APE1 recruits ATRIP to ssDNA in an RPA-dependent and -independent manner to promote the ATR DNA damage response
Figures
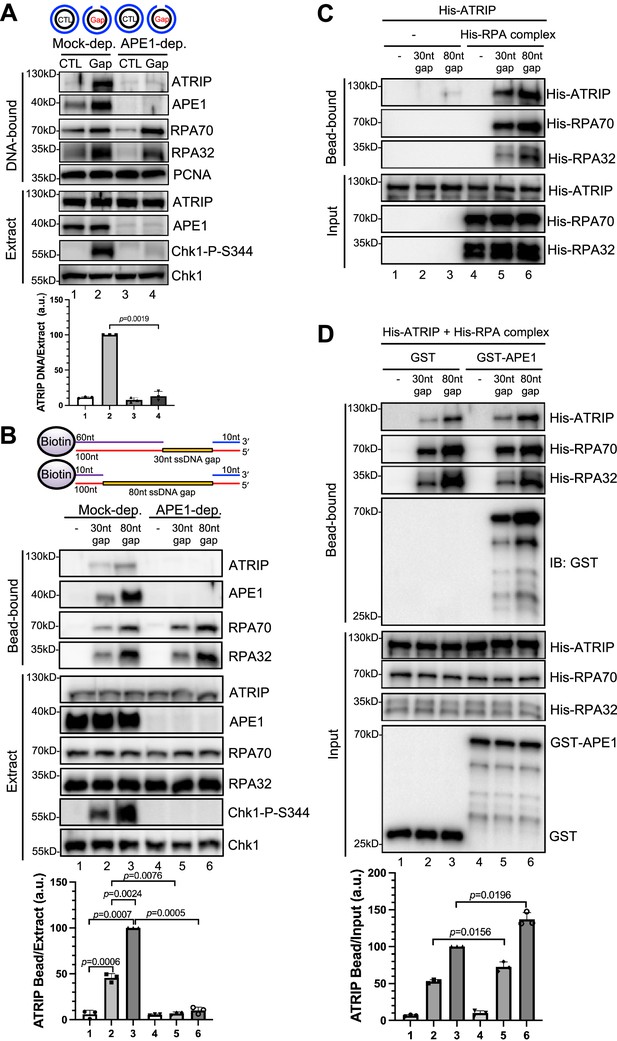
AP endonuclease 1 (APE1) is required for the recruitment of ATRIP to RPA-coated single-stranded DNA (ssDNA) in Xenopus egg extracts.
(A) CTL (control) or Gap plasmid was added to Mock- or APE1-depleted high-speed supernatant (HSS) and incubated for 30 min. The DNA-bound fractions and total egg extract were examined via immunoblotting analysis as indicated. (B) Streptavidin beads coupled with equal moles of biotin-labeled double-stranded DNA (dsDNA) with ssDNA gap structures (30 nt or 80 nt) were added to Mock- or APE1-depleted HSS. After incubation for 30 min at room temperature, the DNA-bound fractions and total egg extract were examined via immunoblotting analysis as indicated. (C) Streptavidin beads coupled with equal moles of biotin-labeled dsDNA with ssDNA gap structures (30 nt or 80 nt) were added to an interaction buffer containing purified His-ATRIP protein with/without His-RPA protein. After incubation for 30 min at room temperature, the DNA-bound fractions and the input were examined via immunoblotting analysis. (D) Streptavidin beads coupled with equal moles of biotin-labeled dsDNA with ssDNA gap structures (30 nt or 80 nt) were added to an interaction buffer containing His-ATRIP and His-RPA, which was supplemented with GST or GST-APE1. After incubation for 30 min at room temperature, the DNA-bound fractions and the input were examined via immunoblotting analysis. (A, B, D) ATRIP intensity was quantified, and the ratio of ATRIP from DNA-bound vs extract/input was examined. a.u., arbitrary unit. Mean ± SD, n=3.
-
Figure 1—source data 1
Raw images of immunoblotting analysis referenced in Figure 1A.
- https://cdn.elifesciences.org/articles/82324/elife-82324-fig1-data1-v1.zip
-
Figure 1—source data 2
Raw images of immunoblotting analysis referenced in Figure 1B.
- https://cdn.elifesciences.org/articles/82324/elife-82324-fig1-data2-v1.zip
-
Figure 1—source data 3
Raw images of immunoblotting analysis referenced in Figure 1C.
- https://cdn.elifesciences.org/articles/82324/elife-82324-fig1-data3-v1.zip
-
Figure 1—source data 4
Raw images of immunoblotting analysis referenced in Figure 1D.
