Enhanced single RNA imaging reveals dynamic gene expression in live animals
Figures
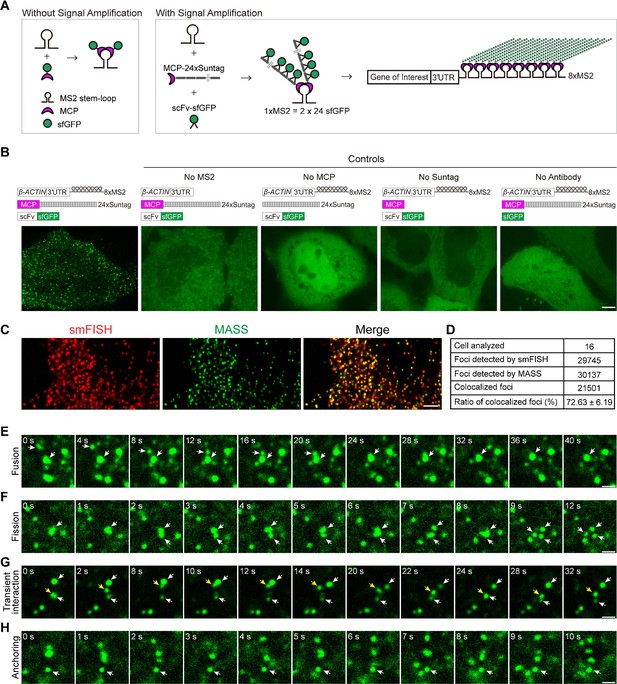
Live-cell imaging of β-ACTIN mRNA with the MS2-based signal amplification with the Suntag system.
(A) Schematic of the classical MS2-MCP system and the MS2-based signal amplification with the Suntag system. (B) Representative images of β-ACTIN-8xMS2 mRNA in live HeLa cells. The sfGFP fluorescence signal is shown. Left panel: Constructs of β-ACTIN-8xMS2, MCP-24xSuntag, and scFv-sfGFP were cotransfected into HeLa cells. Images were taken 12 hr after transfection. Right panels: Where one of the elements was removed (as indicated). Scale bar, 5 µm. (C) Representative confocal images of β-ACTIN-8xMS2 labeled by MASS and single-molecule in situ hybridization (smFISH) with probes against MS2 stem-loops and probes against the linker region between the MS2 stem-loops in HeLa cells. Scale bar, 5 µm. See also Figure 1—figure supplement 1. (D) Quantification of the total and colocalized foci of β-ACTIN-8xMS2 mRNAs detected by smFISH or MASS with tdMCP-24xSuntag in HeLa cells. A total of 16 cells from three independent smFISH experiments were analyzed. See also Figure 1—source data 1. (E–H) Time-lapse imaging of β-ACTIN-8xMS2 mRNA dynamics in HeLa cells. sfGFP foci (β-ACTIN mRNAs) are shown. Constructs of β-ACTIN-8xMS2, MCP-24xSuntag, and scFv-sfGFP were cotransfected into HeLa cells. Images were taken 12 hr after transfection. (E) A fusion event of two sfGFP spots (white arrows). (F) A fission event: with large sfGFP foci split into three spots (white arrows). (G) Transient interactions of an sfGFP spot (yellow arrow) between two spots (white arrows). (H) An sfGFP spot showing no movement over a 10-s period (white arrow). Scale bars, 1 µm.
-
Figure 1—source data 1
qRT-PCR (Quantitative Reverse Transcription PCR), number of foci detected by smFISH and MASS, signal-to-noise ratio, velocity, and intensity of the foci of mRNA detected in HeLa cells.
