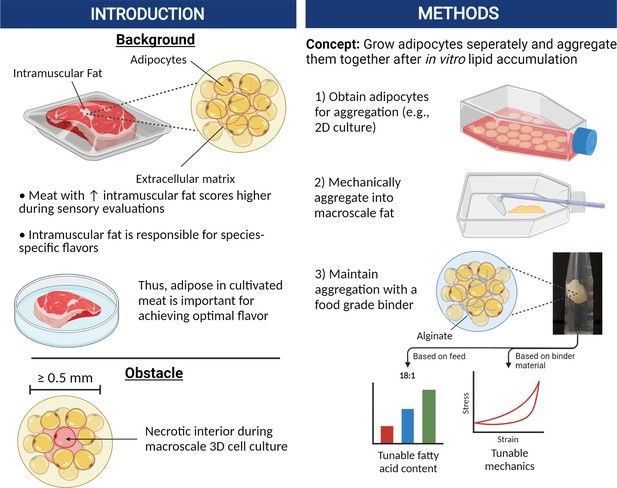
Graphical abstract covering the overall concepts behind producing macroscale volumes of cultured fat in this study.
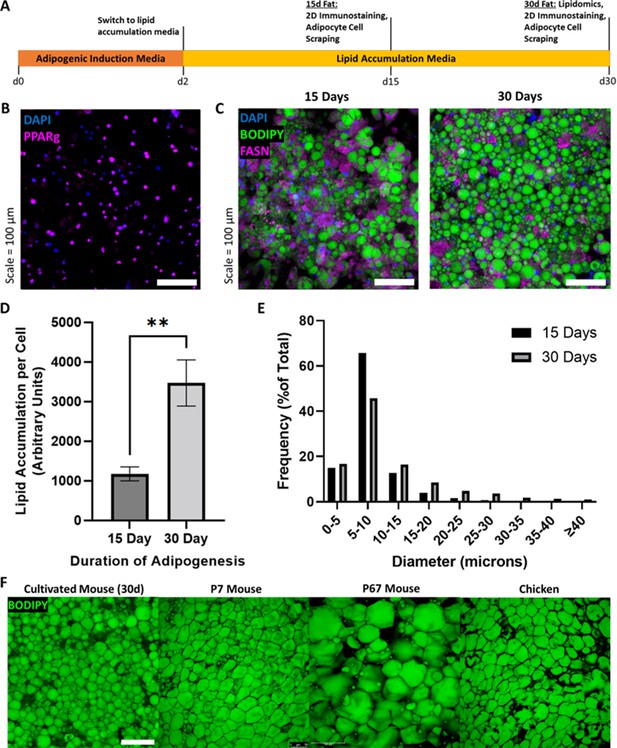
Assessment of in vitro murine adipocyte differentiation.
(A) Timeline of 3T3-L1 adipogenic differentiation. Confluent preadipocytes were grown in adipogenic induction medium for 2 days, then switched to lipid accumulation media for 15 or 30 days, where cells were stained and imaged, or harvested for lipidomics and 3D cultured fat tissue formation. (B) 15-day adipocytes stained for the adipogenic transcription factor peroxisome proliferator-activated receptor gamma (PPARγ) (magenta), as well as nuclei via DAPI (blue). (C) Lipid-stained (BODIPY, green) adipocytes after 15 and 30 days of adipogenesis. The in vitro adipocytes were also stained for DNA via DAPI (blue) and fatty acid synthetase (magenta). (D) The mean degree of lipid accumulation in 15- and 30-day cultured adipocytes, normalized by the number of cells detected via nucleus counting. Sample groups were compared using an unpaired t test with Welch’s correction, where p≤0.01 (**). (E) Frequency distributions of lipid droplet diameters from cultured adipocytes adipogenically grown for 15 and 30 days, compared via chi-square test in Supplementary file 2. (D) represents n=4 technical replicates from one experiment, while (E) represents n=4 technical replicates from a second experiment. (F) Lipid staining images (BODIPY) of 30-day in vitro 3T3-L1s compared to native adipocytes from chicken and mice of two ages. ‘7D Mouse’ and ‘67D Mouse’ refer to 7- and 67-day-old mice, respectively. Scale bar (same for all images) represents 100 µm.
