Host-microbiome metabolism of a plant toxin in bees
Figures
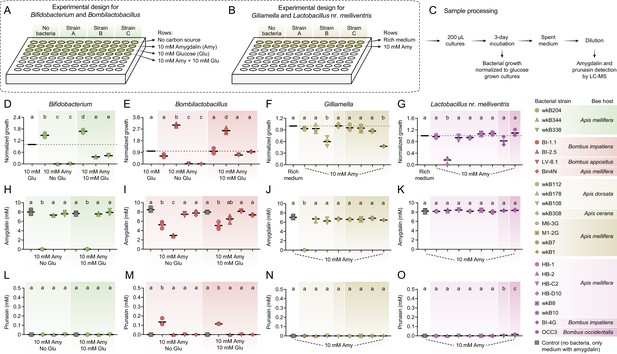
In vitro exposure of bee gut bacteria to amygdalin.
Experimental design in (A) semi-defined or (B) nutritionally rich media in 96-well plates. (C) Sample processing for LC-MS analysis. (D) Bifidobacterium and (E) Bombilactobacillus growth in semi-defined media in the presence of amygdalin (or amygdalin and glucose) normalized to growth in the presence of glucose. (F) Gilliamella and (G) Lactobacillus nr. melliventris growth in nutritionally rich media in the presence of amygdalin normalized to growth in the absence of amygdalin. Bacterial growth was measured as optical density at 600 nm after 3 days of incubation at 35°C and 5% CO2. (H–K) Amygdalin and (L–O) prunasin concentrations in spent medium of amygdalin (or amygdalin and glucose) grown cultures of Bifidobacterium, Bombilactobacillus, Gilliamella, and Lactobacillus nr. melliventris, respectively. Controls consisted of media with amygdalin (or amygdalin and glucose) but no bacteria. Experiments were performed in three biological replicates. Groups with different letters are significantly different (p < 0.01, one-way ANOVA test followed by Tukey’s multiple-comparison test).
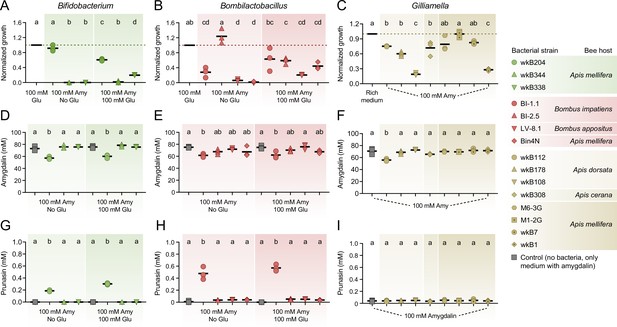
In vitro exposure of bee gut associated bacteria to 100 mM amygdalin.
Growth of (A) Bifidobacterium and (B) Bombilactobacillus strains in semi-defined media in the presence of 100 mM amygdalin (or 100 mM amygdalin and 100 mM glucose) normalized to the bacterial growth in the presence of 100 mM glucose. Growth of (C) Gilliamella strains in nutritionally rich media in the presence of 100 mM amygdalin normalized to the bacterial growth in the absence of amygdalin. Bacterial growth was measured as optical density at 600 nm after 3 days of incubation at 35°C and 5% CO2. (D–F) Amygdalin and (G–H) prunasin concentrations in spent medium of amygdalin (or amygdalin and glucose) grown cultures of Bifidobacterium, Bombilactobacillus, and Gilliamella strains, respectively. Controls consisted of media with amygdalin (or amygdalin and glucose) but no bacteria. Experiments were performed in three biological replicates. Groups with different letters are statistically significantly different (p < 0.01, one-way ANOVA test followed by Tukey’s multiple-comparison test).
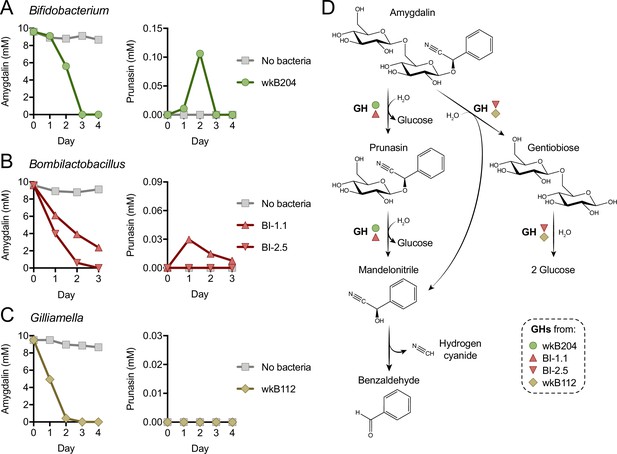
Mechanism of amygdalin degradation by bee gut bacteria.
