Intermediate filament network perturbation in the C. elegans intestine causes systemic dysfunctions
Figures
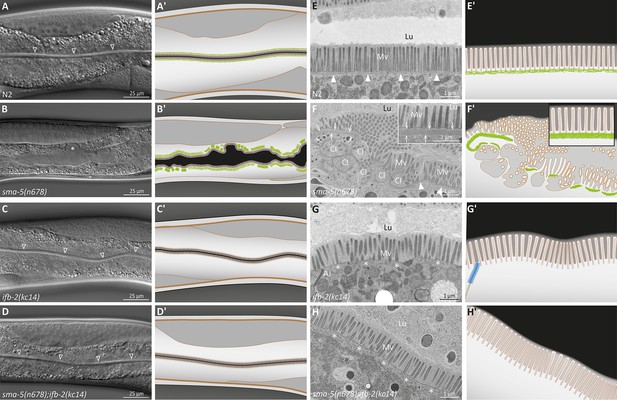
The ifb-2(kc14) knockout allele rescues the luminal widening and cytoplasmic invagination phenotypes and abolishes the aberrant intermediate filament (IF) network of sma-5(n678) mutants.
(A–D′) Differential interference contrast pictures and corresponding schematics of viable wild-type N2 (A, A′), sma-5(n678) (B, B′), ifb-2(kc14) (C, C′), and sma-5(n678);ifb-2(kc14) animals (D, D′). ifb-2 knockout causes only minor intestinal defects. In contrast, sma-5(n678) animals display extensive luminal widening and large cytoplasmic invaginations of the apical plasma membrane in intestinal cells (asterisk in B). This phenotype is effectively reversed by the ifb-2(kc14) knockout allele. Non-filled arrowheads: normal-appearing intestinal lumen. (E–H′) Electron microscopy images of high-pressure frozen samples and corresponding schematics show wild-type N2 (E′, E′), sma-5(n678) (F, F′), ifb-2(kc14) (G, G′), and sma-5(n678);ifb-2(kc14) intestinal cell apices (H, H′). sma-5(n678) animals contain regions with an enlarged endotube consisting of densely packed IFs (arrows) and differently sized cytoplasmic invaginations (CI) with no endotube or a reduced endotube (arrowheads). Additional knockout of ifb-2 almost restores the wild-type morphology with only residual luminal widening and mildly perturbed microvillar arrangement. But the endotube, which is easily detected in the wild type (arrowheads), is completely absent in single ifb-2(kc14) and double sma-5(n678);ifb-2(kc14) mutants (expected position marked by asterisks). Lu, lumen; Mv, microvilli; AJ, C. elegans apical junction.
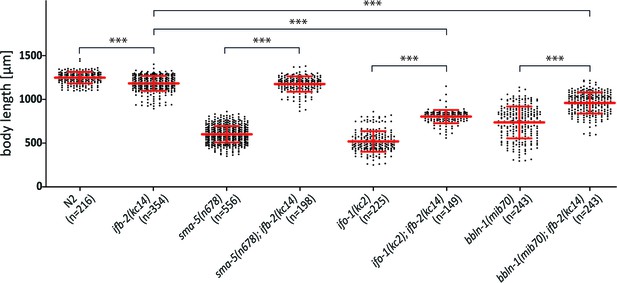
Depletion of IFB-2 rescues growth defects in sma-5(n678), ifo-1(kc2), and bbln-1(mib70) loss-of-function mutants.
The scatter dot blot shows a comparison of body lengths 4 days after egg laying in N2, ifb-2(kc14), sma-5(n678), sma-5(n678);ifb-2(kc14), ifo-1(kc2), ifo-1(kc2);ifb-2(kc14), bbln-1(mib70), and bbln-1(mib70);ifb-2(kc14). Note that loss of IFB-2 rescues the body length phenotype of sma-5(n678) to the level of ifb-2(kc14) single mutants (N2: 1252±65.59 µm; ifb-2(kc14): 1187±84.85 µm; sma-5(n678): 608.2±91.97 µm; sma-5(n678);ifb-2(kc14): 1179±87.57 µm; ***p<0.0001). Rescue of body length is also observed in ifo-1(kc2) and bbln-1(mib70) albeit less efficiently and to different degrees (ifo-1(kc2): 526.7.7±117.2 µm; ifo-1(kc2);ifb-2(kc14): 809.6.3±75.2 µm; ***p<0.0001; bbln-1(mib70): 742.7±181.3 µm; bbln-1(mib70);ifb-2(kc14): 964.3±120.1 µm; ***p<0.0001).