- https://cdn.elifesciences.org/articles/82178/elife-82178-fig1-data1-v3.xlsx
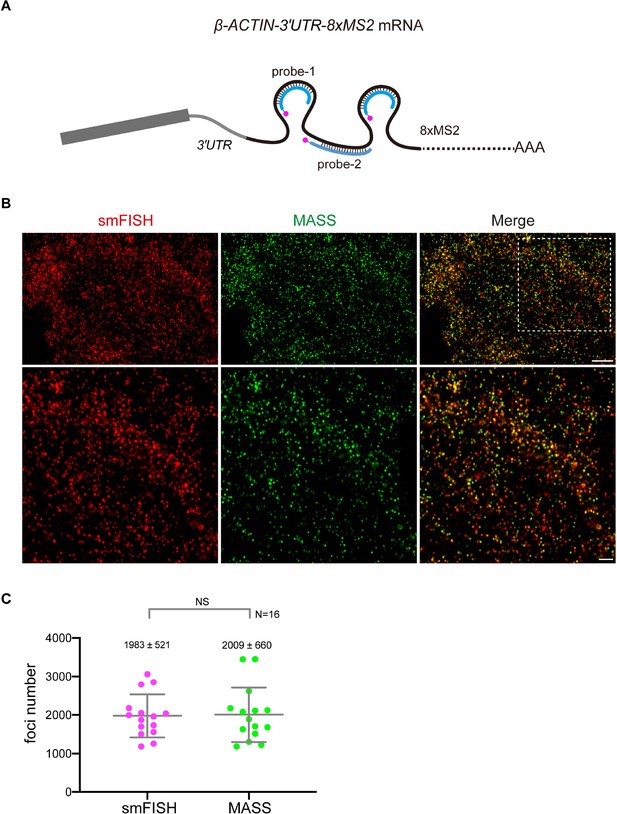
Labeling of β-ACTIN-8xMS2 mRNA by smFISH and MASS with the tdMCP-24xSuntag in HeLa cells.
(A) Schematic of probes targeting the β-ACTIN-8xMS2 tagged mRNA. Probe-1: against the MS2 stem-loop; probe-2: against the linker region. (B) Representative confocal images of β-ACTIN-8xMS2 labeled by MASS and single-molecule in situ hybridization (smFISH) with probes in (A). Scale bar, 5 µm (top) and 2 µm (bottom). (C) Distribution of the foci number of β-ACTIN-8xMS2 mRNAs detected by smFISH and MASS with tdMCP-24xSuntag in a single HeLa cell. Sixteen cells from three independent experiments were analyzed. Bars indicate mean ± standard deviation (SD). Unpaired t-test, p = 0.911. NS: not significant. See also Figure 1—source data 1.
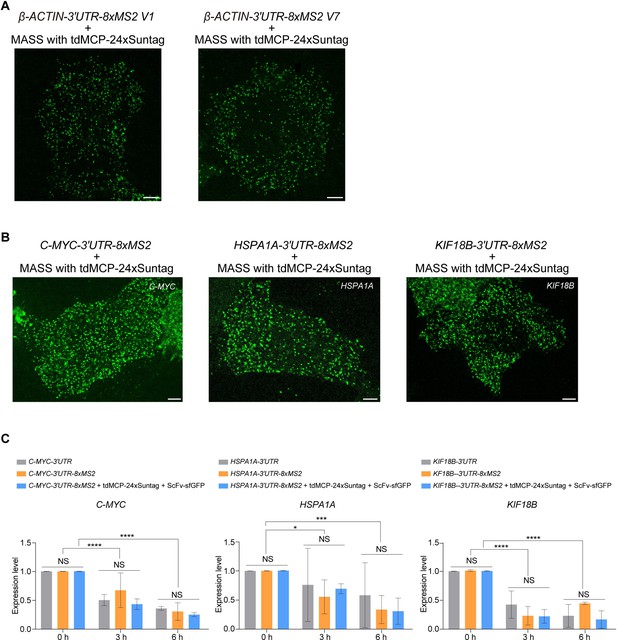
MASS did not affect the stability of mRNAs.
