Large vesicle extrusions from C. elegans neurons are consumed and stimulated by glial-like phagocytosis activity of the neighboring cell
Figures
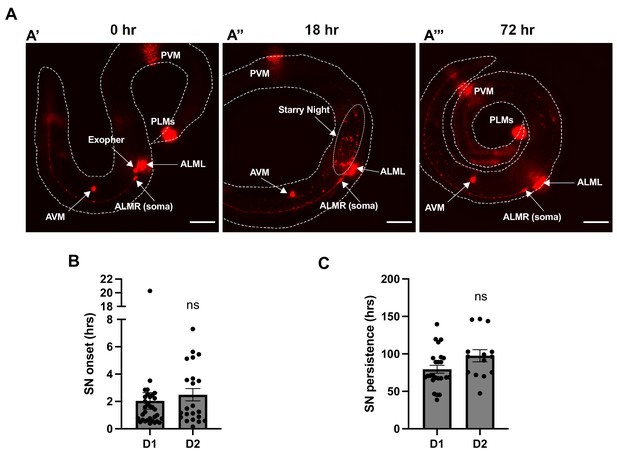
Longitudinal analysis indicates that exophers vesiculate prior to degradation of exopher-derived cargo.
(A) Sequential images captured on a fluorescence dissecting microscope are shown for the same animal at different timepoints. The six touch receptor neurons are marked by mCherry expression. Images focus on the ALMR neuron near the center of the body. The images show exopher production (A’), followed by exopher vesiculation (A”), and finally loss of exopher-derived mCherry signal (A’’’). ALMR, anterior lateral microtube neuron right; ALML, anterior lateral microtube neuron left; AVM, anterior ventral microtube neuron; PVM, posterior ventral microtube neuron; PLM, posterior lateral microtube neuron. (B) Time from exopher identification to first observation of exopher vesiculation is graphed. Each data point represents an individual tracked animal. Data from adult day 1 (D1) exophers and adult day 2 (D2) exophers are graphed separately. SN = starry night. (C) Time from the start of exopher vesiculation to the observed loss of exopher-derived mCherry signal is graphed. Each data point represents an individual tracked animal. Data from adult day 1 (D1) exophers and adult day 2 (D2) exophers are graphed separately. Scale bar = 50 μm.
-
Figure 1—source data 1
Numerical data for Figure 1.
- https://cdn.elifesciences.org/articles/82227/elife-82227-fig1-data1-v2.docx
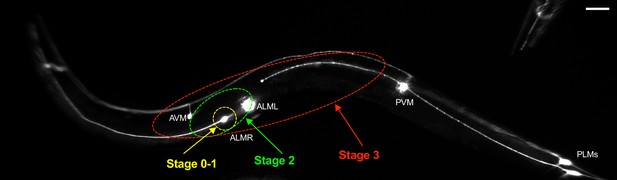
Three exopher zones.
Diagram depicting three zones used to divide exophers into three categories, early (stage 1) within one soma length, fully formed (stage 2) within two soma lengths, and exophers that had moved through the hypodermis to sites distant from the soma (stage 3).
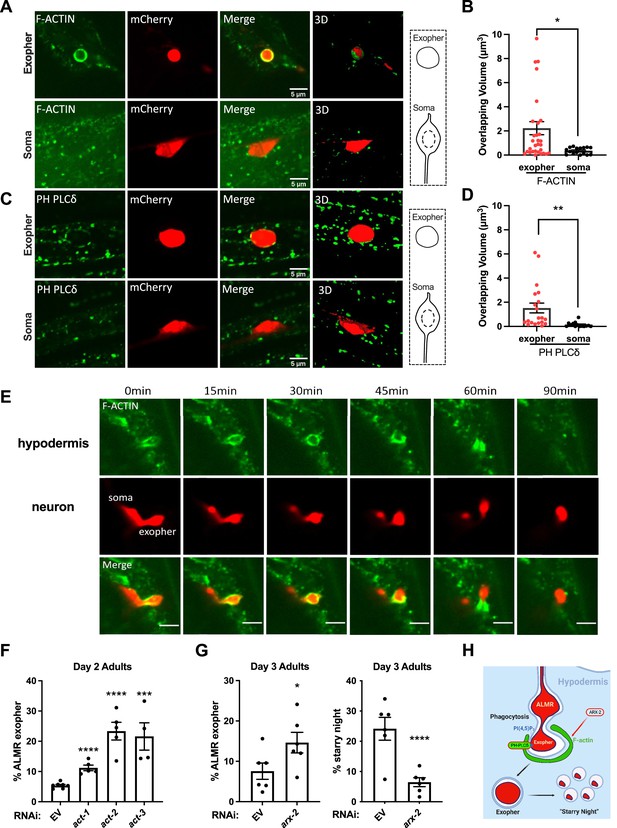
Neuronal exophers are phagocytosed by the adjacent hypodermis in an F-actin-dependent manner.