Amygdalin and prunasin concentrations detected by LC-MS in spent-medium of 3- or 4-day-old cultures of (A) Bifidobacterium strain wkB204, (B) Bombilactobacillus strains BI-1.1 and BI-2.5, and (C) Gilliamella strain wkB112. Concentrations were determined every day for 3–4 days. Controls consisted of medium with amygdalin but no bacteria. Only wkB204 and BI-1.1 produced prunasin as an intermediate. (D) Proposed mechanism of amygdalin degradation by different bacterial species in the bee gut.
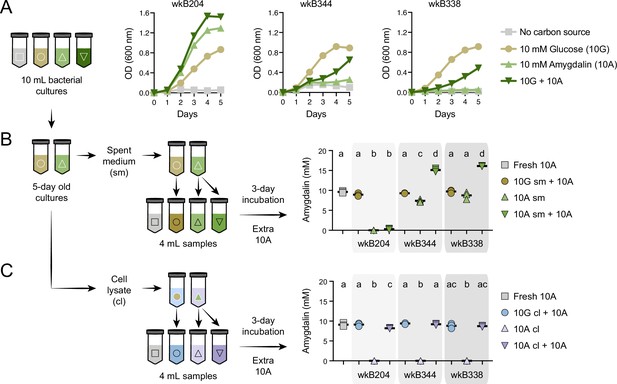
Amygdalin degradation in spent media and cell lysates of Bifidobacterium strains.
(A) Bacterial growth curves of Bifidobacterium strains cultured in semi-defined media (SDM) without a carbon source, with 10 mM glucose (10G), with 10 mM amygdalin (10A), or with both 10 mM glucose and 10 mM amygdalin (10G+10A) as carbon sources at 35°C and 5% CO2. Experiments were performed in three biological replicates. Each data point represents the average optical density (600 nm) measured every day for 5 days. (B–C) For each strain, 10G and 10A grown cultures were separated into (B) spent medium (sm), originating from samples 10G sm and 10A sm, and (C) cell lysate (cl), originating from samples 10G cl and 10 A cl. These samples were used to investigate amygdalin degradation by adding extra 10A to the samples. Controls consisted of 10A grown cultures without adding extra 10A and fresh SDM with 10A. Reactions were incubated at 35°C and 5% CO2 for 3 days, after which amygdalin concentration was determined. Experiments were performed in three biological replicates. Groups with different letters are significantly different (p < 0.01, one-way ANOVA test followed by Tukey’s multiple-comparison test).
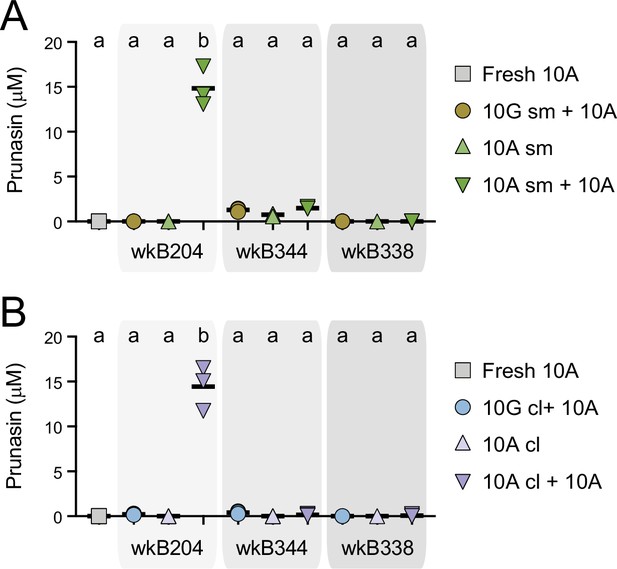
Prunasin concentrations in spent media and cell lysates of Bifidobacterium strains.