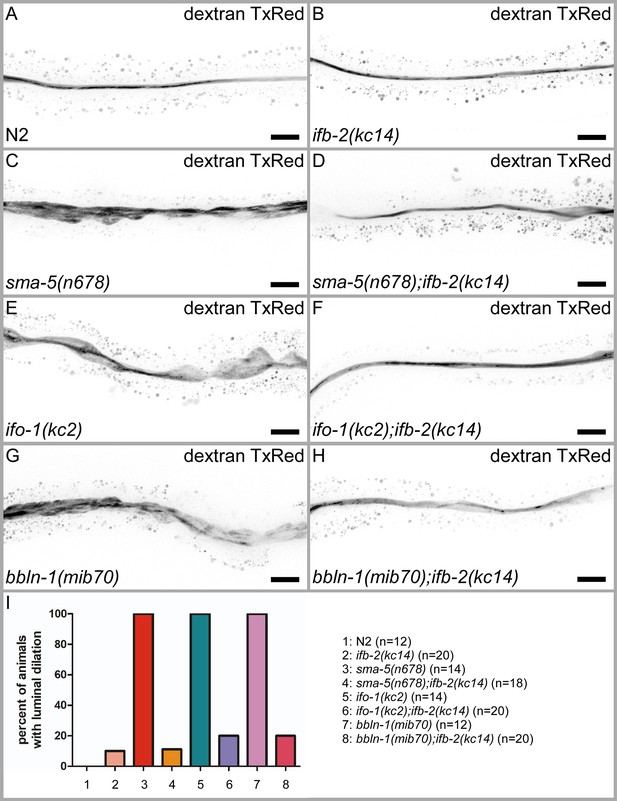
Depletion of IFB-2 rescues the luminal dilation phenotype in sma-5(n678), ifo-1(kc2), and bbln-1(mib70) loss-of-function mutants.
(A–H) Young adult animals were fed with fluorescent dextran Texas Red (70 kDa) and subsequently imaged to visualize the intestinal lumen. Note that the luminal dilation phenotype present in sma-5(n678) (C), ifo-1(kc2) (E), and bbln-1(mib70) animals (G) can be efficiently rescued by depletion of IFB-2 (D, F, H). Scale bars: 20 µm. (I) Quantification of animals with luminal dilation phenotype (N2: 0%, ifb-2(kc14): 10%, sma-5(n678): 100%, sma-5(n678);ifb-2(kc14): 11.1%, ifo-1(kc2): 100%, ifo-1(kc2);ifb-2(kc14): 20%, bbln-1(mib70): 100%, bbln-1(mib70); ifb-2(kc14): 20%).
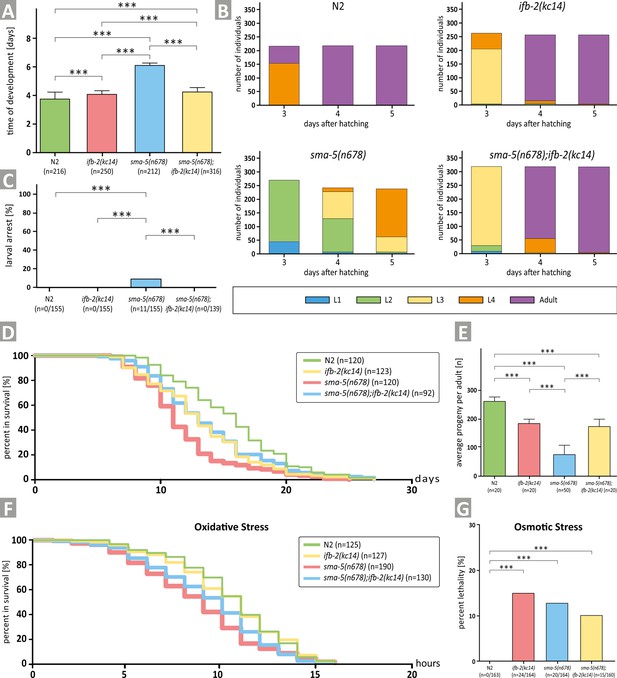
Depletion of IFB-2 (ifb-2(kc14)) rescues developmental retardation, larval arrest, reduced median life span, decreased brood size, and increased sensitivity to oxidative stress of sma-5(n678) mutants.
(A) The histogram shows a comparison of the time of development in N2, ifb-2(kc14), sma-5(n678), and sma-5(n678);ifb-2(kc14) (N2: 3.7±0.5 days; ifb-2(kc14): 4.1±0.2 days; sma-5(n678): 6.0±0.2 days; sma-5(n678);ifb-2(kc14): 4.2±0.4 days; ***p<0.0001). (B) The color-coded histograms depict the number of larval and adult stages detected 3, 4, and 5 days after hatching. (C) The histogram illustrates a complete rescue of the larval arrest phenotype observed in sma-5(n678) by ifb-2(kc14) (N2: 0%; ifb-2(kc14): 0%; sma-5(n678): 7.1%; sma-5(n678);ifb-2(kc14): 0%). (D) The plot shows that the reduced life span of sma-5(n678) is rescued by addition of ifb-2(kc14) to the level encountered in ifb-2(kc14) but not the wild-type level (median survival for N2: 16 days; ifb-2(kc14): 13 days; sma-5(n678): 11 days; sma-5(n678);ifb-2(kc14): 13 days; p=0.0004 ifb-2(kc14) versus N2; p<0.0001 sma-5(n678) versus N2; p=0.0003 sma-5(n678) versus ifb-2(kc14); p<0.0001 sma-5(n678);ifb-2(kc14) versus N2; p<0.0046 sma-5(n678);ifb-2(kc14) versus sma-5(n678)). (E) The histogram reveals that the drastic reduction in progeny observed in sma-5(n678) mutants versus N2 (76±31 vs 263±13; p<0.0001) is rescued in sma-5(n678);ifb-2(kc14) double mutants (175±21 vs 76±31; p<0.001) but does not reach wild-type level (175±21 vs 263±13; p<0.001) and is similar to ifb-2(kc14) (175±21 vs 183±15; p>0.5). (F) The survival plot shows the effect of acute oxidative stress in the wild type (N2), ifb-2(kc14), sma-5(n678) and sma-5(n678);ifb-2(kc14) backgrounds (median survival for N2: 11 hr; ifb-2(kc14): 11 hr; sma-5(n678): 9 hr; sma-5(n678);ifb-2(kc14): 10 hr; p<0.0001 for N2 or ifb-2(kc14) versus sma-5(n678); p<0.05 for sma-5(n678);ifb-2(kc14) versus sma-5(n678); p<0.01 for N2 or ifb-2(kc14) versus sma-5(n678);ifb-2(kc14)). (G) The histogram scores the percentage of dead worms in response to acute osmotic stress for N2 (0%), ifb-2(kc14) (14.6%), sma-5(n678) (12.2%), and sma-5(n678);ifb-2(kc14) (9.4%).