- https://cdn.elifesciences.org/articles/82324/elife-82324-fig1-data4-v1.zip
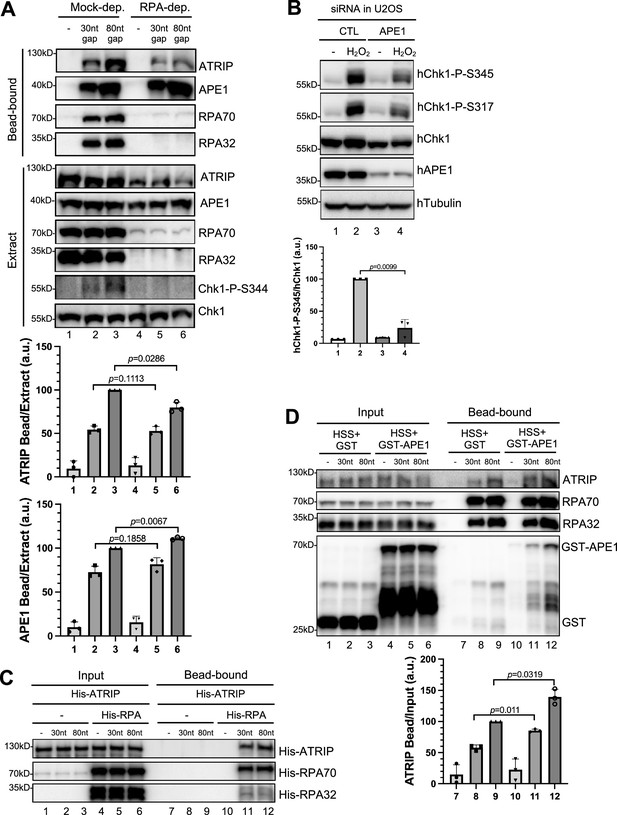
AP endonuclease 1 (APE1) further promote ATRIP’s recruitment to RPA-coated DNA.
(A) Streptavidin beads coupled with equal moles of biotin-labeled double-stranded DNA (dsDNA) with single-stranded DNA (ssDNA) gap structures (30 nt or 80 nt) were added to Mock- or RPA-depleted high-speed supernatant (HSS). After incubation for 30 min at room temperature, the DNA-bound fractions and total egg extract were examined via immunoblotting analysis as indicated. (B) siRNA-mediated APE1 knockdown impaired Chk1 phosphorylation at Ser345 and at Ser317 induced by H2O2 (1.25 mM) in U2OS cells. Total cell lysates were extracted and analyzed via immunoblotting analysis as indicated. (C) Streptavidin beads coupled with same amount of biotin-labeled dsDNA with ssDNA gap structures (30 nt or 80 nt) were added to an interaction buffer containing His-ATRIP, which was supplemented with/without His-RPA. DNA-bound fractions and total extract samples were examined via immunoblotting analysis as indicated. (D) Streptavidin beads coupled with equal moles of biotin-labeled dsDNA with ssDNA gap structures (30 nt or 80 nt) were added to HSS, which was supplemented with GST or WT GST-APE1. DNA-bound fractions and total extract samples were examined via immunoblotting analysis as indicated. (A, B, D) The intensity of ATRIP, APE1, hChk1-P-S345, or hChk1 was quantified, and the ratio of ATRIP or APE1 DNA-bound vs extract/input, or hChk1-P-S345 vs hChk1 was examined. a.u., arbitrary unit. Mean ± SD, n=3.
-
Figure 1—figure supplement 1—source data 1
Raw images of immunoblotting analysis referenced in Figure 1—figure supplement 1A.
- https://cdn.elifesciences.org/articles/82324/elife-82324-fig1-figsupp1-data1-v1.zip
-
Figure 1—figure supplement 1—source data 2
Raw images of immunoblotting analysis referenced in Figure 1—figure supplement 1B.
- https://cdn.elifesciences.org/articles/82324/elife-82324-fig1-figsupp1-data2-v1.zip
-
Figure 1—figure supplement 1—source data 3
Raw images of immunoblotting analysis referenced in Figure 1—figure supplement 1C.
- https://cdn.elifesciences.org/articles/82324/elife-82324-fig1-figsupp1-data3-v1.zip
-
Figure 1—figure supplement 1—source data 4
Raw images of immunoblotting analysis referenced in Figure 1—figure supplement 1D.
- https://cdn.elifesciences.org/articles/82324/elife-82324-fig1-figsupp1-data4-v1.zip
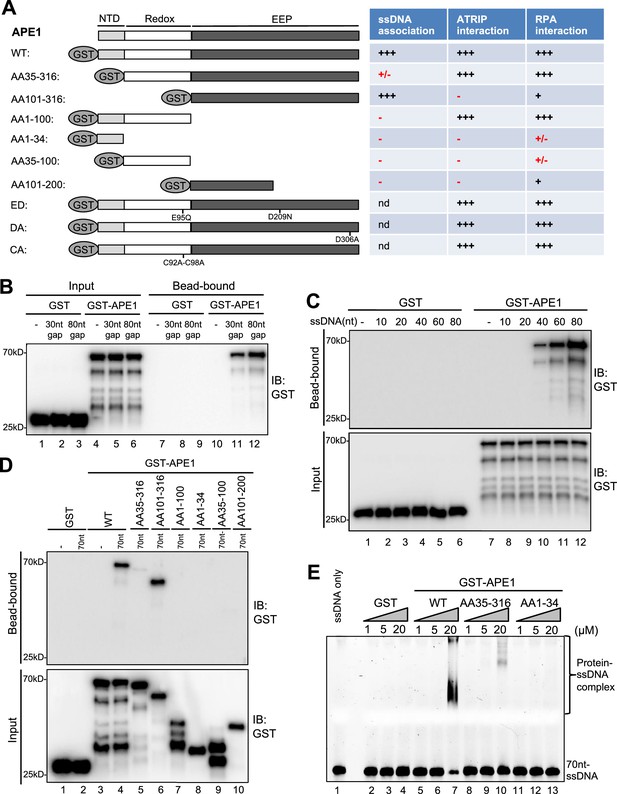
AP endonuclease 1 (APE1) recognizes and binds with single-stranded DNA (ssDNA) in a length-dependent fashion in vitro.