Bifidobacterium strains were cultured in semi-defined media (SDM) without a carbon source, with 10 mM glucose (10G), with 10 mM amygdalin (10A), or with both 10 mM glucose and 10 mM amygdalin (10G+10A) as carbon sources at 35°C and 5% CO2. For each strain, 10G and 10A grown cultures were separated into (A) spent medium (sm), originating the samples 10G-sm and 10A-sm, and (B) cell lysate (cl), originating the samples 10G-cl and 10A-cl. These samples were used to investigate prunasin release by adding extra 10A to the samples. Controls consisted of 10A grown cultures without adding extra 10A and fresh SDM with 10A. Reactions were incubated at 35°C and 5% CO2 for 3 days, after which amygdalin concentration (see Figure 6) and prunasin release were determined. Experiments were performed in three biological replicates. Groups with different letters are statistically significantly different (p < 0.05, one-way ANOVA test followed by Tukey’s multiple-comparison test).
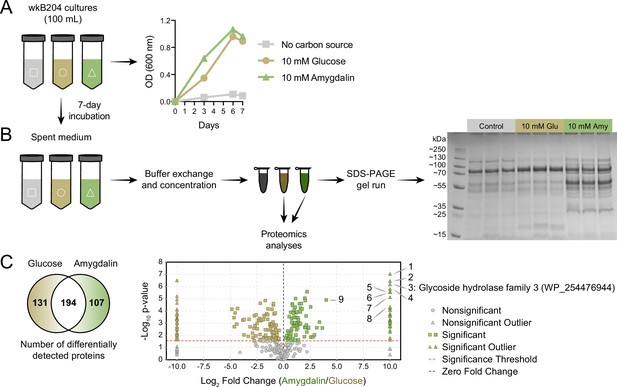
Identification of an amygdalin degrading enzyme from Bifidobacterium.
(A) Large-scale culture of Bifidobacterium strain wkB204 in semi-defined media (SDM) without a carbon source, with 10 mM glucose, or with 10 mM amygdalin at 35°C and 5% CO2. Experiments were performed in three biological replicates and each data point represents the average optical density (600 nm) measured every day for 7 days. (B) Spent medium concentration for running on an SDS-PAGE gel. (C) Venn diagram and volcano plot showing the number of differentially expressed proteins in spent medium of glucose- or amygdalin-grown cultures. Numbers in the volcano plot: 1: alpha/beta fold hydrolase (WP_254477374), 2: nucleoside hydrolase (WP_254477231), 3: glycoside hydrolase family 3 (WP_254476944), 4: beta-galactosidase (WP_254477161), 5: alpha-mannosidase (WP_254477012), 6: Nudix hydrolase (WP_254477413), 7: MFS transporter (WP_254476943), 8: alpha-L-fucosidase (WP_254477430), 9: glycoside hydrolase family 30 (WP_254477160) (p<0.05, t-test followed by Benjamini-Hochberg procedure to control for false discovery rate).
-
Figure 4—source data 1
SDS-PAGE gel run for cultures of Bifidobacterium strain wkB204.
From left to right, columns represent: (1) PageRuler Plus Prestained Protein Ladder; (2–10) Supernatants of cultures (1–4) grown in the absence of a carbon source, (5–7) in the presence of 10 mM glucose as sole carbon source, or (8–10) in the presence of 10 mM amygdalin as sole carbon source. Each sample (30 μL) was mixed with 5 μL of 6× SDS gel-loading buffer (0.35 M Tris-Cl pH 6.8, 10% w/v SDS, 0.012% w/v bromophenol blue, 30% v/v glycerol, 0.6 mM dithiothreitol), denatured at 100°C for 5 min, then run on a Bolt 4–12% Bis-Tris Plus, 1.0 mm, protein gel at 200 V for 22 min.
- https://cdn.elifesciences.org/articles/82595/elife-82595-fig4-data1-v2.zip
-
Figure 4—source data 2
Differential protein expression analysis for amygdalin- and glucose-grown cultures of Bifidobacterium strain wkB204.
- https://cdn.elifesciences.org/articles/82595/elife-82595-fig4-data2-v2.xlsx
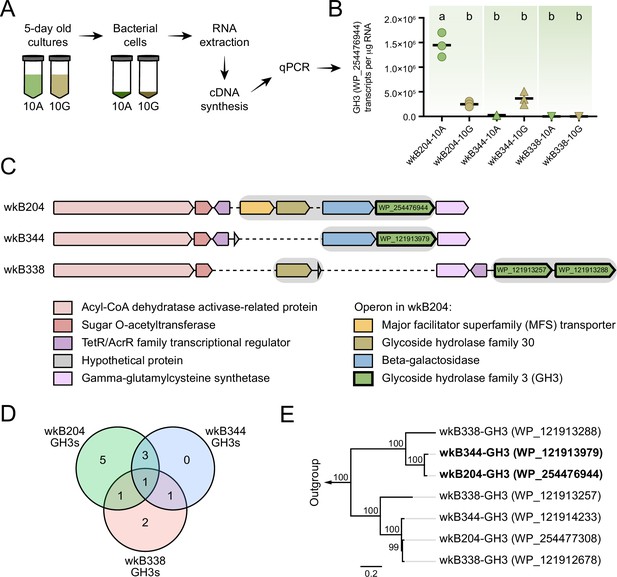
Glycoside hydrolase family 3 (GH3) gene expression in Bifidobacterium cultures.