(A) Representative confocal images of β-ACTIN-8xMS2 V1 or β-ACTIN-8xMS2 V7 mRNAs labeled by MASS with tdMCP-24xSuntag. (B) Representative confocal images of C-MYC-8xMS2, HSPA1A-8xMS2, and KIF18B-8xMS2 mRNAs labeled by MASS with tdMCP-24xSuntag in HeLa cells. Scale bar, 5 µm. (C) Quantitative RT-PCR of the mRNA expression level of C-MYC, HSPA1A, and KIF18B jn HeLa cells transfected with the indicated constructs. Twelve hours after transfection, cells were treated with Actinomycin D (5 μM) for 0, 3, and 6 hr and harvested for RNA extraction. Primers for qPCR were listed in Methods. n = 3 independent experiments; bars indicate mean ± standard deviation (SD). Unpaired t-test and ordinary one-way analysis of variance (ANOVA). NS: not significant, *p < 0.05, ***p < 0.001, ****p < 0.0001. See also Figure 1—source data 1.
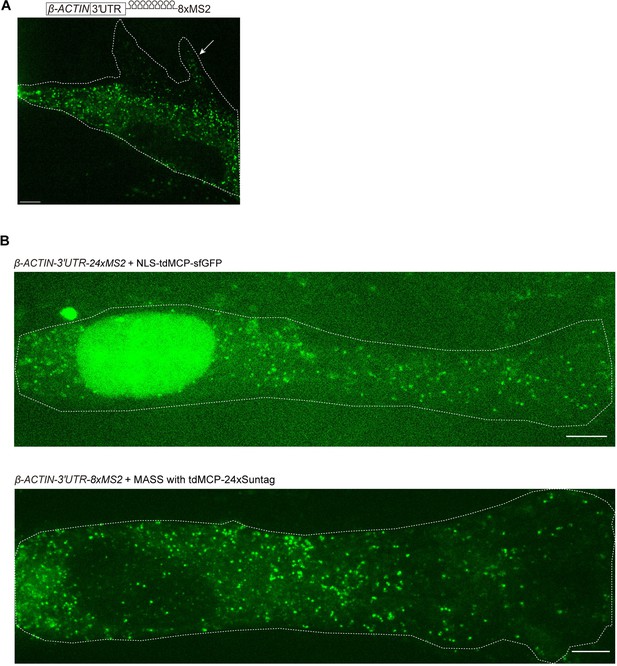
MASS did not affect mRNA subcellular localization.
(A) Confocal image of β-ACTIN-8xMS2 mRNA in live HeLa cells. sfGFP foci are shown. Constructs of β-ACTIN-8xMS2, MCP-24xSuntag, and scFv-sfGFP were cotransfected into HeLa cells. Images were taken 12 hr after transfection. The white dashed line demarcates the cell. The white arrow indicates lamellipodia. Scale bar, 5 µm. (B) Confocal images of β-ACTIN-24xMS2 mRNAs labeled by the conventional 24xMS2 system (top) and β-ACTIN-8xMS2 mRNAs labeled by MASS with tdMCP-24xSuntag (bottom) in live NIH/3T3 cells. The white dashed lines demarcate the cells. Scale bar, 5 µm.