(A) Schematic diagram of APE1 functional domains and a summary of its interactions with ssDNA, ATRIP, and RPA from this study. Various symbols indicate estimates of APE1 interactions: ‘+++’, indicates the strongest interaction; ‘+’ indicates moderate interaction; ‘+/-’ indicates minimal to no interaction; ‘-’ indicates almost no interaction; ‘nd’, not determined. (B) Streptavidin beads coupled with biotin-labeled double-stranded DNA (dsDNA) with ssDNA gap structures (30 nt or 80 nt) were added to an interaction buffer containing GST or GST-APE1. After incubation for 30 min at room temperature, the DNA-bound fractions and the input were examined via immunoblotting analysis as indicated. (C) Streptavidin beads coupled with biotin-labeled ssDNA with different lengths (10 nt, 20 nt, 40 nt, 60 nt, or 80 nt) were added to an interaction buffer containing GST or GST-APE1. After incubation for 30 min at room temperature, the DNA-bound fractions and the input were examined via immunoblotting analysis. (D) Streptavidin beads coupled with biotin-labeled ssDNA (70 nt) were added to an interaction buffer containing (70 nt) GST or WT or fragment of GST-APE1. After incubation for 30 min at room temperature, the DNA-bound fractions and the input were examined via immunoblotting analysis. (E) An electrophoretic mobility shift assay (EMSA) shows the interaction between WT, AA35-316 and AA1-34 GST-APE1, and the 70 nt-ssDNA structure in vitro.
-
Figure 2—source data 1
Raw images of immunoblotting analysis referenced in Figure 2B.
- https://cdn.elifesciences.org/articles/82324/elife-82324-fig2-data1-v1.zip
-
Figure 2—source data 2
Raw images of immunoblotting analysis referenced in Figure 2C.
- https://cdn.elifesciences.org/articles/82324/elife-82324-fig2-data2-v1.zip
-
Figure 2—source data 3
Raw images of immunoblotting analysis referenced in Figure 2D.
- https://cdn.elifesciences.org/articles/82324/elife-82324-fig2-data3-v1.zip
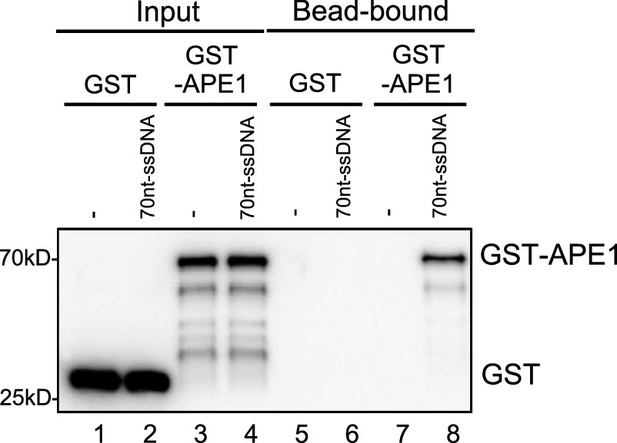
GST- AP endonuclease 1 (APE1) but not GST associated with beads coupled with single-stranded DNA (ssDNA).
Streptavidin beads coupled with biotin-labeled 70 nt-ssDNA were added to interaction buffer, which was supplemented with GST or WT GST-APE1. DNA-bound fractions and input samples were examined via immunoblotting analysis as indicated.
-
Figure 2—figure supplement 1—source data 1
Raw images of immunoblotting analysis referenced in Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/82324/elife-82324-fig2-figsupp1-data1-v1.zip
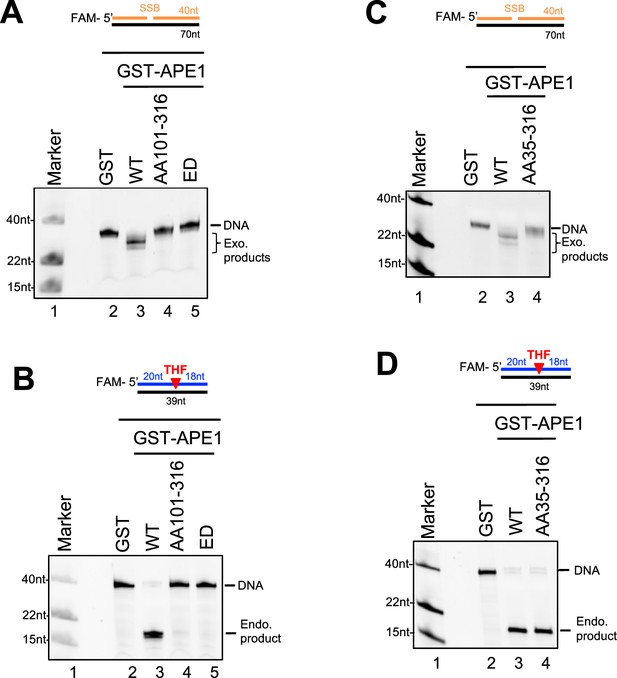
Endo/exonuclease activities of WT or various mutant/fragment of AP endonuclease 1 (APE1) in vitro.
