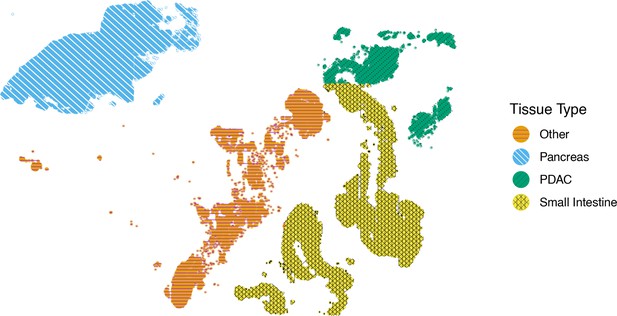
Generating colorblind-friendly scatter plots for single-cell data
Figures
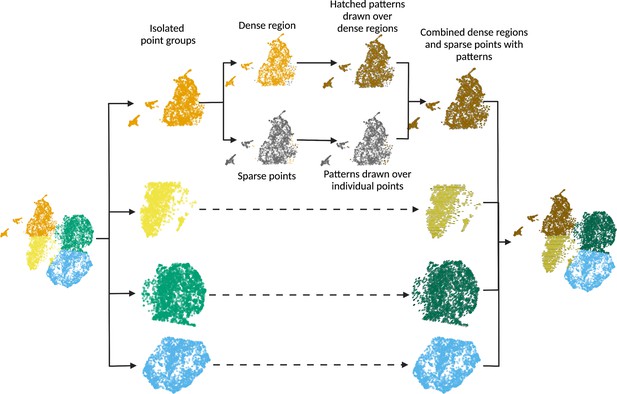
scatterHatch generates accessible scatter plots by redundant coding of point groups using colors and patterns.
For every point group, scatterHatch separates sparsely distributed points from the dense clusters. scatterHatch plots coarse patterns over the dense clusters and individually plots patterns over the sparse points. Created using Biorender.com.
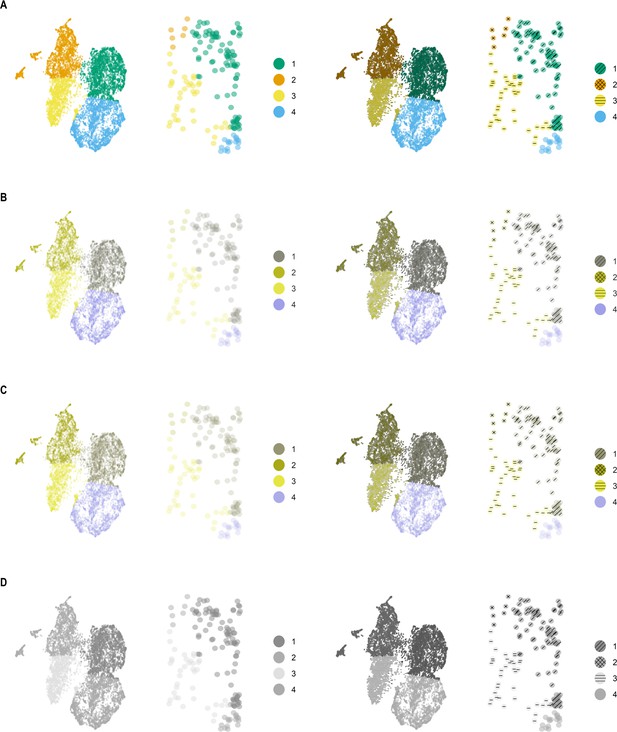
scatterHatch plots are more accessible compared to scatter plots to individuals with CVD.
Simulated perception of a UMAP scatter plot compared with a scatterHatch plot by individuals with (A) normal color vision, (B) deuteranomaly, (C) protanomaly, and (D) monochromacy, with the insets showing a magnified sparse region showing patterns assigned to individual cells. Despite the change in color perception, readers have access to secondary visual information in the form of patterns to help interpret the data. CVD, color-vision deficiency.
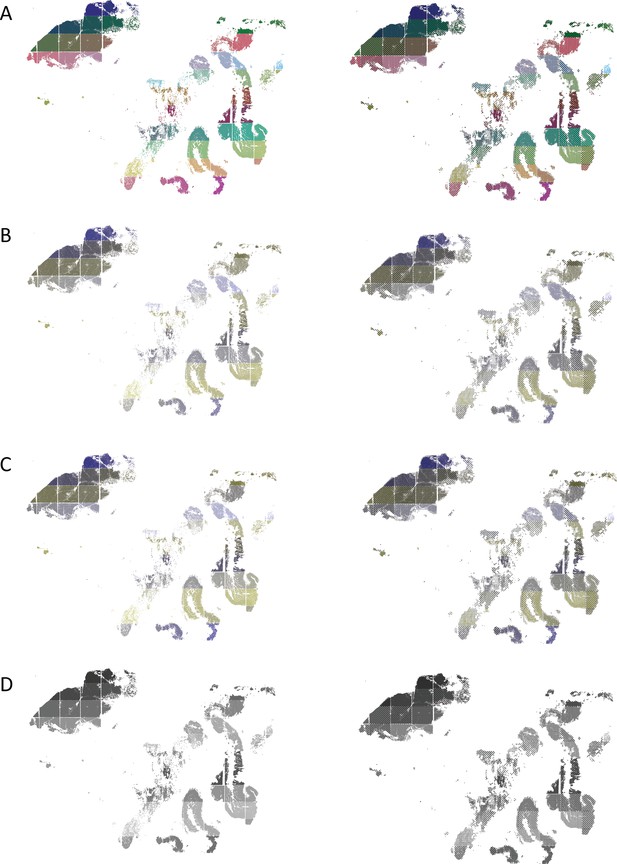
scatterHatch plots are more accessible than scatter plots for all readers when number of cell groups is high.
Perception of a spatial plot of the PDAC data set with 82-cell groups compared with a corresponding scatterHatch plot as simulated for (A) normal color vision, (B) deuteranomaly, (C) protanomaly, and (D) monochromacy. As the number of colors in the scatter plot increases, its interpretability reduces even for normal color vision. The redundant coding used in scatterHatch plots results in increased accessibility. PDAC, Pancreatic Ductal Carcinoma.
Tables
Parameters to enable users to customize pattern aesthetics or to create new patterns.
| Name | Description | Options |
|---|---|---|
| pattern | Specifies the pattern type | Default options are ‘horizontal’, ‘vertical’, ‘positiveDiagonal’, ‘negativeDiagonal’, ‘cross’, ‘checkers’, ‘blank’ E.g.: pattern=‘checkers” |
| angle | Allows users to specify line angles to be included in the pattern (enables users to create new patterns) | Numeric array with values from 0 to 180. E.g.: angle=c(45, 90, 135) |
| lineWidth | Width of the lines in a pattern | Numeric - default value based on point size E.g.: lineWidth=0.1 |
| lineColor | Color of the lines in a pattern | Character string specifying a color E.g.: lineColor=‘white’ |
| lineType | Type of the lines in a pattern | Character string to specify the line type from the ggplot2 package. E.g.: lineType=‘dotted’ |
| LineAlpha | Transparency of the lines in a pattern | Numeric value from 0 to 1. Default is 1. Ex: lineAlpha=0.1 |
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/82128/elife-82128-mdarchecklist1-v2.pdf
-
Supplementary file 1
'scatterHatch user guide'.
- https://cdn.elifesciences.org/articles/82128/elife-82128-supp1-v2.pdf