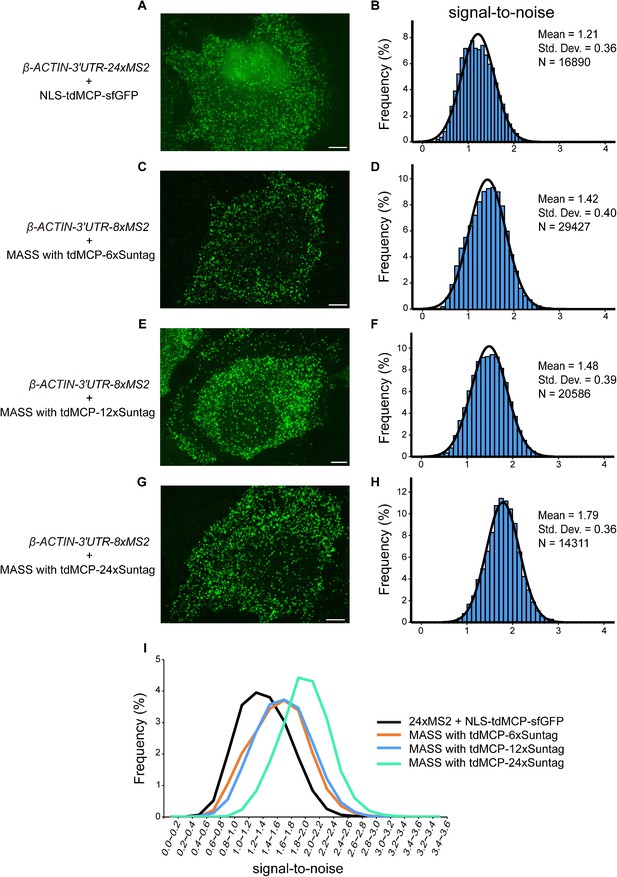
MASS has a higher signal-to-noise ratio than the conventional 24xMS2 method.
(A, C, E, G) Confocal images of β-ACTIN-24xMS2 mRNAs labeled by the conventional 24xMS2 system and β-ACTIN-8xMS2 mRNAs labeled by MASS with tdMCP-6xSuntag, tdMCP-12xSuntag, or tdMCP-24xSuntag. Images were taken 12 hr after transfection. Scale bar, 5 µm. (B, D, F, H, I) Distribution of the signal-to-noise ratio of foci of β-ACTIN mRNAs detected in A, C, E, G. For the conventional 24xMS2 system: N = 16,890 foci from 42 cells in three independent experiments. For MASS with tdMCP-6xSuntag: N = 29,427 foci from 46 cells in three independent experiments. For MASS with tdMCP-12xSuntag, N = 20,586 foci from 36 cells in three independent experiments. For MASS with tdMCP-24xSunta: N = 14,311 foci from 39 cells in three independent experiments. See also Figure 1—source data 1.
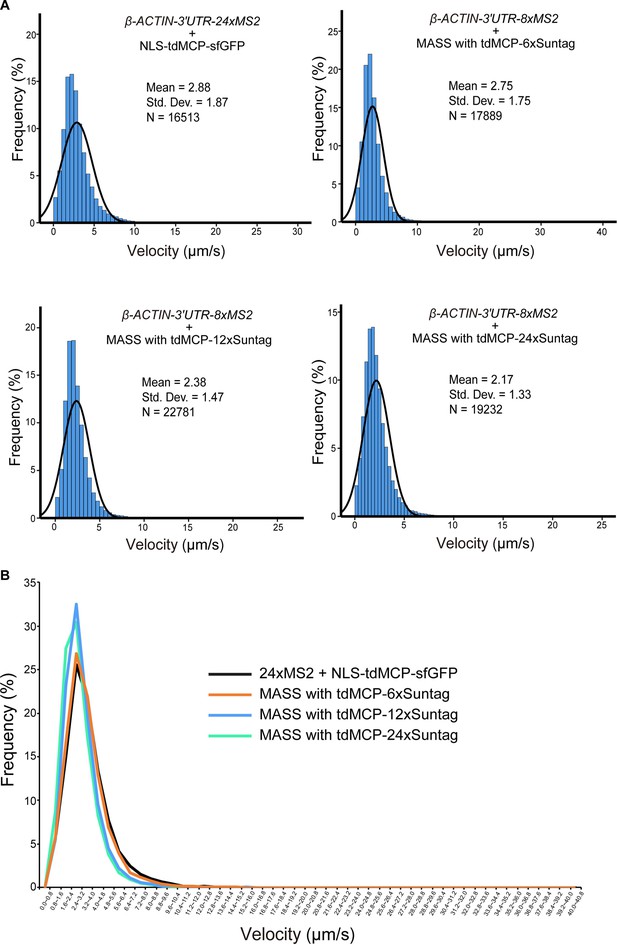
MASS with tdMCP-6xSuntag did not affect the speed of mRNA movement.