(A and C) Various GST or WT/mutant/fragment GST-APE1 was added to nuclease assay buffer containing the 70 bp double-stranded DNA (dsDNA)-single-strand break (SSB) structure for exonuclease activity assays. Samples were examined via denaturing urea PAGE electrophoresis and visualized. (B and D) Various GST or WT/mutant/fragment GST-APE1 was added to nuclease assay buffer containing the 39 bp-dsDNA-AP structure for endonuclease activity assays. Samples were examined via denaturing urea PAGE electrophoresis and visualized.
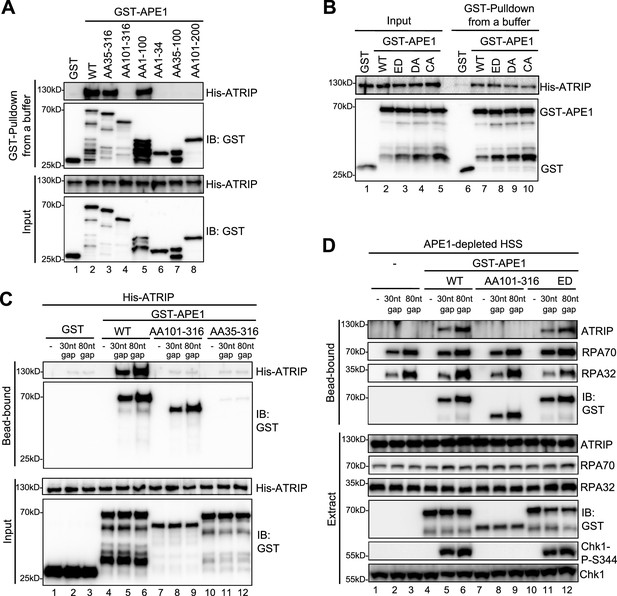
AP endonuclease 1 (APE1) interacts and recruits ATRIP onto single-stranded DNA (ssDNA) in an RPA-independent manner in vitro and promotes the ATR DNA damage response (DDR) pathway in Xenopus egg extracts using a non-catalytic function mechanism.
(A–B) GST pulldown assays with GST, WT, or fragment/mutant GST-APE1 as well as His-ATRIP in an interaction buffer. The input and pulldown samples were examined via immunoblotting analysis. (C) Streptavidin beads coupled with biotin-labeled double-stranded DNA (dsDNA) with ssDNA gap structures (30 nt or 80 nt) were added to an interaction buffer containing His-ATRIP and GST/GST-tagged proteins (WT, AA101-316, or AA35-316 GST-APE1) as indicated. DNA-bound fractions and input samples were examined via immunoblotting analysis as indicated. (D) Streptavidin beads coupled with biotin-labeled dsDNA with ssDNA gap structures (30 nt or 80 nt) were added to APE1-depleted high-speed supernatant (HSS), which was supplemented with GST or GST-tagged proteins (WT, AA101-316, or ED GST-APE1) as indicated. DNA-bound fractions and total extract samples were examined via immunoblotting analysis as indicated.
-
Figure 3—source data 1
Raw images of immunoblotting analysis referenced in Figure 3A.
- https://cdn.elifesciences.org/articles/82324/elife-82324-fig3-data1-v1.zip
-
Figure 3—source data 2
Raw images of immunoblotting analysis referenced in Figure 3B.
- https://cdn.elifesciences.org/articles/82324/elife-82324-fig3-data2-v1.zip
-
Figure 3—source data 3
Raw images of immunoblotting analysis referenced in Figure 3C.
- https://cdn.elifesciences.org/articles/82324/elife-82324-fig3-data3-v1.zip
-
Figure 3—source data 4
Raw images of immunoblotting analysis referenced in Figure 3D.
