Dependence of diffusion in Escherichia coli cytoplasm on protein size, environmental conditions, and cell growth
Figures
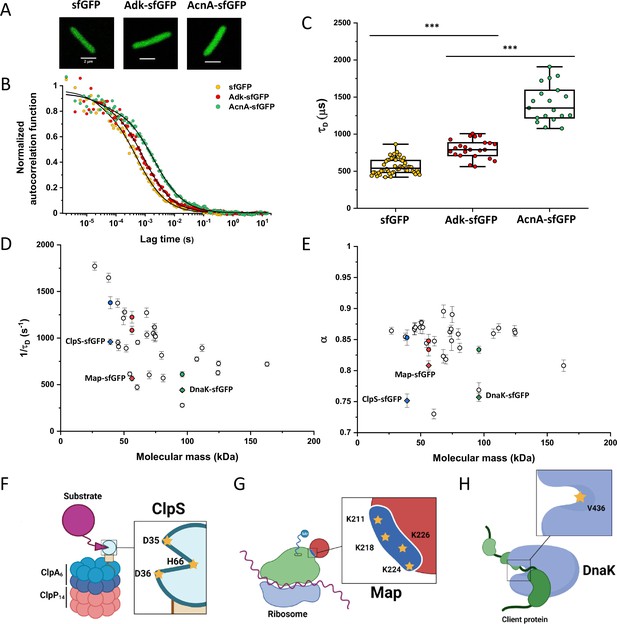
Dependence of protein mobility in bacterial cytoplasm on molecular mass and cellular interactions.
(A) Examples of fluorescence microscopy images of Escherichia coli cells expressing either sfGFP or the indicated sfGFP-tagged cytoplasmic proteins. Scale bars are 2 μm. (B) Representative autocorrelation functions (ACFs) measured by FCS for the indicated protein constructs. Data were fitted using the anomalous diffusion model (solid lines). All ACF curves were normalized to their respective maximal values to facilitate comparison. (C) Diffusion times (τD) measured for the indicated protein constructs. Each dot in the box plot represents the value for one individual cell, averaged over six consecutive acquisitions (Figure 1—figure supplement 3). The numbers of cells measured for each construct are shown in Appendix 6. ***p<0.0001 in a two-tailed heteroscedastistic t-test. Exact p-valuescan be found in Appendix 5. (D, E). Dependence of protein mobility (1/τD; D) and apparent anomaly of diffusion (α; E) on molecular mass. Each symbol represents the average value for all individual cells that have been measured for that particular construct and the error bars represent the standard error of the mean. Individual values are shown in Figure 1—figure supplement 5 and the numbers of measured cells for each construct are shown in Appendix 6. Protein constructs with low mobility for which effects of specific interactions were further investigated are highlighted in color and labeled. The values of 1/τD and α for both the original constructs (diamonds) and the constructs where mutations were introduced to disrupt interactions (circles) are shown. For Map, two alternative amino acid substitutions that disrupt its interaction with the ribosome are shown (see Figure 1—figure supplement 10). (F–H) Cartoons illustrating the cellular interactions that could affect mobility of ClpS (F), Map (G), and DnaK (H). ClpS engages with the ClpAP protease and with substrates, cartoon adapted from Figure 1A from Román-Hernández et al., 2011. Map interacts with the actively translating ribosomes, cartoon adapted from Figure 3A from Sandikci et al., 2013. DnaK interacts with unfolded client protein. Amino acidic residues that were mutated to disrupt interactions are highlighted (see text for details). FCS, fluorescence correlation spectroscopy.
-
Figure 1—source data 1
Individual τD measurements from Figure 1C.