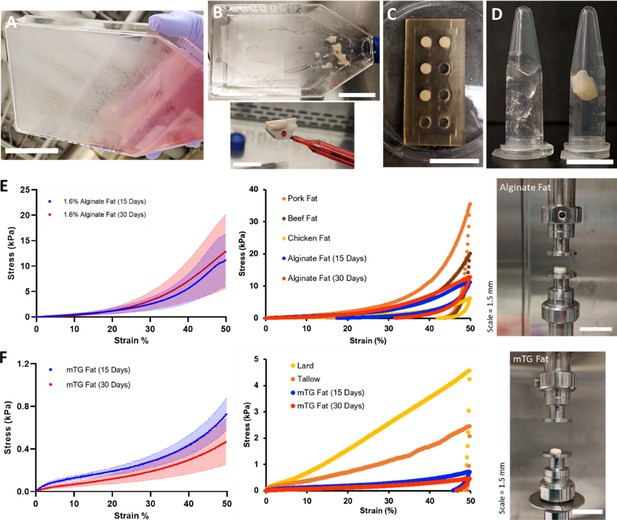
Mechanical characterization of 3D cultured fat tissues.
(A–D) Steps for producing 3D macroscale cultured fat tissues through the aggregation of individual adipocytes grown in vitro. (A) Adipogenesis: Adipocytes were differentiated in vitro for 15–30 days. Lipid-accumulating fat cells turned the bottom of the cell culture flask opaque. Scale bar 5 cm. (B) Cell harvest: Adipocytes were mechanically collected with a cell scraper, which aggregated the cells into masses of cultured fat. Scale bars 5 and 1.5 cm for the top and bottom images, respectively. (C) Binding into 3D tissue: Harvested fat was combined with a binder (e.g., alginate, transglutaminase) in a mold to add structure. Scale bar 3 cm. (D) 3D cultured fat: Cylinders of structured cultured fat tissue after removal from the mold. A piece of 3D cultured fat tissue made with 1.6% alginate is shown in the right tube, while the left tube contains 1.6% alginate without fat cells. Scale bar 1 cm. (E and F) Mechanical testing of cultured and native fat tissues (uniaxial compression). (E, i) shows the compressive strength of alginate-based cultured fat tissues formed using 15- or 30-day adipocytes, while (F, i) shows the same for microbial transglutaminase (mTG)-based cultured fat tissues. Solid lines represent mean values, while the shaded areas represent standard deviations. (E, ii) A compressive strength comparison of alginate-based cultured fat tissues with intact pig, cow, and chicken adipose tissues. Data points represent mean values, and the overall data represent tissue loading from 0% to 50% strain over 30 s, followed by unloading to 0% strain over the same duration. (F, ii) The same compressive strength comparison as (E, ii) but with mTG fat tissues alongside rendered animal fats from pigs (lard) and cows (tallow). (E, iii) depicts macroscale alginate-based cultured fat tissues on the mechanical testing apparatus prior to compression. (F, iii) depicts the same for mTG-based cultured fat tissues. n=4 for all alginate-based cultured fat constructs. n=3 and 5 for 15- and 30-day mTG cultured fat constructs, respectively. n=5, 6, 7 for beef, pork, and chicken adipose samples respectively. n=4 for lard and tallow samples respectively. All ‘n’ values refer to technical replicates.

Photographs of aggregated cell-cultured adipocytes after manual harvest.
(Left) Cell-scraped adipocytes prepared on a glass slide (25×75 mm2) with mounting media for subsequent microscopy. (Right) Cell-scraped adipocytes from three T175 cell culture flasks collected in a 50 ml conical tube. The bulk aggregated cells take on an appearance similar to lipoaspirate.