(A, C) Confocal fluorescence micrographs are shown for an mCherry-labeled ALMR neuron-derived exopher or the similar-sized mCherry-labeled ALMR neuronal soma from the same neuron. Surrounding hypodermis-specific expression of UtrCH::mNeonGreen (an F-actin biosensor) and PH(PLCδ)::mNeonGreen (a biosensor for the lipid PI(4,5)P2) expressed from the hypodermis-specific semo-1 promoter are shown. A merged image and a merged 3-D projection are shown for each example. Scale bar, 5 µm. (B, D) As a measure of hypodermal marker recruitment to the exopher, we measured the volume of overlapping signal in 3-D projections between the hypodermal marker and ALMR-neuron-derived exopher, comparing to the ALMR neuronal soma as a control, *p<0.05 by two-tailed unpaired t-test. N = 24 (F-actin) and N = 18 (PI(4,5)P2). (E) Time-lapse images of hypodermal F-actin dynamics during the engulfment of the exopher. ‘0 min’ indicates the beginning of observed time course. Scale bar, 5 µm. (F) Histogram depicting percentage of exopher-positive animals upon hypodermal-specific RNAi for empty vector control (L4440), act-1, act-2, or act-3 in day 2 adults. ****p<0.0001, *p<0.05 calculated using the Cochran–Mantel Haenszel (CMH) test. Each point represents a trial of n = 50 animals scored, with four trials per condition. (G) Histogram depicting exopher and starry night-positive animal frequency after hypodermal-specific RNAi for empty vector (EV) control, or arx-2 (ARP2/3 subunit) in day 3 adults. ****p<0.0001, *p<0.05 calculated using the CMH test. Each point represents a trial of N = 50 animals scored, with six trials per condition. (H) Diagram summarizing interpretation of events, including branched actin-dependent engulfment of ALMR-neuron exophers by the hypodermis, a prerequisite for later vesiculation of the completed phagosome.
-
Figure 2—source data 1
Numerical data for Figure 2.
- https://cdn.elifesciences.org/articles/82227/elife-82227-fig2-data1-v2.docx
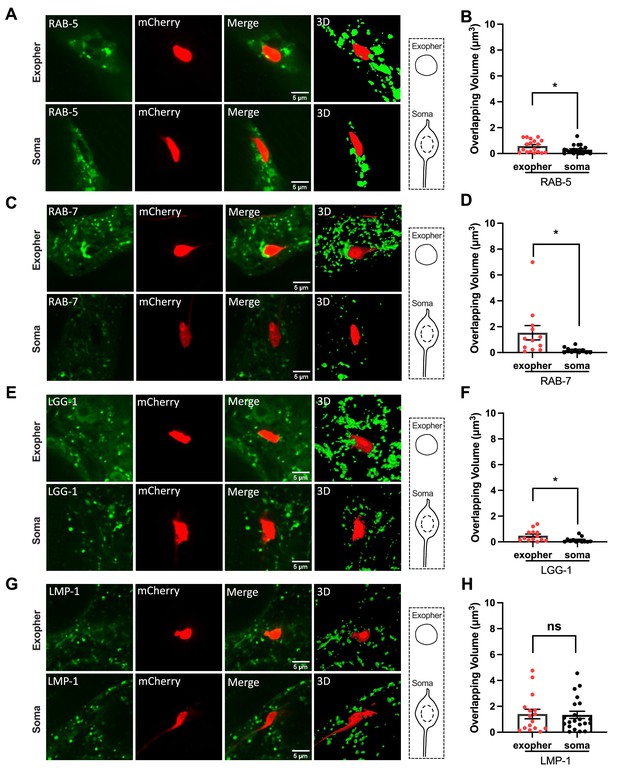
Hypodermal endosomes and autophagosomes, but not lysosomes, begin recruitment prior to exopher-laden phagosome vesiculation.