(A) RNA extraction and complementary DNA (cDNA) synthesis from cultures of Bifidobacterium strains wkB204, wkB344, and wkB338. (B) qPCR data for the transcript levels of GH3 in cells of Bifidobacterium strains cultured in the presence of 10 mM glucose (10G) or 10 mM amygdalin (10A). Experiments were performed in three biological replicates. Groups with different letters are significantly different (p < 0.01, one-way ANOVA test followed by Tukey’s multiple-comparison test). (C) The genomic region containing the GH3 gene with high sequence similarity in wkB204 and wkB344. The corresponding region is included for wkB338 for comparison. Gray shading indicates operons. Dashed lines indicate regions not present in the genome. (D) Venn diagram showing the number of GH3s shared between the strains with amino acid similarity to other annotated GH3s according to the NCBI inference database. (E) Phylogenetic analysis for the GH3s found in the genomic regions shown in C. Outgroup is represented by two amygdalin-degrading GH3s isolated from Rhizomucor miehei strain RmBglu3B (AIY32164.1) and Talaromyces cellulolyticus strain Bgl3B (GAM39187.1).
-
Figure 5—source data 1
dbCAN meta server results for Bifidobacterium strains wkB204, wkB344, and wkB338.
- https://cdn.elifesciences.org/articles/82595/elife-82595-fig5-data1-v2.xlsx
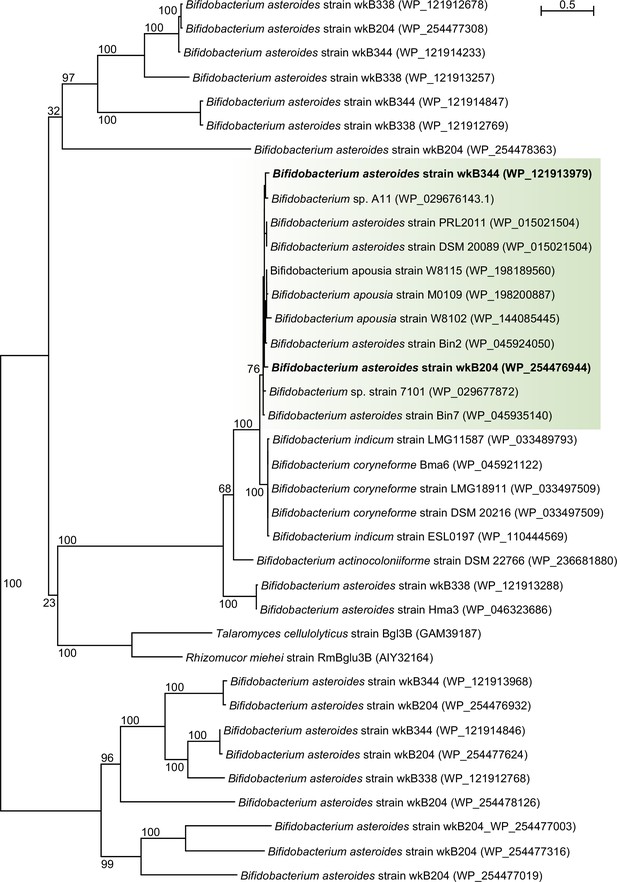
Maximum-likelihood phylogeny based on amino acid sequences of bee associated Bifidobacterium glycoside hydrolases with sequence homology to a glycoside hydrolase family 3 highly expressed in amygdalin-grown cultures of Bifidobacterium strain wkB204 (PhyML 3.1, LG model + Gamma4, 100 bootstrap replicates).
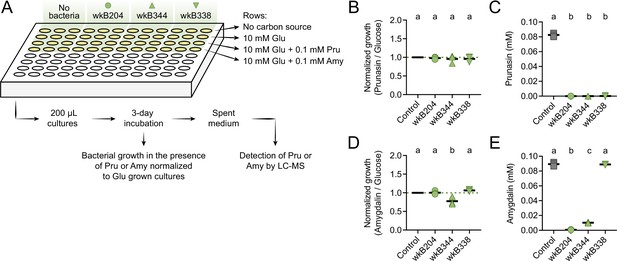
Prunasin degradation by bee gut-associated Bifidobacterium strains.