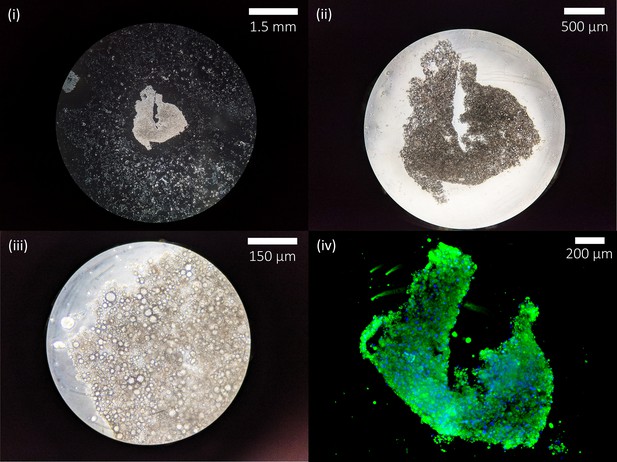
Low magnification micrographs of a group of aggregated adipocytes after manual harvest.
(i–iii) Brightfield micrographs of a small piece of aggregated murine adipocytes after cell scraping to illustrate cultured fat adipocyte and lipid droplet morphology (no binder or crosslinker added). (iv) Fluorescent staining of the cultured fat piece with BODIPY (lipids) and Hoescht 33342 (nuclei).
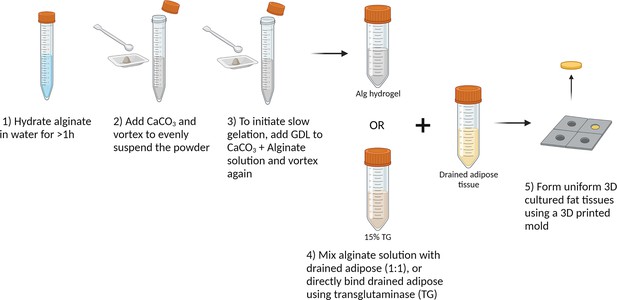
Detailed steps for producing macroscale 3D cultured fat constructs from aggregated adipocytes, using slow-gelling alginate or microbial transglutaminase as binders.
Created using BioRender.com.
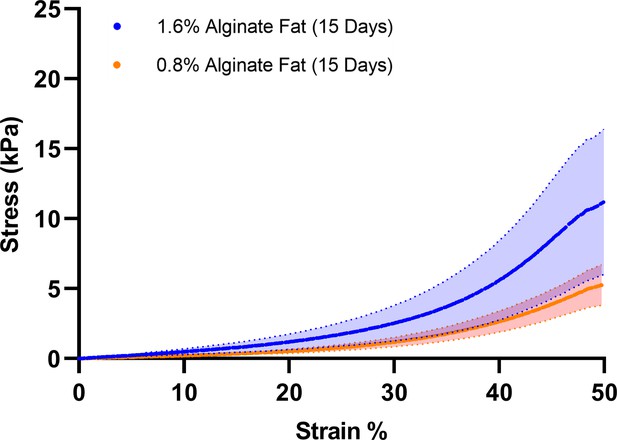
Uniaxial compression testing of macroscale cultured fat tissues produced from 0.8% and 1.6% (final concentration) alginate containing aggregated lipid-laden in vitro mouse adipocytes (15 days of adipogenesis).