(A, C, E, G) Fluorescence micrographs are shown for an mCherry-labeled ALMR neuron-derived exopher, or the similarly sized mCherry-labeled ALMR neuronal soma from the same neuron. Surrounding hypodermis-specific expression of (A) mNeonGreen::RAB-5 (an early endosome marker), (C) mNeonGreen::RAB-7 (a late endosome marker), (E) mNeonGreen::LGG-1/LC3 (an autophagosome marker), and (G) LMP-1::mNeonGreen (a lysosome marker), driven by the hypodermis-specific semo-1 promoter, are shown. A merged image and a merged 3-D projection are shown for each example. Scale bar, 5 µm. (B, D, F, H) As a measure of hypodermal marker recruitment to the exopher, we measured the volume of overlapping signal between the hypodermal marker and ALMR-neuron derived exopher, comparing to the ALMR neuronal soma as a control, *p<0.05 by two-tailed unpaired t-test. N = 13 (LGG-1), N = 20 (RAB-5), N = 12 (RAB-7), and N = 21 (LMP-1).
-
Figure 3—source data 1
Numerical data for Figure 3.
- https://cdn.elifesciences.org/articles/82227/elife-82227-fig3-data1-v2.docx
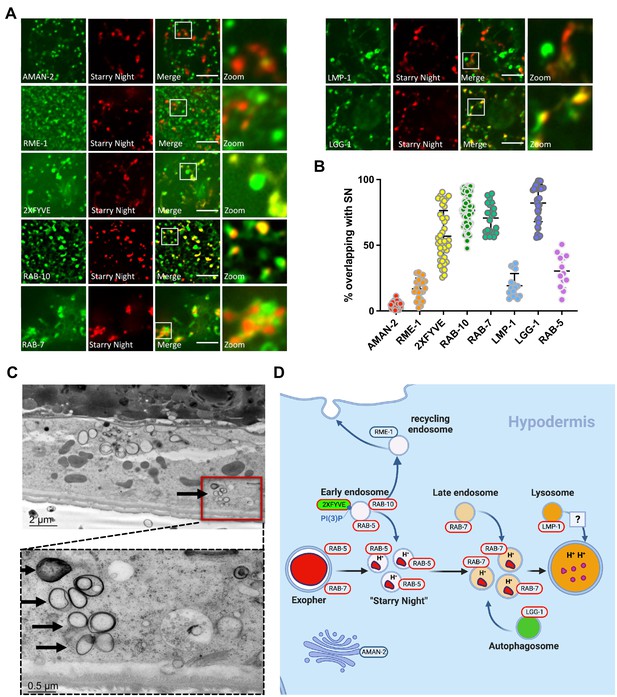
Starry night vesicles are exopher-phagosome vesiculation products that further fuse with hypodermal endosomes.
(A) Fluorescence micrographs show ALMR-exopher-derived mCherry labeling exopher-laden phagosome vesiculation products. Hypodermis-specific expression of AMAN-2::mNeonGreen (a Golgi marker), mNeonGreen::RME-1 (a recycling endosome marker), mNeonGreen::2XFYVE (an early endosome/phagosome marker and PI(3)P biosensor), mNeonGreen::RAB-10 (an early phagosome/endosome marker and recycling regulator), mNeonGreen::RAB-7 (a late endosome/phagosome marker), LMP-1::mNeonGreen (a lysosome marker), and mNeonGreen::LGG-1/LC3 (an autophagosome marker), each driven by the hypodermis-specific semo-1 promoter, are shown. Scale bar, 5 µm. (B) Graph quantifying colocalization of each marker with starry night exopher-phagosome vesiculation products. AMAN-2, N = 22; RME-1, N = 20; 2XFYVE; N = 28; RAB-10, N = 30; RAB-7, N = 21; LMP-1 N = 17; LGG-1, N = 22, RAB-5, N = 12. (C) Thin section electron micrograph showing candidate phagosome-derived vesicles (arrows) within the hypodermis near the ALMR soma. (D) Diagram summarizing interpretation of events, including early endosome, late endosome, and autophagosome fusion with exopher-laden phagosome vesiculation products.
-
Figure 4—source data 1
Numerical data for Figure 4.