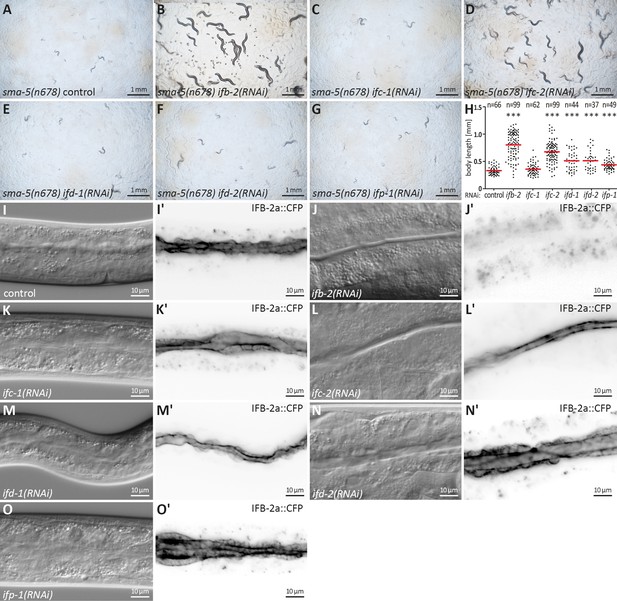
Downregulation of ifb-2 and ifc-2 is most efficient in suppressing developmental retardation, small body size, and cytoplasmic invaginations of sma-5(n678).
(A–G) show bright-field images of agar plates containing F1 sma-5(n678) subjected to RNAi 4 days after egg laying. (H) The scatter dot blot summarizes the results of body length measurements in sma-5(n678) subjected to either empty RNAi vector (control; 331±72.71 µm), ifb-2(RNAi) (802.90±235.10 µm), ifc-1(RNAi) (358.50±103.50 µm), ifc-2(RNAi) (675.60±194.10 µm), ifd-1(RNAi) (511.70±172.90 µm), ifd-2(RNAi) (512.60±178.80 µm) or ifp-1(RNAi) (443.30±109 µm). Note that the strongest rescue is observed for ifb-2 followed by ifc-2 and trailed by ifd-1, ifd-2, and ifp-1 all of which are statistically significant (p<0.0001). No detectable rescue is observed for ifc-1. (I–O') The microscopy images show differential interference contrast at left and corresponding fluorescence detection (inverse presentation) of the IFB-2a::CFP reporter in vital sma-5(n678) animals after RNAi against ifb-2 (J, J'), ifc-1 (K, K'), ifc-2 (L, L'), ifd-1 (M, M'), ifd-2 (N, N'), and ifp-1 (O, O') (RNAi control in I, I'). Note that the knockdown of ifb-2 leads to a loss of reporter fluorescence and efficiently rescues the sma-5(n678) invagination phenotype. A rescue is also detectable after loss of IFC-2 albeit at reduced efficiency. None of the other knockdowns resulted in detectable reduction of the invagination phenotype.
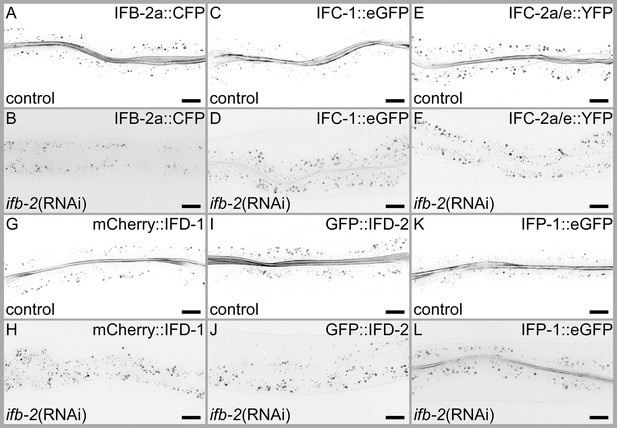
Loss of IFB-2 prevents network formation of IFC-2, IFD-1, and IFD-2, while IFC-1 and IFP-1 network formation is severely reduced.
Fluorescence images of adult animals expressing IFB-2a::CFP (A–B), IFC-1::eGFP (C–D), IFC-2a/e::YFP (E–F), mCherry::IFD-1 (G–H), GFP::IFD-2 (I–J), and IFP-1::eGFP (K–L) after treatment with empty vector RNAi control or ifb-2(RNAi). Downregulation of IFB-2 via RNAi resulted in a complete loss of network-forming capability of IFC-2 (compare E with F), IFD-1 (compare G with H), and IFD-2 (compare I with J). Very limited network-forming capability was noted for IFC-1 (compare C with D) and IFP-1 (compare K with L). Note that in case of ifb-2(RNAi), animals were imaged using higher exposure times compared to the control to check for any residual apical network. Scale bars: 20 µm.