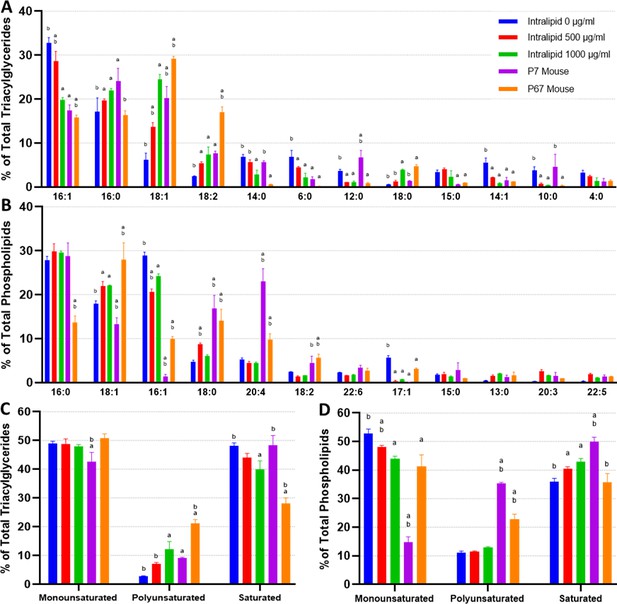
Lipidomics analysis of fatty acid compositions in cultured murine adipocytes.
(A, B) The fatty acid (FA) composition of (A) triacylglycerides (TAGs) and (B) phospholipids from in vitro (30 days of adipogenesis, 0–1000 µg/ml Intralipid) and native murine fats. The stacked graphs on the left illustrate the overall FA composition within each sample group (top 6 FAs color coded). The top-down order of FAs shown are the same in the graph and the legend. The column graphs on the right show the top 8 most prevalent FAs across all the sample groups with error bars (SD). The top-down order in the legend is the same as the left-right order of the samples in the column charts. Tables containing the proportions of all detected FAs, as well as principal component analysis (PCA) graphs, are available (Supplementary files 4 and 5; Figure 4—figure supplement 1). (C, D) FA compositions categorized by degree of saturation are shown for (C) TAGs and (D) phospholipids. The top-down order of monounsaturated (monounsat.), polyunsaturated (polyunsat.), and saturated FAs are the same in the graph and the legend. Column graphs of (C) and (D) with p≤0.05 comparisons are available in Figure 4—figure supplement 3. (E) The 18:3 (TAG fraction) content of 3T3-L1 adipocytes adipogenically cultured for 30 days with various levels of fatty acid (Intralipid) supplementation, versus native mouse fat samples. For all column graphs, ‘a’ and ‘b’ represent a difference of p≤0.05 versus 0 µg/ml Intralipid and 1000 µg/ml Intralipid, respectively (analysis of variance [ANOVA] with Tukey’s post-hoc tests). ‘IN-0’, ‘IN-500’, ‘IN-1000’ stand for Intralipid 0, 500, 1000 µg/ml Intralipid, respectively. ‘7D Mouse’ and ‘7M’ represent 7-day-old mouse adipose, while ‘67D Mouse’ and ‘67M’ represent 67-day-old mouse adipose. n=3 (technical) for all sample groups.

Principal component analyses of (left) triacylglyceride (TAG) and (right) phospholipid fatty acid compositions for in vitro and native murine fats (top 30 fatty acids).
Circles (●) represent in vitro mouse fats while triangles (▲) represent native mouse fats.

The proportion of triacylglycerides and phospholipids as a percentage of total intracellular lipids, for in vitro and native murine fats.
(*) represents p<0.05.

A column graph representation of murine fatty acid (FA) saturation in Figure 4C and D.
‘a’ and ‘b’ represent a difference of p≤0.05 versus 0 µg/ml Intralipid and 1000 µg/ml Intralipid, respectively (analysis of variance [ANOVA] with Tukey’s post-hoc tests).
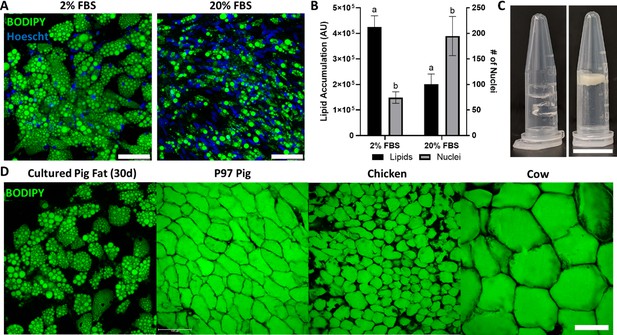
Porcine adipocytes grown to produce macroscale cultured fat.