- https://cdn.elifesciences.org/articles/82227/elife-82227-fig4-data1-v2.docx
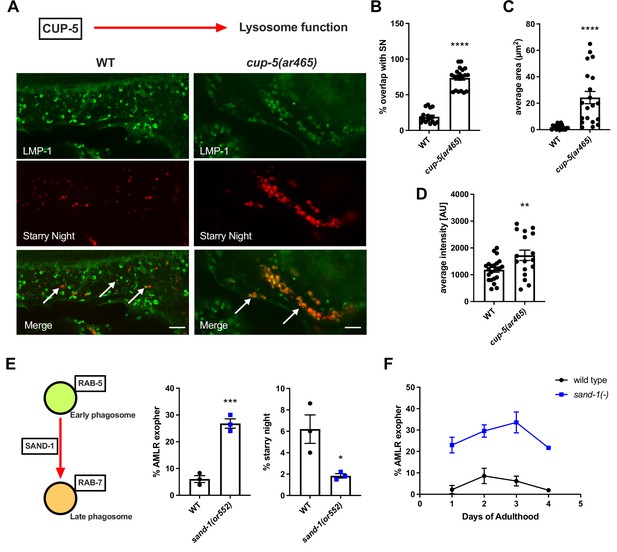
Lysosome function is required for the end-stage degradation of phagocytosed exopher cargo.
(A) Fluorescence micrographs show ALMR-exopher-derived mCherry labeling exopher-phagosome vesiculation products (starry night) in red, with hypodermis-specific expression of LMP-1::mNeonGreen (a lysosome marker) shown in green. Wild-type and cup-5(ar465) mutant images are shown. Arrows indicate LMP-1:mNG that colocalized with neuron-derived mCherry vesicles. Scale bar, 5 µm. (B) Histogram quantifying colocalization of the LMP-1 lysosome marker with ALMR-exopher-derived vesicles in wild-type and cup-5 mutant. (C) Histogram quantifying the average size of ALMR-exopher-derived vesicles in wild-type and cup-5 mutant. (D) Histogram quantifying average the florescence intensity of mCherry signal in ALMR-exopher-derived vesicles in wild-type and cup-5 mutant. (B–D) N = 17 wild-type, N = 20 cup-5(ar465) ****p<0.0001, **p<0.01 by two-tailed unpaired t-test. (E) Histogram quantifying exopher and starry night numbers in wild-type and sand-1(or552) mutant **** p<0.0001, *p<0.05 by the Cochran–Mantel Haenszel (CMH) test, N = 50 per trial over three trials. (F) Line graph quantifying exopher numbers at different days of adulthood in wild-type and sand-1(or552) mutant. N = 50 per trial over three trials.
-
Figure 5—source data 1
Numerical data for Figure 5.
- https://cdn.elifesciences.org/articles/82227/elife-82227-fig5-data1-v2.docx

ARL-8 promotes the vesiculation of exophers-laden phagosomes.
(A) Histogram depicting the frequency ratio of animals displaying day 3 starry night vesicles to those positive for intact exopher-phagosomes on day 2 comparing wild-type and arl-8(wy271) hypomorphic mutants. A reduced ratio suggests delayed vesiculation. **p<0.01 by two-tailed unpaired t-test, n = 10 over three trials. (B) Line graph showing exopher numbers on different days of adulthood in wild-type and arl-8 mutant. ****p<0.0001, ***p<0.001, *p<0.05 by the Cochran–Mantel Haenszel (CMH) test, n = 50 over three trials.
-
Figure 5—figure supplement 1—source data 1
Numerical data for Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/82227/elife-82227-fig5-figsupp1-data1-v2.docx
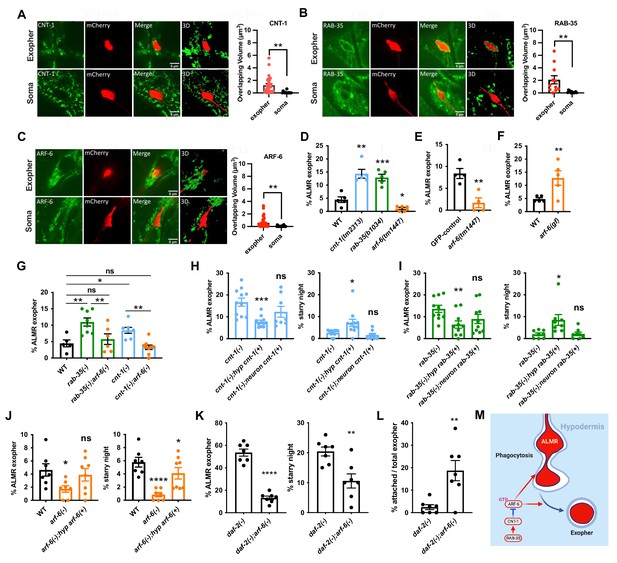
Hypodermal ARF-6 influences neuronal exopher production and interacts with CNT-1 and RAB-35 to complete hypodermal phagocytosis.