- https://cdn.elifesciences.org/articles/82324/elife-82324-fig3-data4-v1.zip
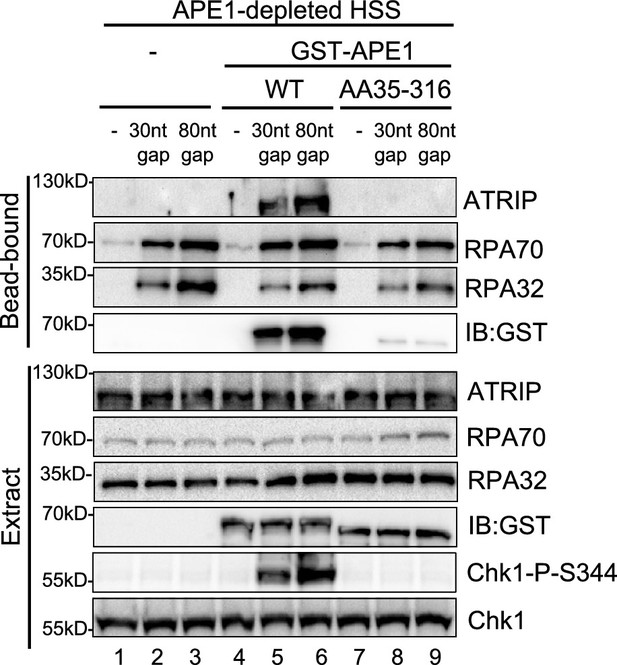
Streptavidin beads coupled with biotin-labeled double-stranded DNA (dsDNA) with single-stranded DNA (ssDNA) gap structures (30 nt or 80 nt) were added to AP endonuclease 1 (APE1)-depleted high-speed supernatant (HSS), which was supplemented with WT or AA35-316 GST-APE1 as indicated.
DNA-bound fractions and total extract samples were examined via immunoblotting analysis as indicated.
-
Figure 3—figure supplement 1—source data 1
Raw images of immunoblotting analysis referenced in Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/82324/elife-82324-fig3-figsupp1-data1-v1.zip
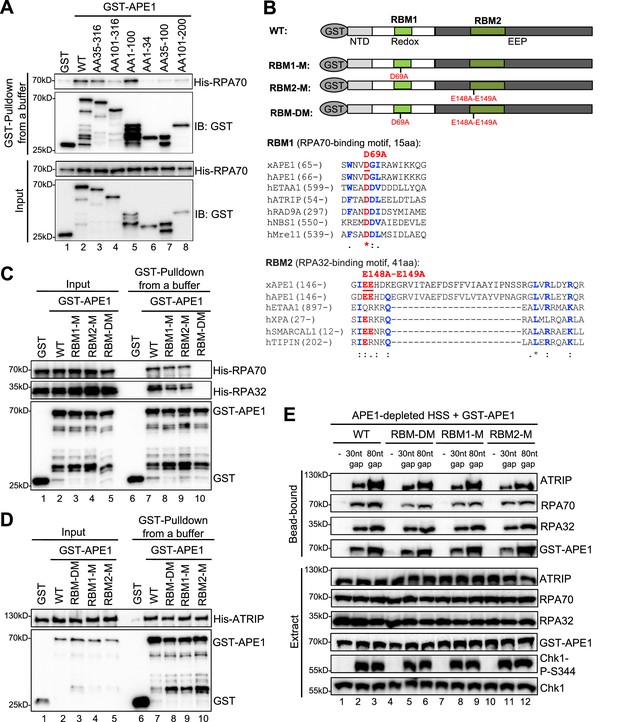
AP endonuclease 1 (APE1) interacts with RPA70 and RPA32 via two distinct binding motifs.
(A) GST pulldown assays with GST, WT, or fragment of GST-APE1 as well as His-RPA in an interaction buffer. The input and pulldown samples were examined via immunoblotting analysis. (B) Schematic diagram of APE1 functional domains and its putative RPA-binding motifs (RBM1 and RBM2), as well as sequence alignment of RBM1 and RBM2 from different RPA-interaction proteins. (C) GST pulldown assays with GST, WT/mutant GST-APE1, as well as His-RPA protein complex in an interaction buffer. The input and pulldown samples were examined via immunoblotting analysis. (D) GST pulldown assays with GST, WT/mutant GST-APE1, as well as His-ATRIP protein in an interaction buffer. The input and pulldown samples were examined via immunoblotting analysis. (E) Streptavidin beads coupled with biotin-labeled double-stranded DNA (dsDNA) with single-stranded DNA (ssDNA) gap structures (30 nt or 80 nt) were added to APE1-depleted high-speed supernatant (HSS), which was supplemented with WT or RBM mutant GST-APE1 (WT, RBM1-M, RBM2-M or RBM-DM GST-APE1) as indicated. DNA-bound fractions and total extract samples were examined via immunoblotting analysis as indicated.
-
Figure 4—source data 1
Raw images of immunoblotting analysis referenced in Figure 4A.
- https://cdn.elifesciences.org/articles/82324/elife-82324-fig4-data1-v1.zip
-
Figure 4—source data 2
Raw images of immunoblotting analysis referenced in Figure 4C.
- https://cdn.elifesciences.org/articles/82324/elife-82324-fig4-data2-v1.zip
-
Figure 4—source data 3
Raw images of immunoblotting analysis referenced in Figure 4D.
- https://cdn.elifesciences.org/articles/82324/elife-82324-fig4-data3-v1.zip
-
Figure 4—source data 4
Raw images of immunoblotting analysis referenced in Figure 4E.