(A) Pig DFAT cells cultured under adipogenic conditions containing 2% or 20% fetal bovine serum (FBS) for 30 days. Fat cells are stained for lipid using BODIPY (green) and for cell nuclei using Hoescht 33342 (blue). Scale bars 100 μm. (B) Lipid and cell number quantification of 30-day pig adipocytes, based on BODIPY and Hoescht 33342 staining, respectively. AU stands for arbitrary units. Results tagged with ‘a’ represent a difference of p≤0.0001, while ‘b’ represents p≤0.01. n=4 (technical) for both groups. (C, Left) Cell-free 1.6% alginate gels on the left. (C, Right) Porcine adipocytes (differentiated in 2% FBS media) mixed with alginate (1.6% final concentration) to form bulk cultured fat. Scale bar represents 1 mm. (D) Lipid staining images (BODIPY) of 30-day in vitro porcine adipocytes, juxtaposed with native adipocytes from a 97-day old (97D) pig, as well as a chicken and a cow. Scale bars represent 100 µm.

Pig DFAT cells cultured under adipogenic conditions with 0% fetal bovine serum (FBS) containing media for 30 days.
Cells were grown similarly to adipocytes cultured in adipogenic media containing 2% FBS, except the media did not contain GlutaMAX or FBS. The induction and lipid accumulation phases lasted 6 and 24 days, respectively. Lipid staining is BODIPY (green) and nuclei staining is Hoescht 33342 (blue). Scale bar represents 100 μm.
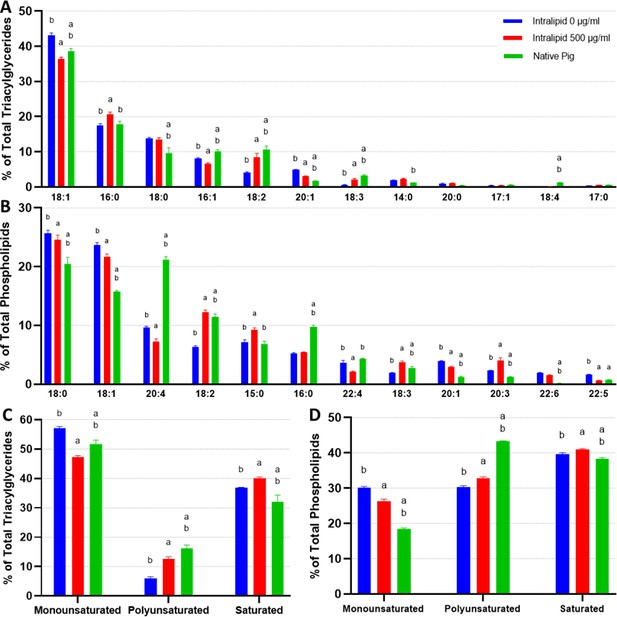
Lipidomics analysis of fatty acid compositions in cultured porcine adipocytes.
(A, B) The fatty acid (FA) composition of (A) triacylglycerides (TAGs) and (B) phospholipids from in vitro (12 days of adipogenesis, 0–500 µg/ml Intralipid) and native porcine fats. The stacked graphs on the left illustrate the overall FA composition within each sample group (top 6 FAs color coded). The top-down order of FAs shown are the same in the graph and the legend. The column graphs on the right show the top 8 most prevalent FAs across all the sample groups with error bars (SD). ‘a’ and ‘b’ represent a difference of p≤0.05 versus 0 µg/ml Intralipid and 500 µg/ml Intralipid, respectively (analysis of variance [ANOVA] with Tukey’s post-hoc tests). The top-down order in the legend is the same as the left-right order of the samples in the column charts. Tables containing the proportions of all detected FAs are available in Supplementary files 6 and 7. (C, D) FA compositions categorized by degree of saturation are shown for (C) TAGs and (D) phospholipids, displayed similarly as in (A) and (B). ‘IN-0’ and ‘IN-500’ stand for Intralipid 0 and 500 µg/ml Intralipid groups, respectively. ‘P’ represents native pig adipose. n=3 (technical) for all sample groups. TAGs and phospholipids versus total lipids are available in Figure 6—figure supplement 3.