(A–C) Fluorescence micrographs showing hypodermally expressed CNT-1/ARF-GAP, GTPase RAB-35, and GTPase ARF-6 associated with the periphery of intact exophers. Histograms indicate overlapping volume of hypodermal marker with exopher and neuronal soma periphery, **p<0.01 by two-tailed unpaired t-test. CNT-1 N = 22; RAB-35 N = 12; ARF-6 N = 24. Scale bar, 5 µm. (D) Histogram depicting exopher frequency in wild-type, cnt-1(tm2313), rab-35(b1034), and arf-6(tm1447) mutants. ***p<0.001, **p<0.01, *p<0.05 by the Cochran–Mantel Haenszel (CMH) test, n = 50 over five trials. (E) Histogram depicting exopher frequency in animals expressing touch neuron-specific GFP from the mec-17 promoter (uIs31), compared with uIs31; arf-6(tm1447) animals. **p<0.01 by the CMH test, N = 50 per trial over four trials. (F) Histogram depicting exopher frequency in wild-type and arf-6(ns388) gain-of-function mutant. **p<0.01 by the CMH test, N = 50 per trial over five trials. (G) Histogram depicting exopher frequency in wild-type, cnt-1 and rab-35 single mutants and double mutants with arf-6(tm1447). **p<0.01, *P<0.05 by the CMH test, N = 50 per trial over five trials. (H–J) Rescue experiments showing exopher and starry night frequency rescue by hypodermis-specific expression of rab-35, cnt-1, and arf-6 in their cognate mutants. (K) Histogram depicting exopher and starry night numbers in daf-2 mutant and in the double mutant with daf-2 and arf-6. ****p<0.0001, **p<0.01 by the CMH test, N = 50, over seven trials. (L) Histogram depicting attached exopher rate in daf-2(e1370) mutant background to potentiate baseline exophergenesis and double mutant with arf-6. **p<0.01 by two-tailed unpaired t-test. N = 50 per trial over seven trials. ****p<0.0001, ***p<0.001, **p<0.01, *p<0.05 by the CMH test, N = 50 per trial. (M) Diagram indicating hypodermal ARF-6 influence on exopher budding and engulfment, and negative regulation of ARF-6 by CNT-1 and RAB-35.
-
Figure 6—source data 1
Numerical data for Figure 6.
- https://cdn.elifesciences.org/articles/82227/elife-82227-fig6-data1-v2.docx
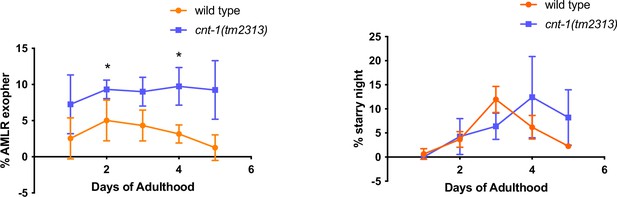
CNT-1 is required for exopher-laden phagosome maturation.
Line graph depicting exopher and starry night frequency on different days of adulthood in wild-type and cnt-1(tm2313) mutants. N = 50 per trial with three trials.
-
Figure 6—figure supplement 1—source data 1
Numerical data for Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/82227/elife-82227-fig6-figsupp1-data1-v2.docx
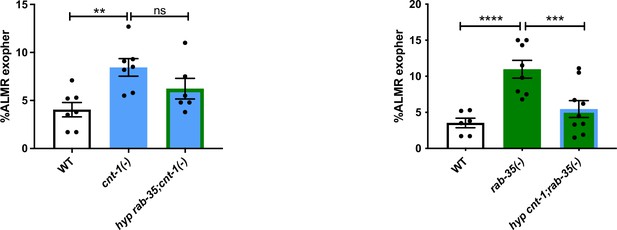
Hypodermal overexpression of CNT-1 can suppress exopher accumulation due to loss of RAB-35, but not vice versa.