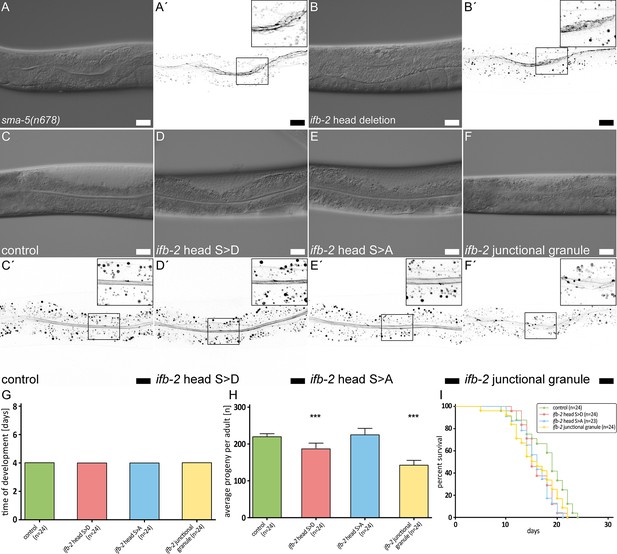
The IFB-2 aminoterminus is involved in intestinal intermediate filament (IF) network morphogenesis, progeny production, and life span.
(A–F′) The microscopy images show differential interference contrasts (A–F) and corresponding fluorescence recordings of the IFC-2a/e::YFP reporter (inverse presentation; A′–F′) in vital sma-5(n678) (A–A′), ifb-2 head deletion (B–B′, ifb-2(mib169[D2-M43 deleted])II), control (C–C′), phosphomimetic ifb-2 head S>D (D–D′, ifb-2(kc22[S2D;S5D;S7D;S16-19D;S24D;S35D])II), phosphodeficient ifb-2 head S>A (E–E′, ifb-2(kc26[S2A;S5A;S7A;S16-19A;S24A;S35A])II) and ‘junctional granule’ mutants (F–F′, ifb-2(kc27[S2A;S5A;S7A;S16-19A;E31-A184 deleted])II). Scale bars: 20 µm. (G–H) The histograms present a comparison of time of development (G) and average progeny (H) of control, phosphomimetic ifb-2 head S>D, phosphodeficient ifb-2 head S>A and ‘junctional granule’ mutants. None of the mutants shows a prolonged development (control: 4.0 days; ifb-2 head S>D: 4.0 days; ifb-2 head S>A: 4.0 days; ‘junctional granule’ mutant: 4.0 days). The average progeny per adult, however, is reduced in ifb-2 head S>D but not in ifb-2 head S>A mutants in comparison to control (control: 227±9; ifb-2 head S>D: 193±16; ifb-2 head S>A: 232±18; p<0.0001 control versus ifb-2 head S>D). Note the even higher reduction in progeny observed in ‘junctional granule’ mutants versus control (control: 227±9; junctional granule mutant: 145±16; p<0.0001). (I) The survival plot shows that mutants with genetic modifications of the ifb-2 head domain have a reduced life expectancy albeit to different degrees with the ifb-2 head S>D mutant showing the most severe phenotype (median survival for control: 19 days; ifb-2 head S>D: 15 days; ifb-2 head S>A: 16 days; junctional granule mutant: 15.5 days; p=0.0471 ifb-2 head S>D versus control; p=0.0206 ifb-2 head S>A versus control; p=0.0468 junctional granule mutant versus control; p<0.0001).
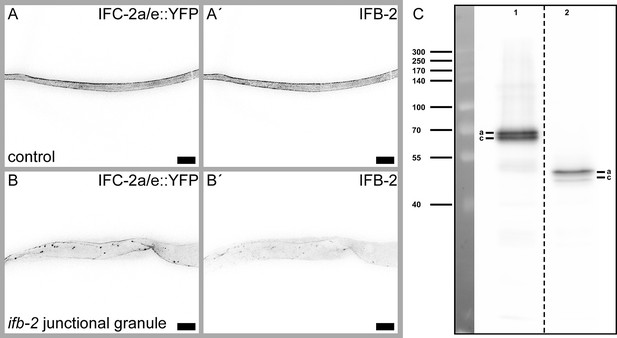
ifb-2(kc27) animals show IFC-2 positive junctional granules in concert with an overall reduced IFB-2/IFC-2-containing network.
(A–B′) The fluorescence micrographs show IFC-2a/e::YFP (A, B) and corresponding IFB-2 immunostainings (A′, B′) of isolated control (A–A′) and ifb-2(kc27[S2A;S5A;S7A;S16-19A;E31-A184 deleted])II intestines (B-B'). Note the reduced IFC-2a/e::YFP in the apical cytoplasm with additional IFC-2-positive junctional granules. In contrast, IFB-2 is also diffusely localized in the cytoplasm. Scale bars: 20 µm. (C) Immunoblot detecting IFB-2 isoforms a and c of control (lane 1) and ifb-2(kc27) animals (lane 2). The IFB-2 deletion in ifb-2(kc27) results in a truncated protein of about 50 kDa.