(A) Experimental design. (B) Bacterial growth, and (C) prunasin degradation after 3 days of incubation in the presence of 0.1 mM prunasin. (D) Bacterial growth and (E) amygdalin degradation after 3 days of incubation in the presence of 0.1 mM amygdalin. Experiments were performed in three biological replicates. Groups with different letters are significantly different (p < 0.01, one-way ANOVA test followed by Tukey’s multiple-comparison test).
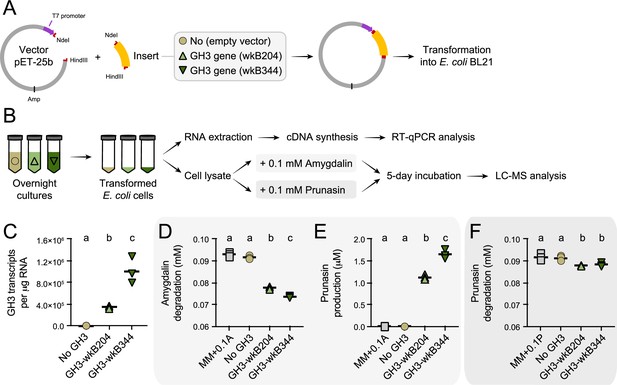
Heterologous expression of Bifidobacterium glycoside hydrolase family 3 (GH3) enzyme in Escherichia coli.
(A) E. coli Rosetta BL21 competent cells were transformed with the vector pET-25b carrying the gene that encodes the wkB204-GH3 or wkB344-GH3, or only the empty vector as a control. (B) Bacterial cells from overnight cultures were lysed to extract RNA and investigate the expression levels of cloned genes by RT-qPCR. In parallel, bacterial cells from similar overnight cultures were lysed and used in incubation assays with 0.1 mM amygdalin or 0.1 mM prunasin in minimal medium at 37°C. Samples were submitted for LC-MS analysis along with amygdalin and prunasin standards. (C) Transcript levels of Bifidobacterium-related GH3 genes expressed in E. coli. (D) Amygdalin degradation and (E) prunasin production levels after 5 days of incubation in the presence of 0.1 mM amygdalin. (F) Prunasin degradation levels after 5 days of incubation in the presence of 0.1 mM prunasin. Experiments were performed in three biological replicates. Groups with different letters are significantly different (p < 0.01, one-way ANOVA test followed by Tukey’s multiple-comparison test).
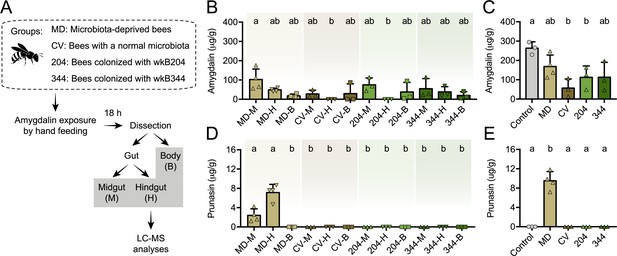
Amygdalin metabolism in honey bees.
(A) Five-day old bees either lacking a microbiota (microbiota deprived, MD, n=4), with a normal microbiota (conventionalized, CV, n=3), or monocolonized with Bifidobacterium strains wkB204 (n=3) or wkB344 (n=3), were exposed to 5 μL of 1 mM amygdalin and dissected 24 hr later to determine the concentrations of (B) amygdalin and (D) prunasin in different bee body compartments (midgut: M, hindgut: H, and body without gut: B) by LC-MS. (C) Amygdalin and (E) prunasin concentrations detected in M, H, and B samples were summed for each group and compared to a control group of unexposed bees that were mixed with 5 μL of 1 mM amygdalin at the beginning of sample processing. Groups with different letters are significantly different (p < 0.05, one-way ANOVA test followed by Tukey’s multiple-comparison test).