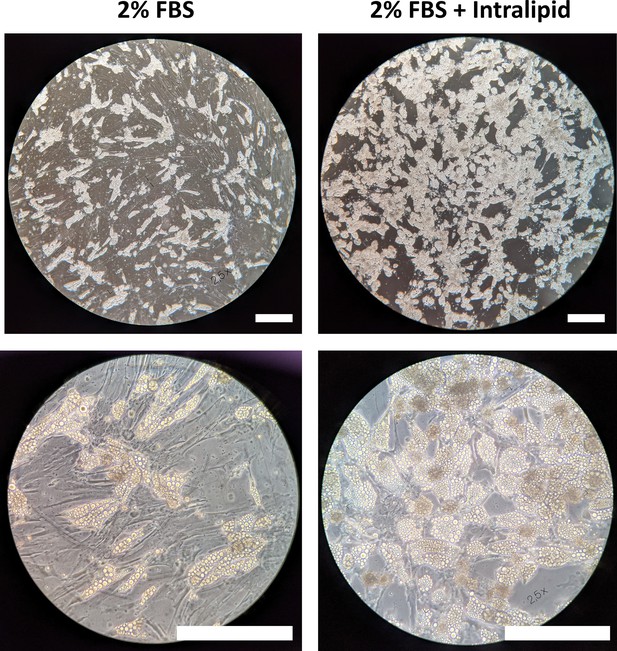
Phase contrast micrographs showing the degree of lipid accumulation in primary porcine adipocytes (DFAT cells) adipogenically differentiated with and without 500 µg/ml Intralipid for 11 days.
Scale bars represent 250 µm.
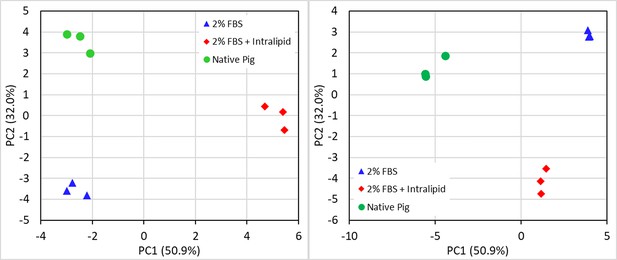
Principal component analyses of (left) triacylglyceride (TAG) and (right) phospholipid fatty acid compositions for in vitro and native porcine fats (all fatty acids).
Circles (●) represent native pig fats, triangles, (▲) represent in vitro adipocytes sans Intralipid, and diamonds (⬥) represent in vitro adipocytes with Intralipid.

The proportion of triacylglycerides and phospholipids as a percentage of total intracellular lipids, for in vitro and native porcine fats.
(***) represents p<0.001 and (****) represents p<0.0001.
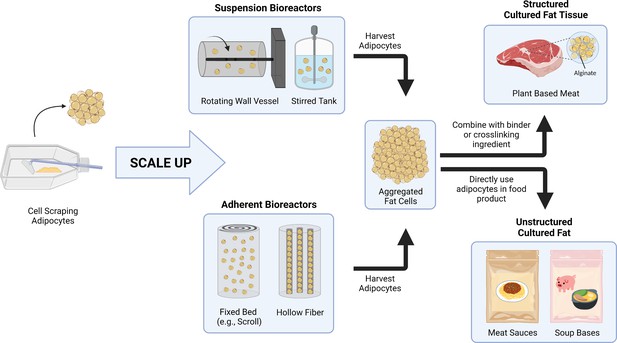
A conceptual schematic for scaling up cultured fat production via adipocyte aggregation.