(A, B) Distribution of the velocity of mRNA movement of β-ACTIN-24xMS2 mRNAs labeled by the conventional 24xMS2 system and β-ACTIN-8xMS2 mRNAs labeled by MASS with tdMCP-6xSuntag, tdMCP-12xSuntag, or tdMCP-24xSuntag. For the conventional 24xMS2 system: N = 16,513 foci from 42 cells in three independent experiments. For MASS with tdMCP-6xSuntag: N = 17,889 foci from 54 cells in three independent experiments. For MASS with tdMCP-12xSuntag, N = 22,781 foci from 47 cells in three independent experiments. For MASS with tdMCP-24xSunta: N = 19,232 foci from 44 cells in three independent experiments. See also Figure 1—source data 1.
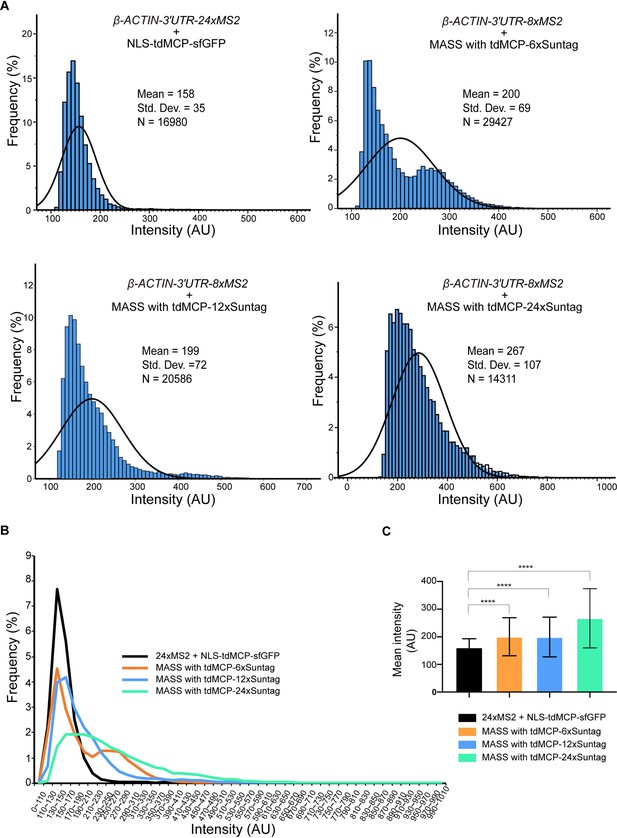
MASS labels mRNAs with a higher intensity than the conventional 24xMS2 imaging system.
(A, B) As Figure 1—figure supplement 5, but the distribution of the average intensity of each spot was shown. (C) Mean intensity of the average intensity of each spot in (A). Bars indicate mean ± standard deviation (SD). Unpaired t-test. ****p < 0.0001. See also Figure 1—source data 1.