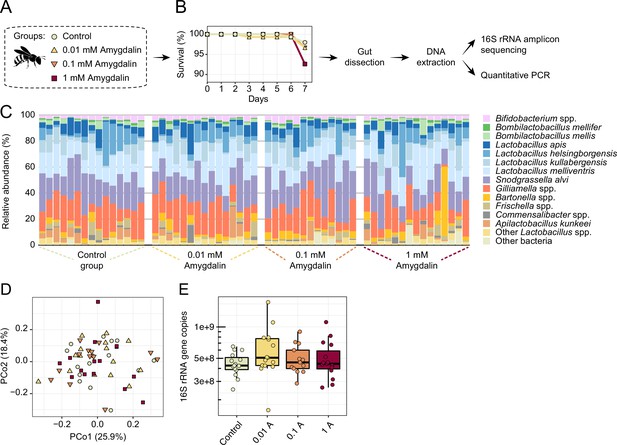
Amygdalin effects on the honey bee gut microbiota.
(A) Experimental design and (B) survival rates of honey bees exposed to different concentrations of amygdalin. (C) Stacked column graphs showing the relative abundance of bee gut bacterial species in control bees (n=15), 0.01 mM amygdalin (n=15), 0.1 mM amygdalin (n=13), and 1 mM amygdalin (n=15) exposed bees. (D) Principal coordinate analysis of gut community compositions of control and amygdalin exposed bees using Bray-Curtis dissimilarity (p>0.5, Permanova test with 9999 permuations). (E) Boxplot of total bacterial 16S rRNA gene copies estimated by qPCR for control and amygdalin exposed bees. Box-and-whisker plots show high, low, and median values, with lower and upper edges of each box denoting first and third quartiles, respectively. No significant differences were observed in total bacterial abundance between control and amygdalin exposed bees (p>0.05, Kruskal-Wallis test).
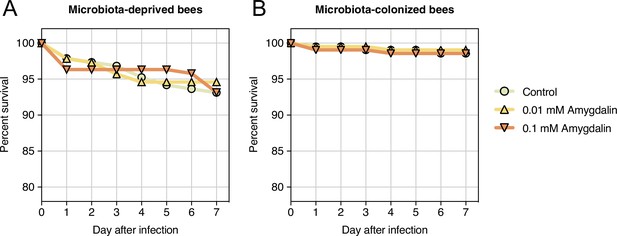
Survival rates of honey bees exposed to different concentrations of amygdalin.
(A) Microbiota-deprived bees were exposed to sterile sucrose syrup (control, n=189 bees), 0.01 mM amygdalin in sterile sucrose syrup (n=185 bees) or 0.1 mM amygdalin in sucrose syrup (n=190 bees). (B) Microbiota-colonized bees were exposed to sterile sucrose syrup (control, n=210 bees), 0.01 mM amygdalin in sterile sucrose syrup (n=210 bees) or 0.1 mM amygdalin in sucrose syrup (n=210 bees). Bees from each group were split into 6 cup cages. No significant effects were found between control and treatment groups.
Tables
Glycoside hydrolases family 3 (GH3) detected in the genomes of Bifidobacterium strains wkB204, wkB344, and wkB338.
Protein ID refers to the unique identification of each GH3 in the NCBI Reference Sequence Database. Inference refers to the closest related GH3 present in the NCBI Reference Sequence Database. Same colors and superscript letters indicate GH3s with similar amino acid sequence. This information was used to make the Venn diagram in Figure 5D.
| Strain | GH3 loci number | Protein ID (NCBI RefSeq) | Inference (NCBI RefSeq) |
|---|---|---|---|
| wkB204 | 10 | WP_254476932a | WP_007147852 |
| WP_254476944b | WP_015021504 | ||
| WP_254477003 | WP_003842825 | ||
| WP_254477019 | – | ||
| WP_254477308c | WP_015022086 | ||
| WP_254477316 | – | ||
| WP_254477624d | WP_016461981 | ||
| WP_254477626e | WP_004221005 | ||
| WP_254478126 | – | ||
| WP_254478363 | – | ||
| wkB344 | 5 | WP_121913968a | WP_007147852 |
| WP_121913979b | WP_015021504 | ||
| WP_121914233c | WP_015022086 | ||
| WP_121914846d | WP_016461981 | ||
| WP_121914847 | WP_004221005 | ||
| wkB338 | 5 | WP_121912678c | WP_015022086c |
| WP_121912768 | WP_003838412 | ||
| WP_121912769e | WP_004221005e | ||
| WP_121913257 | WP_003839235 | ||
| WP_121913288 | WP_015450023 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Bifidobacterium asteroides) | wkB204 | NCBI Reference Sequence | Locus: WP_254476944 | |
| Gene (Bifidobacterium asteroides) | wkB344 | NCBI Reference Sequence | Locus: WP_121913979 | |
| Strain, strain background (Escherichia coli) | DH5-alpha | New England BioLabs | Cat#: C2987H | NEB 5-alpha competent cells |
| Strain, strain background (Escherichia coli) | BL21 (DE3) | New England BioLabs | Cat#: C2527H | Electrocompetent cells |