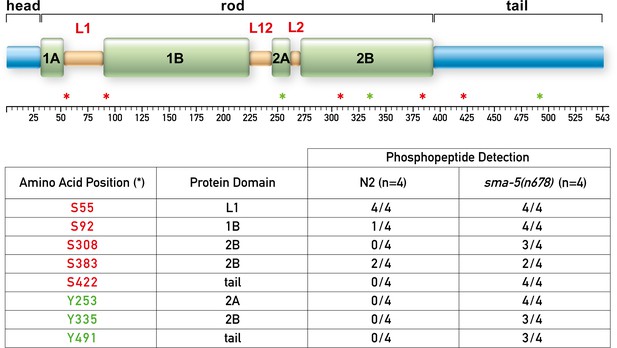
IFB-2 is hyperphosphorylated at multiple sites in the absence of SMA-5.
The upper panel shows a domain model of IFB-2 with its aminoterminal head, alpha helical central rod (coil 1A, linker L1, coil 1B, linker L12, coil 2A, linker L2, and coil 2B), and carboxyterminal tail. Positions of phosphorylated amino acids are marked by asterisks (serines in red, tyrosines in green). The table below lists the number of samples, in which the respective phosphopeptides were identified, and their domain localization.
-
Figure 5—source data 1
Identified phosphopeptides in sma-5(n678) and their corresponding domain localization.
- https://cdn.elifesciences.org/articles/82333/elife-82333-fig5-data1-v2.xlsx
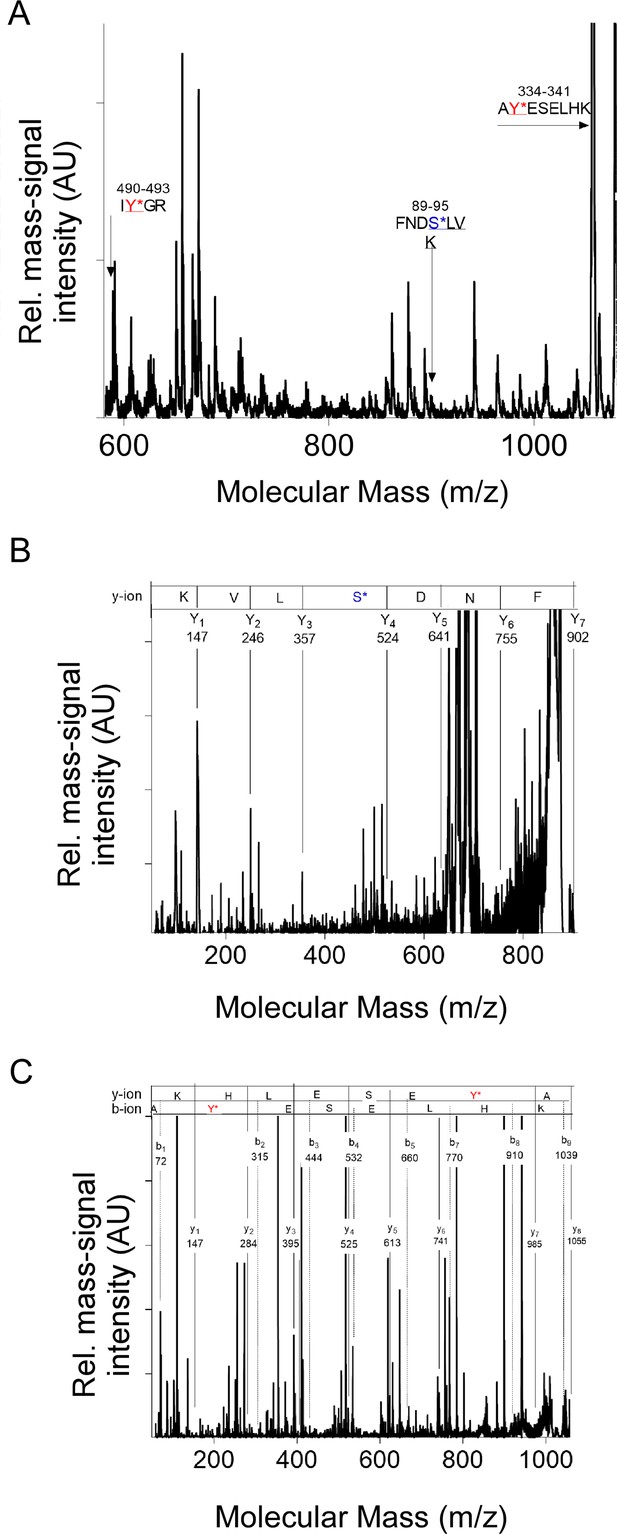
Examples of mass fingerprint spectra of IFB-2 in sma-5(n678) showing the specific phosphate modification of phosphorylated peptides.
(A) Characteristic mass fingerprint spectrum obtained from a sample after gel electrophoresis followed by trypsin digest of proteins within the range of 50–150 kDa. The arrows indicate the molecular mass of IFB-2 peptides. The arrows indicate the phosphorylated peptide with the molecular mass 588 Dalton (Da) with the modification on tyrosine (IY*GR), the molecular mass of 901 Da with the amino sequence FNDS*LVK modified with phosphate on serine and the molecular mass of 1055 Da with the sequence AY*ESELHK and the modification on tyrosine. (B) Characteristic fragment mass spectrum of the molecular mass 901 Da. The asterisk indicates the phosphate modification on serine S92. (C) Characteristic fragment mass spectrum of the molecular mass 1055 Da. The asterisk indicates the phosphate modification on tyrosine Y335.