Histograms depict the results of exopher frequency experiments testing the ability of hypodermis-specific overexpression of rab-35(+) to suppress cnt-1(tm2313), and hypodermis-specific overexpression of cnt-1(+) to suppress rab-35(b1034). ****p<0.0001, ****p<0.001, *p<0.05 by the Cochran–Mantel Haenszel (CMH) test, N = 50 day 2 animals per trial over six trials.
-
Figure 6—figure supplement 2—source data 1
Numerical data for Figure 6—figure supplement 2.
- https://cdn.elifesciences.org/articles/82227/elife-82227-fig6-figsupp2-data1-v2.docx
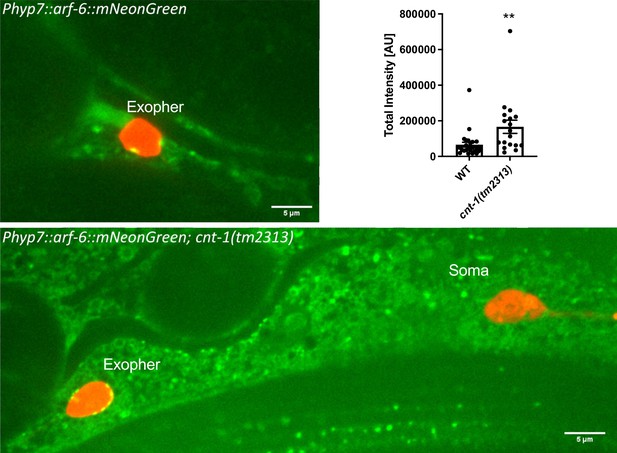
Hypodermal ARF-6 puncta decorate the periphery of engulfed exophers and are increased upon loss of CNT-1.
Fluorescence micrographs displaying mCherry-labeled ALMR exophers and/or soma and hypodermally expressed ARF-6::mNeonGreen in wild-type and cnt-1(tm2313) mutant backgrounds. Histogram depicts the total intensity of hypodermal ARF-6::mNeonGreen signal around the exopher periphery, comparing wild-type and cnt-1(tm2313) mutants. **p<0.01 by two-tailed unpaired t-test. n = 26 (WT) and n = 18 (cnt-1 mutant). Scale bar, 5 µm.
-
Figure 6—figure supplement 3—source data 1
Numerical data for Figure 6—figure supplement 3.
- https://cdn.elifesciences.org/articles/82227/elife-82227-fig6-figsupp3-data1-v2.docx
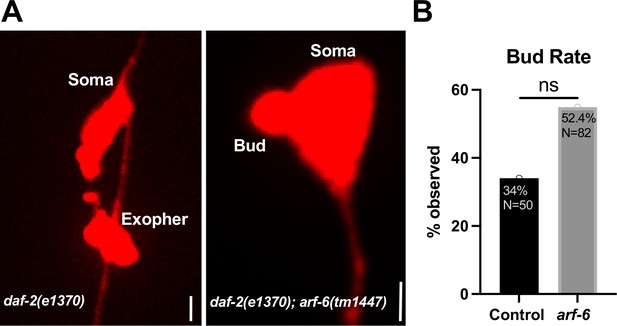
arf-6 mutants maintain early ALM budding.
(A) Fluorescence micrographs show a successful exopher produced in the daf-2(e1370) mutant, and an apparent arrested bud in a daf-2(e1370); arf-6(tm1447) double mutant. Scale Bar = 2.8 µm (B) Histograms depict the results of bud frequency observed in daf-2(e1370) (Control) and daf-2; arf-6(tm1447) double mutants (arf-6). NS = p>0.05 by the Cochran–Mantel Haenszel (CMH) test.
-
Figure 6—figure supplement 4—source data 1
Numerical data for Figure 6—figure supplement 4.