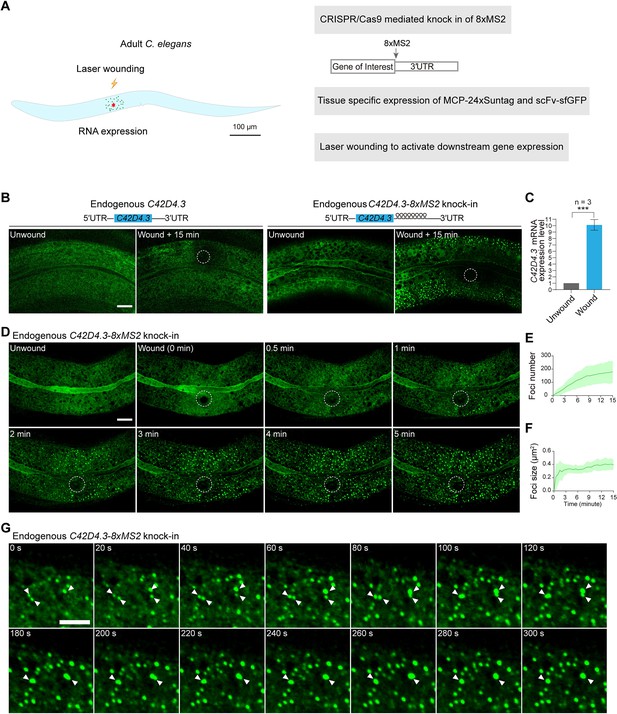
Live imaging of endogenous mRNAs in the epidermis of C. elegans using the MS2-based signal amplification with the Suntag system.
(A) Schematic of the strategy for live imaging of endogenous mRNAs in the epidermis of C. elegans. (B) Representative images of endogenous C42D4.3-8xMS2 mRNA in the epidermis of live C. elegans using the strategy described in (A). Left: C42D4.3 without 8xMS2. Right: C42D4.3 with 8xMS2. Images were taken before and 15 min after wounding. White dashed circles indicate wound sites. Scale bar, 10 μm. (C) Quantitative RT-PCR showing the expression level of endogenous C42D4.3 mRNA in C. elegans before and 15 min after wounding. n = 3 independent experiments; bars indicate mean ± standard deviation (SD). Mann–Whitney test, ***p < 0.001. See also Figure 2—source data 1. (D) Time-lapse imaging of endogenous C42D4.3-8xMS2 mRNA in the epidermis of live C. elegans before and after wounding. White dashed circles indicate the wound sites. Scale bar, 10 μm. Shown are mean ± SD of quantification of the number (E) and size (F) of sfGFP foci (endogenous C42D4.3 mRNA) formed in the epidermis as measured 15 min after wounding. n = 10. (G) Time-lapse imaging showing fusion of endogenous C42D4.3 foci (white arrows) after laser wounding. Scale bar, 5 μm.
-
Figure 2—source data 1
qRT-PCR (Quantitative Reverse Transcription PCR), number, and size of foci of mRNAs detected by MASS in C. elegans.
- https://cdn.elifesciences.org/articles/82178/elife-82178-fig2-data1-v3.xlsx
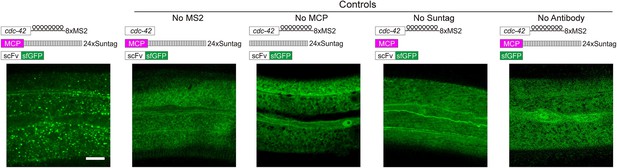
Live imaging of cdc42 mRNA in C. elegans using MS2-based signal Amplification with Suntag System.
Representative images of cdc42-8xMS2 mRNA in live C. elegans. sfGFP fluorescence signals are shown. Left panel: Constructs of cdc42-8xMS2, MCP-24xSuntag, and scFv-sfGFP were coexpressed in the epidermis of C. elegans. Right panels: Where one of the elements was removed as indicated. Scale bar, 10 µm.
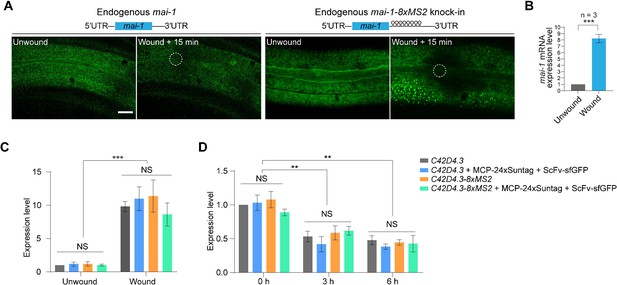
Live imaging of endogenous mRNA in the epidermis of C. elegans using the MS2-based signal amplification with the Suntag system.