- https://cdn.elifesciences.org/articles/82324/elife-82324-fig4-data4-v1.zip
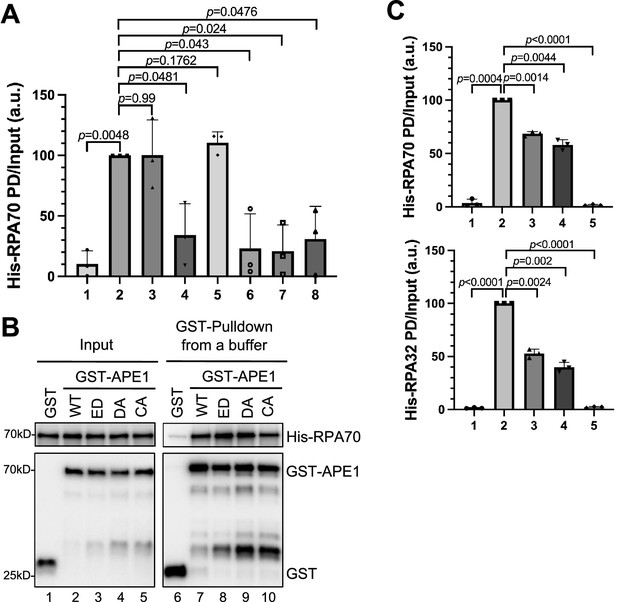
More characterization of the AP endonuclease 1 (APE1)-RPA interaction.
(A) The intensity of His-RPA70 in Figure 4A was quantified, and the ratio of His-RPA70 in PD (pulldown) vs input was analyzed. a.u., arbitrary unit. (B) GST pulldown assays with GST, or WT/mutant GST-APE1 in an interaction buffer. The input and pulldown samples were examined via immunoblotting analysis. (C) The intensity of His-RPA70 and His-RPA32 in Figure 4C was quantified, and the ratio of His-RPA70 or His-RPA32 in PD vs input was analyzed. a.u., arbitrary unit. Mean ± SD, n=3.
-
Figure 4—figure supplement 1—source data 1
Raw images of immunoblotting analysis referenced in Figure 4—figure supplement 1B.
- https://cdn.elifesciences.org/articles/82324/elife-82324-fig4-figsupp1-data1-v1.zip
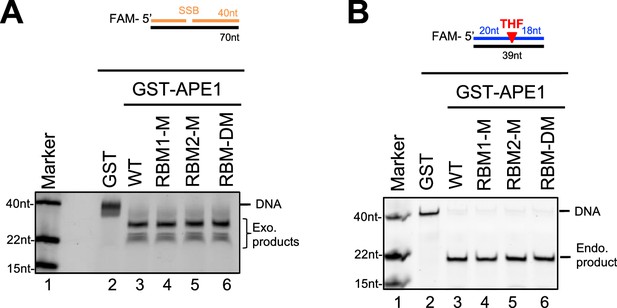
Endo/exonuclease activities of WT or various mutant/fragment of AP endonuclease 1 (APE1) in vitro.
(A) Various GST or WT/mutant GST-APE1 was added to nuclease assay buffer containing the 70 bp double-stranded DNA (dsDNA)-single-strand break (SSB) structure for exonuclease activity assays. Samples were examined via denaturing urea PAGE electrophoresis and visualized. (B) Various GST or WT/mutant GST-APE1 was added to nuclease assay buffer containing the 39 bp-dsDNA-AP structure for endonuclease activity assays. Samples were examined via denaturing urea PAGE electrophoresis and visualized.
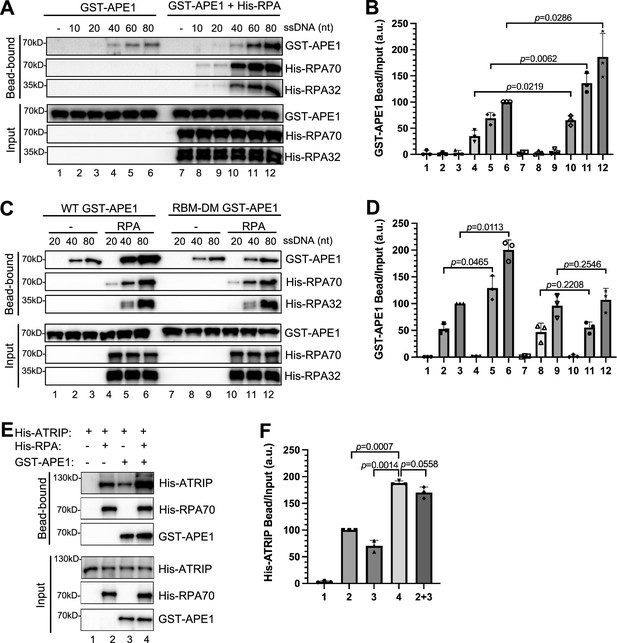
The RPA- AP endonuclease 1 (APE1) interaction promotes APE1 retention on single-stranded DNA (ssDNA).