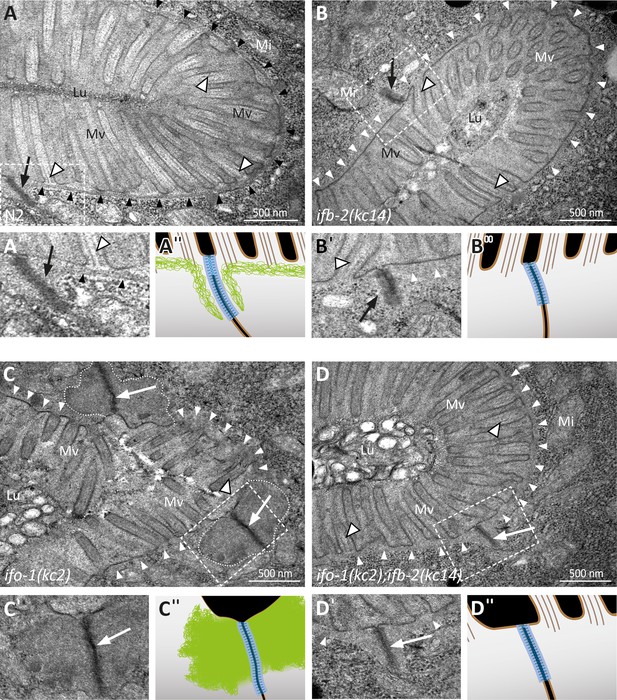
Depletion of IFB-2 partially rescues the ifo-1(kc2) phenotype.
(A–D) The electron micrographs show a comparison of the intestinal cell apices surrounding the lumen (Lu) of wild-type N2 (A, enlarged section in A′, corresponding scheme in A′′), ifb-2(kc14) (B, enlarged section in B′, corresponding scheme in B′′), ifo-1(kc2) (C, enlarged section in C′, corresponding scheme in C′′), and ifo-1(kc2);ifb-2(kc14) (D, enlarged section in D′, corresponding scheme in D′′). Note the distinct endotube in N2 (black arrowheads) and its absence in ifb-2(kc14), ifo-1(kc2), and ifo-1(kc2);ifb-2(kc14) (white arrowheads). The pathognomonic large junctional aggregates of ifo-1(kc2) are delineated by broken white lines. Note also the improved brush border morphology in (D) compared to (C) (Mv, microvilli). Arrows, C. elegans apical junction (CeAJ); outlined white arrowheads, microvillar actin bundles; Mi, mitochondrion.
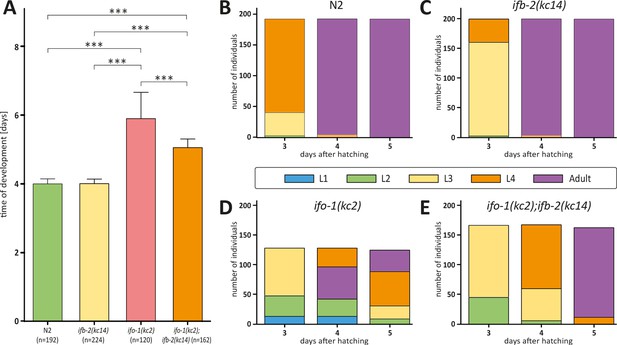
Depletion of IFB-2 partially rescues the ifo-1(kc2) growth defects.
(A–E) The histograms depict the time of development in (A) and the number of staged worms at different times after hatching in N2 (B), ifb-2(kc14) (C), ifo-1(kc2) (D), and ifo-1(kc2);ifb-2(kc14) (E). Note the partial rescue in the double mutants. (N2: 4.0±0.1 days; ifb-2(kc14): 4.0±0.1 days; ifo-1(kc2): 5.9±0.8 days; ifo-1(kc2);ifb-2(kc14): 5.1±0.3 days; ***p<0.0001).
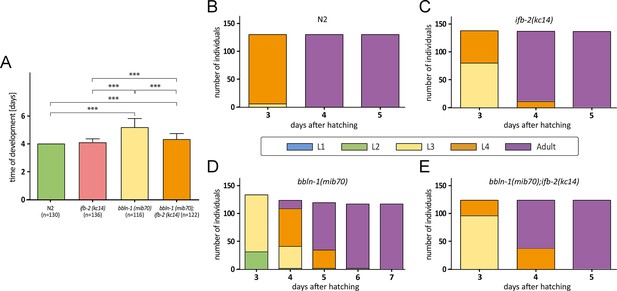
Depletion of IFB-2 partially rescues the bbln-1(mib70) phenotype.
(A–E) The histograms depict the time of development in (A) and the number of staged worms at different times after hatching in N2 (B), ifb-2(kc14) (C), bbln-1(mib70) (D), and bbln-1(mib70);ifb-2(kc14) (E). Note the partial rescue in the double mutants (N2: 4.0 days; ifb-2(kc14): 4.1±0.3 days; bbln-1(mib70): 5.2±0.6 days; bbln-1(mib70);ifb-2(kc14): 4.3±0.5 days; ***p<0.0001).