(A) Representative images of endogenous mai-1-8xMS2 mRNA in the epidermis of live C. elegans using the strategy described in Figure 2A. Left: mai-1 without 8xMS2. Right: mai-1 with 8xMS2. Images were taken before and 15 min after wounding. White dashed circles indicate the wound sites. Scale bar, 10 μm. (B) Quantitative RT-PCR showing the expression level of endogenous mai-1 mRNA in C. elegans before and 15 min after wounding. n = 3 independent experiments; bars indicate mean ± standard deviation (SD). Mann–Whitney test, ***p < 0.001. See also Figure 2—source data 1. (C) Quantitative RT-PCR showing the expression level of endogenous C42D4.3 mRNA before and 15 min after wounding in wild-type, C42D4.3 8xMS2 knock-in animals and animals expressing the MASS imaging system. n = 3 independent experiments; bars indicate mean ± SD. One-way analysis of variance (ANOVA). NS: no significance, ***p < 0.001. See also Figure 2—source data 1. (D) Quantitative RT-PCR showing the expression level of endogenous C42D4.3 mRNA treatment with Actinomycin D (30 μM) for 0, 3, and 6 hr in wild-type, C42D4.3 8xMS2 knock-in animals and animals expressing the MASS imaging system. n = 3 independent experiments; bars indicate mean ± SD. One-way ANOVA. NS: not significant, **p < 0.01, ***p < 0.001. See also Figure 2—source data 1.
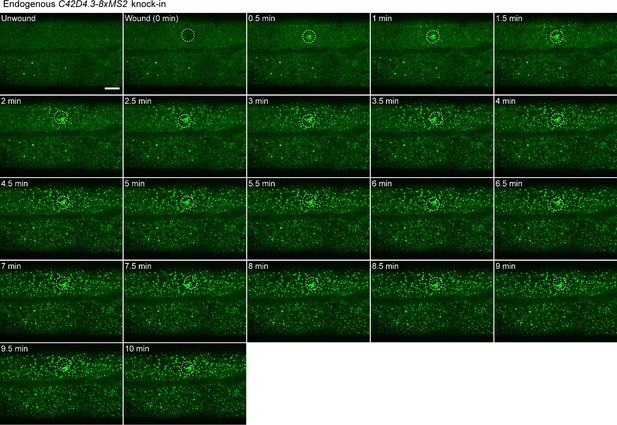
Fast activation and spreading of endogenous gene expression in the epidermis of C. elegans.
Time-lapse imaging of endogenous C42D4.3-8xMS2 mRNA in the epidermis of live C. elegans before and after wounding. White dashed circles indicate the wound sites. Scale bar, 10 μm.
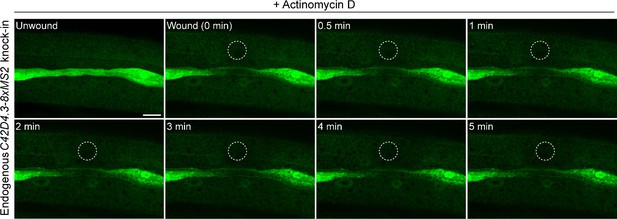
Treatment of Actinomycin D blocks the formation of C42D4.3 mRNA foci.
Time-lapse imaging of endogenous C42D4.3-8xMS2 mRNA in the epidermis of live C. elegans before and after wounding. C. elegans were treated with Actinomycin D (30 μM) for 3 hr before wounding. White dashed circles indicate the wound sites. Scale bar, 10 μm.