| Strain, strain background (Bifidobacterium asteroides) | wkB204 | This paper | JAFMNU020000000 | Bacterial isolate |
| Strain, strain background (Bifidobacterium asteroides) | wkB344 | doi:10.1073/pnas.1916224116 | NPOQ00000000 | Bacterial isolate |
| Strain, strain background (Bifidobacterium asteroides) | wkB338 | doi:10.1073/pnas.1916224116 | NPOR00000000 | Bacterial isolate |
| Strain, strain background (Bombilactobacillus bombi) | BI-2.5 | This paper | CP031513 | Bacterial isolate |
| Strain, strain background (Bombilactobacillus bombi) | BI-1.1 | This paper | QOCR00000000 | Bacterial isolate |
| Strain, strain background (Bombilactobacillus bombi) | LV-8.1 | This paper | QOCS00000000 | Bacterial isolate |
| Strain, strain background (Bombilactobacillus mellifer) | Bin4N | doi:10.1099/ijs.0.059600–0 doi:10.1099/ijsem.0.004107 | JXJQ00000000 | Bacterial isolate |
| Strain, strain background (Lactobacillus bombicola) | OCC3 | This paper | QOCV00000000 | Bacterial isolate |
| Strain, strain background (Lactobacillus bombicola) | BI-4G | This paper | QOCU00000000 | Bacterial isolate |
| Strain, strain background (Lactobacillus nr. melliventris) | HB-1 | This paper | OQ216581 | Bacterial isolate |
| Strain, strain background (Lactobacillus nr. melliventris) | HB-2 | This paper | OQ216582 | Bacterial isolate |
| Strain, strain background (L. nr. melliventris) | HB-C2 | This paper | OQ216583 | Bacterial isolate |
| Strain, strain background (Lactobacillus nr. melliventris) | HB-D10 | This paper | OQ216584 | Bacterial isolate |
| Strain, strain background (Lactobacillus helsingborgensis) | wkB8 | doi:10.1128/genomeA.01176–14 | CP009531 | Bacterial isolate |
| Strain, strain background (Lactobacillus kullabergensis) | wkB10 | doi:10.1128/genomeA.01176–14 | JRJB00000000 | Bacterial isolate |
| Strain, strain background (Gilliamella apicola) | wkB1 | doi:10.1073/pnas.1405838111 | CP007445 | Bacterial isolate |
| Strain, strain background (Gilliamella apicola) | wkB7 | doi:10.1128/mBio.01326–16 | LZGG00000000 | Bacterial isolate |
| Strain, strain background (Gilliamella apis) | M1-2G | doi:10.1128/mBio.01326–16 | LZGQ00000000 | Bacterial isolate |
| Strain, strain background (Gilliamella sp.) | wkB112 | doi:10.1128/mBio.01326–16 | LZGL00000000 | Bacterial isolate |
| Strain, strain background (Gilliamella sp.) | wkB178 | doi:10.1128/mBio.01326–16 | LZGK00000000 | Bacterial isolate |
| Strain, strain background (Gilliamella sp.) | wkB108 | doi:10.1128/mBio.01326–16 | LZGM00000000 | Bacterial isolate |
| Strain, strain background (Gilliamella sp.) | wkB308 | doi:10.1128/mBio.01326–16 | LZGN00000000 | Bacterial isolate |
| Strain, strain background (Gilliamella sp.) | M6-3G | doi:10.1128/mBio.01326–16 | MCIU00000000 | Bacterial isolate |
| Biological sample (Apis mellifera) | Western honey bee Apis mellifera | Collected from hives at UT-Austin | ||
| Recombinant DNA reagent | pGEM-T Easy vector (plasmid) | Promega | Cat#: A1360 | |
| Recombinant DNA reagent | pET25b (plasmid) | Novagen | Cat#: 69753 | |
| Recombinant DNA reagent | pET25b-wkB204-GH3 (plasmid) | This study | pET25b expressing wkB204- GH3 (WP_254476944) | |
| Recombinant DNA reagent | pET25b-wkB344-GH3 (plasmid) | This study | pET25b expressing wkB344- GH3 (WP_121913979) | |
| Sequence-based reagent | B-GH3-F | This paper | PCR primers | ctaccgcaatcccgacct |