(A) Streptavidin beads coupled with biotin-labeled ssDNA with different lengths were added to interaction buffer containing GST-APE1, which was supplemented with or without His-RPA protein. The input and pulldown samples were examined via immunoblotting analysis. (B) The intensity of GST-APE1 in (A) was quantified, and the ratio of GST-APE1 in bead vs input was analyzed. a.u., arbitrary unit. Mean ± SD, n=3. (C) Streptavidin beads coupled with biotin-labeled ssDNA with different lengths were added to interaction buffer containing WT/RBM-DM GST-APE1, which was supplemented with/without His-RPA protein complex. The input and pulldown samples were examined via immunoblotting analysis. (D) The intensity of GST-APE1 in (C) was quantified, and the ratio of GST-APE1 in bead vs input was examined. a.u., arbitrary unit. Mean ± SD, n=3. (E) Streptavidin beads coupled with biotin-labeled double-stranded DNA (dsDNA) with 80 nt-ssDNA gap structure were added to an interaction buffer containing purified His-ATRIP protein (20 μg) with/without His-RPA protein complex (5 μg) and/or GST-APE1 protein (5 μg). After incubation, the bead-bound fractions and the input were examined via immunoblotting analysis. (F) The intensity of His-ATRIP in (E) was quantified, and the ratio of His-ATRIP in bead vs input was examined. a.u., arbitrary unit. Mean ± SD, n=3.
-
Figure 4—figure supplement 3—source data 1
Raw images of immunoblotting analysis referenced in Figure 4—figure supplement 3A.
- https://cdn.elifesciences.org/articles/82324/elife-82324-fig4-figsupp3-data1-v1.zip
-
Figure 4—figure supplement 3—source data 2
Raw images of immunoblotting analysis referenced in Figure 4—figure supplement 3C.
- https://cdn.elifesciences.org/articles/82324/elife-82324-fig4-figsupp3-data2-v1.zip
-
Figure 4—figure supplement 3—source data 3
Raw images of immunoblotting analysis referenced in Figure 4—figure supplement 3E.
- https://cdn.elifesciences.org/articles/82324/elife-82324-fig4-figsupp3-data3-v1.zip
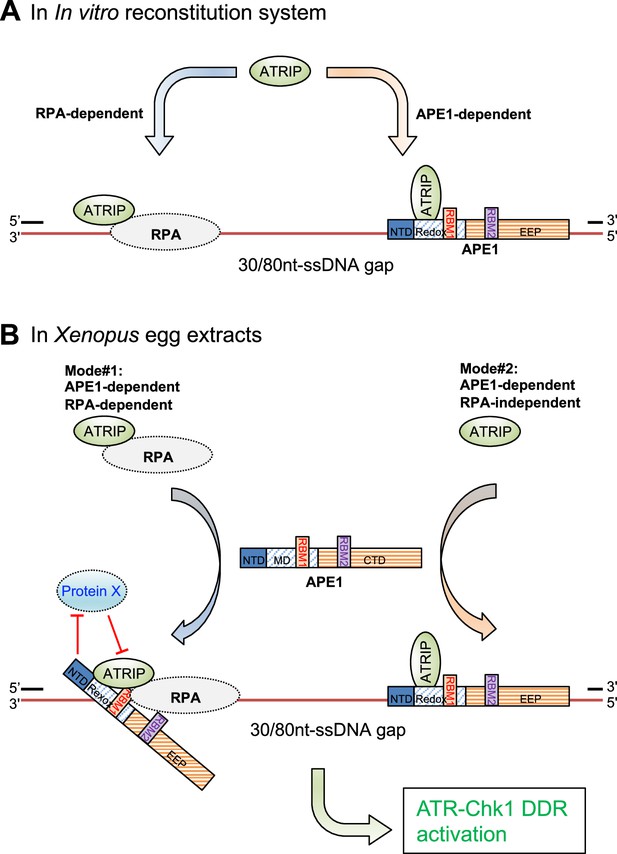
A working model of the distinct mechanism of how AP endonuclease 1 (APE1) directly interacts and recruits ATRIP onto single-stranded DNA (ssDNA) independently in vitro and in a concerted fashion in Xenopus egg extracts for ATR DNA damage response (DDR) pathway.