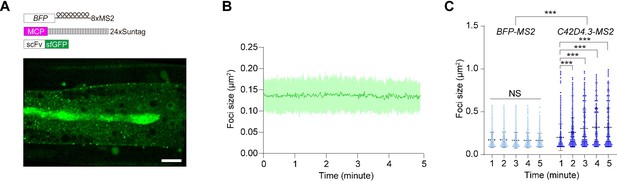
Foci of endogenous C42D4.3-8xMS2 mRNAs showed a larger size than that of BFP-8xMS2 mRNAs.
(A) Time-lapse imaging of BFP-8xMS2 mRNA in the epidermis of C. elegans using MASS with MCP-24xSuntag. Scale bar, 10 μm. (B) Quantification of the puncta size of the BFP-8xMS2 mRNA foci for 5 min, bars indicate the mean ± standard deviation (SD); n = 5. See also Figure 2—source data 1. (C) Quantification of the puncta size of the BFP-8xMS2 mRNA foci at 1, 2, 3, 4, and 5 min in unwounded animals and the puncta size of endogenous C42D4.3-8xMS2 mRNA foci at 1, 2, 3, 4, and 5 min after laser wounding. Bars indicate mean ± SD. One-way analysis of variance (ANOVA). NS: not significant, ***p < 0.001. At each time point, at least 550 mRNA foci from n = 10 worms for C42D4.3-8xMS2 and n = 5 worms for BFP-8xMS2 were analyzed. See also Figure 2—source data 1.
Videos
Time-lapse imaging of β-ACTIN-8xMS2 mRNA in live HeLa cells.
The sfGFP foci of the β-ACTIN-3′UTR-8xMS2 mRNA localize to the lamellipodia in HeLa cells. The white dashed line demarcates the cell. The white arrow indicates lamellipodia. Time interval, 2 s. Scale bar, 5 µm.
Time-lapse imaging of sfGFP foci of β-ACTIN-8xMS2 mRNA with a time interval of 1 s in HeLa cells.
Scale bar, 5 μm.
Time-lapse imaging showing fusion events of sfGFP foci of β-ACTIN-8xMS2 mRNA in HeLa cells where small sfGFP spots (white and yellow arrows) fuse into a single more prominent spot.
Time interval, 2 s. Scale bar, 5μm.
Time-lapse imaging showing fission events of sfGFP foci of β-ACTIN-8xMS2 mRNA in HeLa cells where large sfGFP foci split into smaller spots (white and yellow arrows).
Time interval, 1 s. Scale bar, 1 μm.
Time-lapse imaging showing transient interactions of an sfGFP spot (yellow arrow) between two spots (white arrows) of β-ACTIN-8xMS2 mRNA in HeLa cells.
Time interval, 2 s. Scale bar, 1 μm.
Time-lapse imaging in HeLa cells showing an sfGFP spot of β-ACTIN-8xMS2 mRNA showing no movement over a 10-s period.
Time interval, 1 s. Scale bar, 1 μm.
Time-lapse imaging of endogenous C42D4.3-8xMS2 mRNA dynamics in the epidermis of C. elegans after laser wounding.
Time interval, 5 s. Scale bar, 10 μm.
Time-lapse imaging of endogenous C42D4.3-8xMS2 mRNA dynamics in the epidermis of C. elegans after laser wounding.
A different worm was shown. Time interval, 5 s. Scale bar, 10 μm.
Time-lapse imaging of endogenous mai-1-8xMS2 mRNA dynamics in the epidermis of C. elegans after laser wounding.
Time interval, 2 s. Scale bar, 10 μm.
Time-lapse imaging showing fusion events of sfGFP foci of endogenous C42D4.3-8xMS2 mRNA in the epidermis of C. elegans where small sfGFP spots (white arrows) fused into a more prominent spot.
Time interval, 5 s. Scale bar, 5 μm.
Time-lapse imaging of exogenous BFP-8xMS2 mRNA dynamics in the epidermis of C. elegans.
Time interval, 0.5 s. Scale bar, 10 μm.