| Sequence-based reagent | B-GH3-R | This paper | PCR primers | cacctccttgtccactccc |
| Sequence-based reagent | GH3-NdeI-F | This paper | PCR primers | ttgtttaactttaagaaggagatatacatatggcatcaaggaagttgacagagg |
| Sequence-based reagent | GH3-HindIII-R | This paper | PCR primers | agcccgtttgatctcgagtgcggccgcaagcttacccacggtcaccgtca |
| Commercial assay or kit | Quick-RNA Miniprep kit | Zymo Research | Cat#: R1055 | |
| Commercial assay or kit | iTaq Universal SYBR Green Supermix | Bio-Rad | Cat#: 172–5125 | |
| Commercial assay or kit | Monarch Plasmid Miniprep Kit | New England BioLabs | Cat#: T1010L | |
| Commercial assay or kit | qScript cDNA Synthesis Kit | QuantBio | Cat#: 95047–500 | |
| Chemical compound, drug | Amygdalin | Chem-Impex International | Cat#: 22029 | Lot#: 002681–16112001 |
| Chemical compound, drug | Prunasin | Toronto Research Chemicals | Cat#: P839000 | Lot#: 6-EQJ-155–1 |
| Chemical compound, drug | Ampicillin | Fisher Bioreagents | Cat#: BP1760-5 | |
| Chemical compound, drug | Isopropyl β-D-1-thiogalactopyranoside (IPTG) | Gold Biotechnology | Cat#: I2481C25 | |
| Chemical compound, drug | Antarctic Phosphatase | New England BioLabs | Cat#: M0289S | Enzyme |
| Chemical compound, drug | NdeI | New England BioLabs | Cat#: R0111S | Restriction enzyme |
| Chemical compound, drug | HindIII-HF | New England BioLabs | Cat#: R3104S | Restriction enzyme |
| Software, algorithm | SeaView | http://pbil.univ-lyon1.fr/software/seaview3.html | RRID:SCR_015059 | |
| Other | Insectagro DS2 media | Corning | Cat#: 13-402-CV | Lot#: 12818007 |
| Other | Difco Lactobacilli MRS broth | BD | Cat#: 288130 | Lot#: 9211338 |
| Other | Heart Infusion Agar | Criterion | Cat#: C5822 | Lot#: 491030 |
| Other | Defibrinated Sheep Blood | HemoStat Laboratories | Cat#: DSB1 | Lot#: 663895–2 |
| Other | Protein extraction reagent (B-PER) | Thermo Scientific | Cat#: 78248 | Lot#: LJ148147A |
| Other | Bolt 4–12% Bis- Tris Plus Gel | Thermo Scientific | Cat#: NW04120BOX | Lot#: 21022470 |
Composition of a semi-defined medium (SDM) recipe used to culture Bifidobacterium and Lactobacillus strains.
Specific carbon sources (amygdalin and/or glucose) were added according to the experiments. Recipe was adapted from Walker et al., 2014.
| Ingredient | Amount (g/L) |
|---|---|
| Defined | |
| Ammonium chloride | 2 |
| Cysteine hydrochloride | 0.4 |
| Magnesium chloride | 0.08 |
| Manganese chloride | 0.08 |
| Nicotinic acid | 0.5 |
| Pantothenic acid | 0.5 |
| Potassium phosphate monobasic | 2 |
| Pyridoxine hydrochloride | 0.1 |
| Sodium acetate | 5 |
| Undefined | |
| Yeast extract | 4 |
| SC, synthetic complete supplement (Sunrise Scientific Products, Knoxville, TN, USA) | 2 |
| Tween 80 | 1 |
Composition of a minimum medium (MM, pH 6.8) recipe used to culture transformed Escherichia coli strains.
Specific carbon sources (amygdalin, prunasin, or glucose) were added according to the experiments. Recipe was adapted from Li et al., 2014.
| Ingredient | Amount (g/L) |
|---|---|
| Ammonium iron (III) citrate | 0.1 |
| Ammonium phosphate tetrahydrate | 4 |
| Boric acid | 0.003 |
| Citric acid | 1.55 |
| Cobalt (II) chloride hexahydrate | 0.003 |
| Copper (II) chloride dihydrate | 0.002 |
| Magnesium sulfate | 0.59 |
| Manganese (II) chloride tetrahydrate | 0.015 |
| Potassium phosphate monobasic | 13.3 |
| Sodium molybdate dihydrate | 0.002 |
| Zinc sulfate heptahydrate | 0.034 |