As opposed to cell scraping methods with cell culture flasks, large-scale adipose production could utilize different types of bioreactors, including suspension bioreactors (e.g., rotating wall vessel and stirred tank) and adherent bioreactors (e.g., fixed bed, hollow fiber) for cell proliferation and differentiation. The adipocytes are then harvested and aggregated for incorporation into food products. For products requiring structured cultured fat tissues, fat cells are combined with binders or crosslinking ingredients to form a rigid construct. Adipocytes can also be directly included in food products that do not require the cultured fat to be structured.
Videos
Methods for minimizing adipocyte cell detachment during cell culture (when changing culture media).
Inversion during media (liquid) aspiration (0:06). Dispensing to sidewall of flask during inversion (1:18). Reinverting flask carefully from inverted position (1:42).
Harvesting cultured fat by cell scraping in vitro murine adipocytes.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Mus musculus) | 3T3-L1; murine adipocytes | American Type Culture Collection | CL-173; RRID: CVCL_0123 | |
| Biological sample (Sus scrofa domesticus) | DFAT cells; porcine adipocytes | This paper | Subcutaneous fat (pork belly) | Isolation technique outlined in Materials and methods section |
| Biological sample (Bos taurus) | Beef fat | This paper | Pectoralis minor (brisket point cut) | Obtained from local butcher shop |
| Biological sample (Gallus gallus domesticus) | Chicken fat | This paper | Chicken thigh | Obtained from local butcher shop |
| Biological sample (S. scrofa domesticus) | Pork fat | This paper | Subcutaneous fat (pork belly) | Obtained from local butcher shop |
| Biological sample (S. scrofa domesticus) | Lard (Porcine) | Goya | – | |
| Biological sample (B. taurus) | Tallow (Bovine) | Epic Provisions | – | |
| Other | 300 µm cell strainer | pluriSelect | 43-50300-03 | For primary cell isolation (separating cells from debris) |
| Other | Collagenase | Worthington Biochemical | LS004176 | For primary cell isolation (digesting ECM) |
| Chemical compound, drug | DMEM | Thermo Fisher | 10569044 | |
| Chemical compound, drug | Advanced DMEM/F12 | Thermo Fisher | 12634-028 | |
| Chemical compound, drug | GlutaMAX | Thermo Fisher | 35050-061 | |
| Other | Bovine calf serum | Sigma-Aldrich | 12,133C | Culture media component |
| Other | Fetal bovine serum (FBS) | Thermo Fisher | A31606-01; Lot: 2129571 | Culture media component |
| Chemical compound, drug | Antibiotic/antimycotic | Thermo Fisher | 15240062 | |
| Chemical compound, drug | Primocin | InvivoGen | ant-pm-1 | |
| Other | Accumax | Innovative Cell Technologies | AM105 | Cell detachment reagent |
| Other | NucleoCounter | Chemometec | NC-200 | Cell counter |
| Peptide, recombinant protein | Insulin | Sigma-Aldrich | I0516 | |
| Chemical compound, drug | Dexamethasone | Sigma-Aldrich | D4902 | |
| Chemical compound, drug | IBMX | Sigma-Aldrich | I5879 | |
| Chemical compound, drug | Rosiglitazone | TCI America | R0106 | |
| Chemical compound, drug | Intralipid | Sigma-Aldrich | I141 | |
| Chemical compound, drug | Biotin | TCI America | B04631G | |
| Chemical compound, drug | Calcium-D-pantothenate | TCI America | P001225G | |
| Other | Stereolithography 3D Printer | Formlabs | Form 2 | For making custom molds when producing 3D cultured fats |
| Chemical compound, drug | Biocompatible 3D printing resin | Formlabs | Surgical guide resin | |
| Chemical compound, drug | Medical device adhesive | Henkel Adhesives | Loctite 3556 | |