- https://cdn.elifesciences.org/articles/82227/elife-82227-fig6-figsupp4-data1-v2.docx
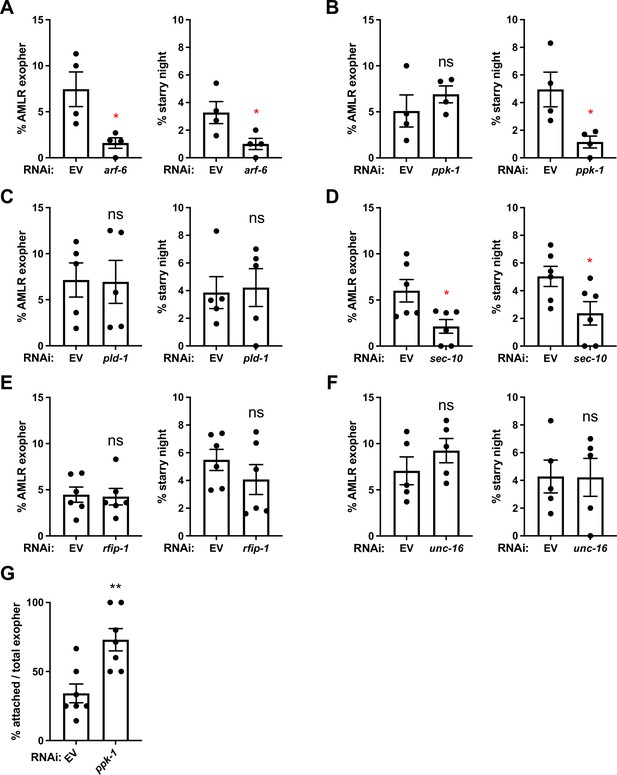
Hypodermis-specific RNAi identifies SEC-10/exocyst and PPK-1/PI-5 kinase as likely ARF-6 effectors in exopher production and phagocytosis.
(A–F) Histograms depict exopher and starry night numbers after hypodermis-specific RNAi for candidate ARF-6 effectors. EV indicates empty vector control, arf-6 encodes GTPase ARF-6/Arf6, ppk-1 encodes phosphatidylinositol-4-phosphate 5-kinase, rfip-1 encodes the C. elegans homolog of Rab11-FIP3/Arfophilin, pld-1 encodes the C. elegans homolog of phospholipases D1 and D2, unc-16 encodes the C. elegans homolog of C-Jun-amino-terminal kinase-interacting protein 3 (JIP3), and sec-10 encodes the C. elegans homolog of Exocyst subunit Sec10. **p<0.01, *p<0.05 by the Cochran–Mantel Haenszel (CMH) test, N = 50 per trial over four trials. (G) Histogram depicting attached exopher rate in hypodermis-specific RNAi for ppk-1. **p<0.01 by two-tailed unpaired t-test. N = 50 per trial over seven trials.
-
Figure 7—source data 1
Numerical data for Figure 7.
- https://cdn.elifesciences.org/articles/82227/elife-82227-fig7-data1-v2.docx
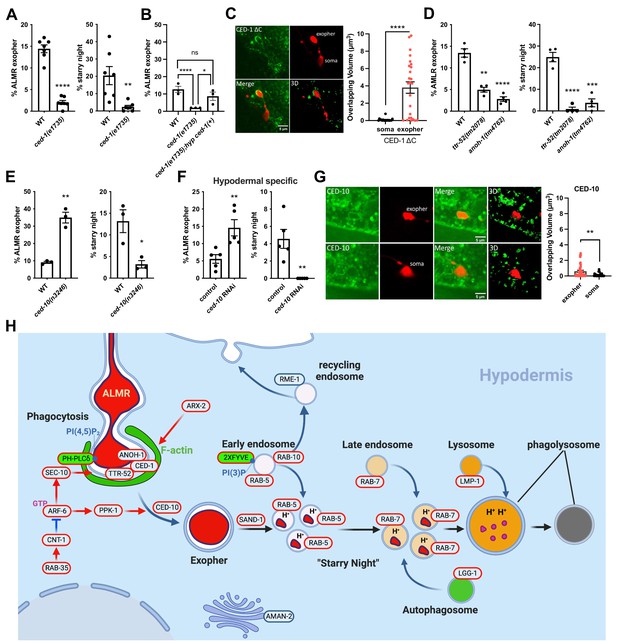
CED-1 functions in exopher recognition and exopher production.