(A) In in vitro reconstitution system, RPA can recruit ATRIP to ssDNA gaps. In parallel, APE1 promotes the recruitment of ATRIP to ssDNA gaps in vitro via APE1 direct interaction with ssDNA and ATRIP protein. The RPA-dependent and APE1-dependent recruitment of ATRIP onto ssDNA is likely independent of each other in in vitro reconstitution system. (B) APE1 is required for the recruitment of ATRIP onto ssDNA gaps for the ATR/Chk1 DDR activation in the Xenopus high-speed supernatant (HSS) system via two modes: (#1) APE1-dependent and RPA-dependent; and (#2) APE1-dependent but RPA-independent. In Mode #1: RPA interacts with ATRIP protein and recruits ATRIP onto RPA-ssDNA in HSS. However, this Mode #1 may be inhibited by a currently uncharacterized Protein X in HSS, and such inhibitory effect by Protein X can be reversed by APE1. In Mode #2, APE1 can interact with ATRIP that is not in complex with RPA protein complex, and recruits ATRIP onto ssDNA gaps independent of RPA in HSS. Other significant DDR proteins are omitted from this diagram for a simplified illustration. NTD, N-terminal domain; Redox, redox domain; EEP, exonuclease-endonuclease-phosphatase domain; RBM1, RPA70-binding motif; RBM2, RPA32-binding motif.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | U2OS | ATCC | HTB-96 | NA |
| Strain, strain background (Escherichia coli) | BL21(DE3) | Sigma-Aldrich | CMC0015 | Electrocompetent cells |
| Transfected construct (human) | siRNA to APEX1 (ON-TARGETplus SMARTpool) | Dharmacon/Horizon Discovery Lts. and Li et al., 2022 | L-010237-00-0005 | Transfected construct (human) |
| Sequence-based reagent (oligonucleotides) | 70 nt FAM-ssDNA structure | This study and Lin et al., 2020 | Oligo#1 | FAM-5'-TCGGTACCCGGGGATCCTCTAGAGTCGACCTGCAGGCATGCAAGCTTGGCGTAATCATGGTCATAGCTGT-3' |
| Sequence-based reagent (oligonucleotides) | 60 nt Biotin-labeled top strand | This study | Oligo#2 | Biotin-5'-GGGTAACGCCAGGGTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGTGAATTCGAGC-3' |
| Sequence-based reagent (oligonucleotides) | 10 nt top strand | This study | Oligo#3 | 5’-TGCAGGCATG-3' |
| Sequence-based reagent (oligonucleotides) | 100 nt bottom strand | This study | Oligo#4 | 5'-CATGCCTGCAGGTCGACTCTAGAGGATCCCCGGGTACCGAGCTCGAATTCACTGGCCGTCGTTTTACAACGTCGTGACTGGGAAAACCCTGGCGTTACCC- 3' |
| Sequence-based reagent (oligonucleotides) | 10 nt Biotin-labeled top strand | This study | Oligo#5 | Biotin-5'-GGGTAACGCC-3' |
| Sequence-based reagent (oligonucleotides) | 70 nt Biotin-ssDNA structure | This study | Oligo#6 | Bioin-5'- ACAGCTATGACCATGATTACGCCAAGCTTGCATGCCTGCAGGTCGACTCTAGAGGATCCCCGGGTACCGA-3' |
| Sequence-based reagent (oligonucleotides) | 10 nt Biotin-ssDNA structure | This study and Ha et al., 2020 | Oligo#7 | Bioin-5'-GGTCGACTCT-3' |
| Sequence-based reagent (oligonucleotides) | 20nt Biotin-ssDNA structure | This study and Ha et al., 2020 | Oligo#8 | Bioin-5'- GGTCGACTCTAGAGGATCCC-3' |
| Sequence-based reagent (oligonucleotides) | 40 nt Biotin-ssDNA structure | This study and Ha et al., 2020 | Oligo#9 | Bioin-5'- GGTCGACTCTAGAGGATCCCCGGGTACCGAGCTCGAATTC-3' |
| Sequence-based reagent (oligonucleotides) | 60 nt Biotin-ssDNA structure | This study and Ha et al., 2020 | Oligo#10 | Bioin-5'-GGTCGACTCTAGAGGATCCCCGGGTACCGAGCTCGAATTCACTGGCCGTCGTTTTACAAC-3' |
| Sequence-based reagent (oligonucleotides) | 80 nt Biotin-ssDNA structure | This study | Oligo#11 | Bioin-5'- GGTCGACTCTAGAGGATCCCCGGGTACCGAGCTCGAATTCACTGGCCGTCGTTTTACAACGTCGTGACTGGGAAAACCCT-3' |
| Antibody | Anti-Xenopus APE1 (Rabbit polyclonal) | Lin et al., 2020 | IB (1:2000) | |
| Antibody | Anti-Xenopus ATRIP (Rabbit polyclonal) | Willis et al., 2013 | IB (1:2000) | |
| Antibody | Anti-Xenopus RPA70 (Rabbit polyclonal) | Acevedo et al., 2016 | IB (1:5000) | |
| Antibody | Anti-Xenopus RPA32 (Rabbit polyclonal) | Acevedo et al., 2016 | IB (1:5000) | |
| Antibody | Anti-Chk1-P-S345 (Rabbit monoclonal) | Cell Signaling Technology | Cat#2348 | IB (1:2000) |
| Antibody | Anti-Chk1-P-S317 (Rabbit monoclonal) | Cell Signaling Technology | Cat#12302 | IB (1:1000) |
| Antibody | Anti-Chk1 (Mouse monoclonal) | Santa Cruz Biotechnology | Cat#sc-8408 | IB (1:2000) |
| Antibody | Anti-GST (Mouse monoclonal) | Santa Cruz Biotechnology | Cat#sc-138 | IB (1:5000) |
| Antibody | Anti-His (Mouse monoclonal) | Santa Cruz Biotechnology | Cat#sc-8036 | IB (1:1000) |
| Antibody | Anti-human APE1 (Mouse monoclonal) | Santa Cruz Biotechnology | Cat#sc-17774 | IB (1:2000) |
| Antibody | Anti-PCNA (Mouse monoclonal) | Santa Cruz Biotechnology | Cat#sc-56 | IB (1:4000) |