| Chemical compound, drug | Sodium alginate | Modernist Pantry | 1007-50 | |
| Chemical compound, drug | Calcium carbonate | Modernist Pantry | 1505-50 | |
| Chemical compound, drug | Glucono delta-lactone | Modernist Pantry | 1159-50 | |
| Peptide, recombinant protein | Microbial transglutaminase (mTG) | Ajinomoto | Activa TI | |
| Other | Goat serum | Thermo Fisher | 16210-064 | For use during immunofluorescent staining |
| Chemical compound, drug | BODIPY 493/503 | Invitrogen | D3922 | |
| Chemical compound, drug | DAPI | Thermo Fisher | 62248 | |
| Chemical compound, drug | Hoescht 33342 | Invitrogen | H3570 | |
| Antibody | Anti-fatty acid synthetase (rabbit, monoclonal) | Invitrogen | MA5-14887; RRID: AB_10980075 | (1:50) |
| Antibody | Anti-peroxisome proliferator-activated receptor gamma (rabbit, polyclonal) | Abcam | ab45036; RRID: AB_1603934 | (1:500) |
| Antibody | Anti-rabbit Alexa Fluor plus 647 (goat, polyclonal) | Invitrogen | A32733; RRID: AB_2633282 | (1:500) |
| Chemical compound, drug | Mounting media | Vector Laboratories | H-1700 | |
| Software, algorithm | Image analysis/quantification | CellProfiler (https://cellprofiler.org) | CellProfiler 4.2.1; RRID: SCR_007358 | |
| Other | Cell scraper (large) | Sarstedt | 83.3952 | For harvest and aggregation of differentiated adipocytes |
| Chemical compound, drug | Methyl tert-butyl ether | Alfa Aesar | 41839AK | |
| Chemical compound, drug | Methanol | Thermo Fisher | BPA4544 | |
| Software, algorithm | Graphing and statistical software | GraphPad Prism (https://www.graphpad.com) | GraphPad Prism 9.3.0; RRID: SCR_002798 |
Additional files
-
Supplementary file 1
Proportion of nuclei positive for the adipogenic transcription factor peroxisome proliferator-activated receptor gamma (PPARγ) in 3T3-L1 adipocytes grown under adipogenic conditions for 15 days, n=4 technical replicates.
- https://cdn.elifesciences.org/articles/82120/elife-82120-supp1-v1.xlsx
-
Supplementary file 2
A frequency distribution table of 15- and 30-day cultured adipocyte lipid droplet diameters compared using a chi-square test (degrees of freedom =8).
- https://cdn.elifesciences.org/articles/82120/elife-82120-supp2-v1.xlsx
-
Supplementary file 3
The major fatty acids present in soybean oil (the major component of Intralipid, other than water), according to the package insert supplied with Intralipid 20% (intravenous fat emulsion).
- https://cdn.elifesciences.org/articles/82120/elife-82120-supp3-v1.xlsx
-
Supplementary file 4
The full fatty acid compositions (mean % of total of triacylglycerides measured, n=3) of in vitro- (30 days of adipogenesis) and in vivo-grown murine fats.
- https://cdn.elifesciences.org/articles/82120/elife-82120-supp4-v1.xlsx
-
Supplementary file 5
The full fatty acid compositions (mean % of total of phospholipids measured, n=3) of in vitro- (30 days of adipogenesis) and in vivo-grown murine fats.
- https://cdn.elifesciences.org/articles/82120/elife-82120-supp5-v1.xlsx
-
Supplementary file 6
The full fatty acid compositions (mean % of total of triacylglycerides measured, n=3) of in vitro- (30 days of adipogenesis) and in vivo-grown porcine fats.
- https://cdn.elifesciences.org/articles/82120/elife-82120-supp6-v1.xlsx
-
Supplementary file 7
The full fatty acid compositions (mean % of total of phospholipids measured, n=3) of in vitro- (30 days of adipogenesis) and in vivo-grown porcine fats.
- https://cdn.elifesciences.org/articles/82120/elife-82120-supp7-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/82120/elife-82120-mdarchecklist1-v1.pdf