(A) Histogram depicting exopher and starry night numbers in wild-type and ced-1(e1735) mutant. N = 22. ****p<0.0001, **p<0.01 by the Cochran–Mantel Haenszel (CMH) test, N = 50 per trial over seven trials. (B) Histogram depicting exopher numbers in wild-type, ced-1(e1735), and ced-1(e1735) expressing CED-1(+) from a hypodermis-specific promoter. N = 22. ****p<0.0001, **p<0.01 by the CMH test, N = 50 per trial over three trials. (C) Fluorescence micrographs showing intact exophers interacting with hypodermal CED-1ΔC::GFP. Scale bar, 5 µm. Histogram depicts overlapping volume of hypodermal CED-1ΔC::GFP with exopher. ****p<0.0001 by two-tailed unpaired t-test. (D) Histogram depicting exopher and starry night numbers in wild-type, ttr-52(tm2078), and anoh-1(tm4762) mutants. ****p<0.0001, ***p<0.001, **p<0.01 by the CMH test, N = 50 per trial over four trials. (E) Histogram depicting exopher and starry night numbers in wild-type and ced-10(n3246) mutants. ****p<0.0001, **p<0.01 by the CMH test, N = 50 per trial over three trials. (F) Histogram depicting exopher and starry night numbers after hypodermis-specific knockdown in empty vector control and ced-10(RNAi) animals. ****p<0.0001, **p<0.01 by the CMH test, N = 50 per trial over three trials. (G) Fluorescence micrographs showing intact exophers interacting with hypodermal mNeonGreen::CED-10. Scale bar, 5 µm. Histogram depicts overlapping volume of hypodermal mNeonGreen::CED-10 with exopher and soma. **p<0.01 by two-tailed unpaired t-test. (H) Model of ALMR-neuron derived exopher recognition, engulfment, and processing by the surrounding hypodermis.
-
Figure 8—source data 1
Numerical data for Figure 8.
- https://cdn.elifesciences.org/articles/82227/elife-82227-fig8-data1-v2.docx
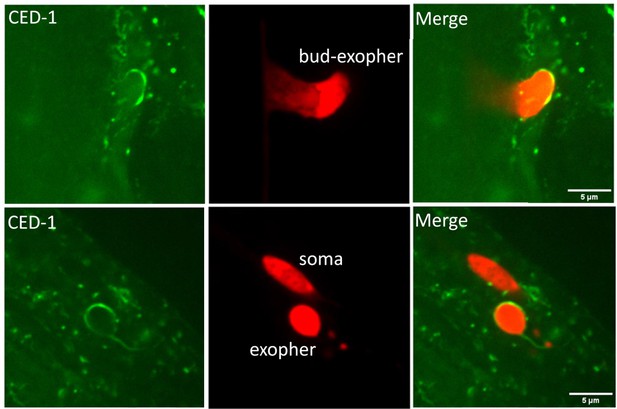
Hypodermal CED-1 distinguishes between exopher and soma.
Fluorescence micrographs display an early stage, or fully formed but attached, ALMR-derived exophers decorated by hypodermal CED-1ΔC::GFP. Note lack of CED-1 labeling of attached neuronal soma. Scale bar, 5 µm.

arf-6 and ced-1 mutants display reduced touch sensitivity in old age.
Synchronized WT, arf-6(tm1447), and ced-1(e1735) animals were touch tested on adult days 5 and 10 and scored for reversal of direction. Three biological replicates of 30 animals/replicate.
-
Figure 8—figure supplement 2—source data 1
Numerical data for Figure 8—figure supplement 2.
- https://cdn.elifesciences.org/articles/82227/elife-82227-fig8-figsupp2-data1-v2.docx
Videos
Exophers tubulate and vesiculate to form starry night vesicles.
mCherry-filled ALMR neuron-derived exopher tubulates and vesiculates after release from the neuron. Video depicts a 3 hr 17 min time course, 5 min per frame. Scale Bar = 3 µm.
Hypodermal F-actin dynamics mark engulfment of the neuronal exopher.
An mCherry-filled ALMR neuron soma is shown during exopher production, with hypodermis-specific expression of mNeonGreen::UtrCH marking F-actin accumulation and dynamics during engulfment. Video depicts a 90 min time course, 15 min per frame. Scale Bar = 3 µm.
Exopher budding in a daf-2 mutant.
An mCherry-filled ALMR neuron soma in a daf-2(e1370) mutant is shown during exopher production. Video depicts a 4 hr time course, 4 min per frame. Scale Bar = 3 µm.
Early bud formation in daf-2; arf-6 double mutant.
An mCherry-filled ALMR neuron soma in a daf-2(e1370); arf-6(tm1447) double mutant is shown producing a small bud from the soma. Video depicts a 4 hr time course, 4 min per frame. Scale Bar = 3 µm.
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/82227/elife-82227-mdarchecklist1-v2.docx
-
Supplementary file 1
Table of strains used in this study.
- https://cdn.elifesciences.org/articles/82227/elife-82227-supp1-v2.docx