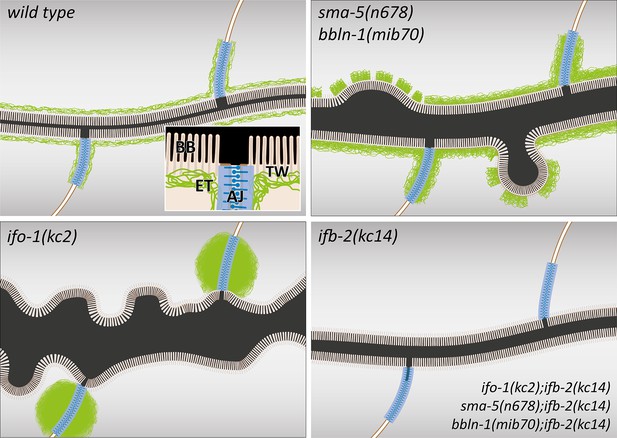
The schematic drawings highlight changes in the intestinal lumen and intermediate filament (IF) network organization in the different mutant backgrounds.
The wild-type scheme (upper left) depicts the adluminal brush border (BB) consisting of microvilli with bundled actin filaments, the terminal web (TW) with traversing microvillar actin rootlets that rest on the IF-rich endotube (ET) together with the C. elegans apical junction (AJ), which serves as an anchorage site for the IF network. Note the luminal enlargement and cytoplasmic invaginations in sma-5(n678) and bbln-1(mib70) presenting a thickened and discontinuous endotube with slightly disordered microvilli (upper right). ifo-1(kc2) is characterized by endotube loss and formation of intermediate filament aggregates at the C. elegans apical junction (lower left). Luminal widening, cytoplasmic invaginations and microvillar disorder are also observed. The mildest phenotype is detectable in ifb-2(kc14) and in the double mutants ifo-1(kc2);ifb-2(kc14), sma-5(n678);ifb-2(kc14), and bbln-1(mib70);ifb-2(kc14).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-IFB-2 (mouse monoclonal) | Developmental Studies Hybridoma Bank | AB_528311 MH33 | (1:100-1:1000), (Francis and Waterston, 1991) |
| Antibody | Anti-mouse IgG coupled to horseradish peroxidase (goat polyclonal) | DAKO | #P0447 | (1:5000) |
| Antibody | Anti-GFP (rabbit polyclonal) | Invitrogen | #A-11122 | (1:1000) |
| Antibody | Anti-mouse IgG coupled to Alexa Fluor 488 (goat polyclonal) | Invitrogen | #A-11029 | (1:200) |
| Antibody | Anti-mouse IgG coupled to Alexa Fluor 555 (goat polyclonal) | Invitrogen | #A-21424 | (1:200) |
| Strain, strain background (Escherichia coli) | OP50 | CGC | N/A | Strain can be obtained from the Caenorhabditis Genetics Center |
| Strain, strain background (Escherichia coli) | HT115 | CGC | N/A | Strain can be obtained from the Caenorhabditis Genetics Center |
| Strain, strain background (Escherichia coli) | Vidal full-length HT115 RNAi feeding library | SourceBio-Science | 3320_Cel_ORF_RNAi | |
| Strain, strain background (Escherichia coli) | Ahringer fragment HT115 RNAi feeding library | SourceBio-Science | 3318_Cel_RNAi_complete | |
| Chemical compound, drug | Dextran, Texas Red, 70,000 MW | Invitrogen | #D1830 | |
| Chemical compound, drug | Alt-R S.p. Cas9 Nuclease V3 | IDT | #1081058 | |
| Strain, strain background (Caenorhabditis elegans) | Wild type (Bristol) | CGC | N2 | Strain can be obtained from the Caenorhabditis Genetics Center |
| Strain, strain background (Caenorhabditis elegans) | sma-5(n678)X | CGC | FK312 | Strain can be obtained from the Caenorhabditis Genetics Center |
| Strain, strain background (Caenorhabditis elegans) | kcIs6[ifb-2p::ifb-2a::cfp]IV | Hüsken et al., 2008 | BJ49 | Strain can be obtained from the Leube lab |
| Strain, strain background (Caenorhabditis elegans) | ifo-1(kc2)IV | Carberry et al., 2012 | BJ142 | Strain can be obtained from the Leube lab |
| Strain, strain background (Caenorhabditis elegans) | sma-5(n678)X;kcIs6[ifb- 2p::ifb-2a::cfp]IV | Geisler et al., 2016 | OLB18 | Strain can be obtained from the Leube lab |
| Strain, strain background (Caenorhabditis elegans) | ifb-2(kc14)II | Geisler et al., 2019 | BJ309 | Strain can be obtained from the Leube lab |
| Strain, strain background (Caenorhabditis elegans) | erm-1(mib40[erm-1::AID::mCherry])I; Pelt-2::TIR::tagBFP-2::tbb-2–3'UTR (mib58[Pelt-2::TIR-1::tagBFP-2 -Lox511::tbb-2–3'UTR])IV;C15C7.5 (mib70[pC15C7.5::GFP1-3, C15C7.5 0-914del])X | Remmelzwaal et al., 2021 | BOX415 | Strain can be obtained from the Boxem lab |
| Strain, strain background (Caenorhabditis elegans) | kcEx78[WRM0639B_D12(pRedFlp-Hgr) (ifc-1[24863] ::S0001_pR6K_Amp_2xTY1ce_ EGFP_FRT_ rpsl_neo_FRT_3xFlag) dFRT ::unc-119-Nat];unc-119(ed3)III | Geisler et al., 2020 | BJ324 | Strain can be obtained from the Leube lab |
| Strain, strain background (Caenorhabditis elegans) | ifc-2(kc16)X | Geisler et al., 2020 | BJ316 | Strain can be obtained from the Leube lab |
| Strain, strain background (Caenorhabditis elegans) | kcIs40[ifp-1p::ifp-1::egfp]IV | Geisler et al., 2020 | BJ312 | Strain can be obtained from the Leube lab |
| Strain, strain background (Caenorhabditis elegans) | erm-1(mib40[erm-1::AID::mCherry]) I; ifd-2(mib94[GFP::ifd-2])X | Remmelzwaal et al., 2021 | BOX614 | Strain can be obtained from the Boxem lab |
| Strain, strain background (Caenorhabditis elegans) | sma-5(n678)X;ifb-2(kc14)II | This study | BJ346 | Strain can be obtained from the Leube lab |
| Strain, strain background (Caenorhabditis elegans) | ifo-1(kc2)IV;ifb-2(kc14)II | This study | BJ328 | Strain can be obtained from the Leube lab |
| Strain, strain background (Caenorhabditis elegans) | bbln-1(mib70) | This study | BJ411 | Strain can be obtained from the Leube lab |
| Strain, strain background (Caenorhabditis elegans) | bbln-1(mib70);ifb-2(kc14) | This study | BJ412 | Strain can be obtained from the Leube lab |
| Strain, strain background (Caenorhabditis elegans) | erm-1(mib40[erm-1::AID::mCherry])I; Pelt-2::TIR::tagBFP-2 ::tbb-2–3'UTR (mib58[Pelt-2:: TIR-1:: tagBFP-2-Lox511::tbb-2–3'UTR])IV; C15C7.5(mib70[pC15C7.5 ::GFP1-3, C15C7.5 0-914del])X;ifb-2(kc14)II | This study | BJ364 | Strain can be obtained from the Leube lab |
| Strain, strain background (Caenorhabditis elegans) | ifd-2(mib94[GFP::ifd-2])X | This study | BJ405 | Strain can be obtained from the Leube lab |
| Strain, strain background (Caenorhabditis elegans) | ifb-2(mib155[S2D;S5D; S7D;S16-19D])II; ifc-2a/e::yfp(kc16)X | This study | BOX793 | Strain can be obtained from the Leube lab |
| Strain, strain background (Caenorhabditis elegans) | ifb-2(kc22[S2D;S5D; S7D;S16-19D;S24D; S35D])II;ifc-2a/e::yfp(kc16)X | This study | BJ427 | Strain can be obtained from the Leube lab |
| Strain, strain background (Caenorhabditis elegans) | ifb-2(mib156[S2A;S5A; S7A;S16-19A])II; ifc-2a/e::yfp(kc16)X | This study | BOX794 | Strain can be obtained from the Leube lab |
| Strain, strain background (Caenorhabditis elegans) | ifb-2(kc26[S2A;S5A; S7A; S16-19A;S24A;S35A])II; ifc-2a/e::yfp(kc16)X | This study | BJ431 | Strain can be obtained from the Leube lab |
| Strain, strain background (Caenorhabditis elegans) | ifb-2(kc27[S2A;S5A;S7A; S16-19A;E31-A184 deleted])II; ifc-2a/e::yfp(kc16)X | This study | BJ432 | Strain can be obtained from the Leube lab |
| Strain, strain background (Caenorhabditis elegans) | ifb-2(mib169[S2-M43 deleted])II;ifc-2a/e::yfp(kc16)X | This study | BOX827 | Strain can be obtained from the Leube lab |
| Strain, strain background (Caenorhabditis elegans) | ifd-1(mib95[mCherry::ifd-1])X | This study | BOX540 | Strain can be obtained from the Boxem lab |
| Sequence- based reagent | See Supplementary file 1 and ’’RNAi and body length determination’’ section for sequence details | IDT | ||
| Recombinant DNA reagent | L4440 | Addgene | #1654 | |
| Recombinant DNA reagent | ifd-1 (L4440) | Remmelzwaal et al., 2021 | pSMR35 | Reagent can be obtained from the Boxem lab |
| Recombinant DNA reagent | ifp-1 (L4440) | Remmelzwaal et al., 2021 | pSMR34 | Reagent can be obtained from the Boxem lab |
| Recombinant DNA reagent | rol-6(su1006) | Mello et al., 1991 | pRF4 | Reagent can be obtained from the Leube lab |
| Software, algorithm | ImageJ | Rasband, W.S. (NIH) | RRID: SCR_003070 | |
| Software, algorithm | Zen Blue | Zeiss | RRID:SCR_013672 | |
| Software, algorithm | GraphPad Prism | GraphPad | RRID: SCR_002798 | |
| Software, algorithm | SnapGene | Insightful Science | RRID: SCR_015052 | |
| Software, algorithm | Clone Manager | Scientific & Educational Software | RRID:SCR_014521 | |
| Software, algorithm | Bruker Bio-Tool 3.2 and the Mascot 2.3 search engine | Matrix Science Ltd | N/A |
Additional files
-
Supplementary file 1
Summary of DNA/RNA-based reagents used for CRISPR-Cas9.
- https://cdn.elifesciences.org/articles/82333/elife-82333-supp1-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/82333/elife-82333-mdarchecklist1-v2.pdf