- https://cdn.elifesciences.org/articles/82654/elife-82654-fig1-data1-v3.xlsx
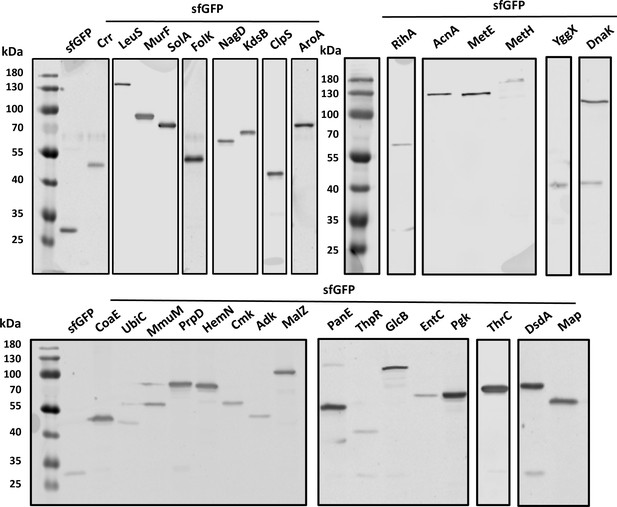
Expression analysis for all Escherichia coli protein constructs made in this study.
Expression of proteins was analyzed by SDS-PAGE and immunoblotting using a primary antibody specific for GFP. All fusion proteins displayed a dominant band corresponding to the expected molecular mass of the full-length fusion (Table 1).
-
Figure 1—figure supplement 1—source data 1
Uncropped western blot images for Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/82654/elife-82654-fig1-figsupp1-data1-v3.zip
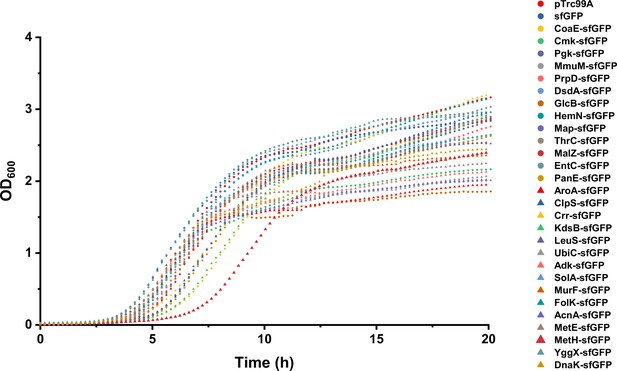
Growth curves of Escherichia coli strains expressing tested sfGFP-tagged proteins.
Growth curves of strains expressing sfGFP, sfGFP-tagged proteins, or carrying the control empty vector pTrc99A were measured at 37°C. The optical density of cultures was monitored at 600 nm (OD600) and the measured values were normalized for an optical path of 1 cm.
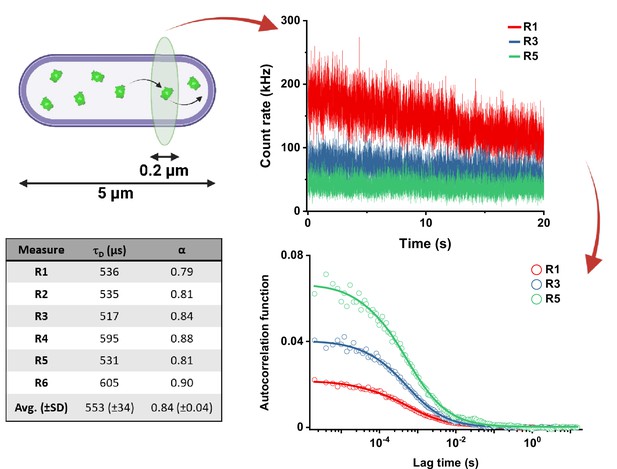
Workflow of a typical FCS experiment.
The focus of the confocal microscope is positioned near one pole in the cell of interest, and six subsequent acquisitions of the fluorescence intensity of 20 s each are performed. The example for sfGFP shows only traces for R1, R3, and R5 measurements (different colors). The autocorrelation functions (ACFs) are then calculated independently for each acquisition. The values for τD and α are extracted independently from the fit to each ACF using the anomalous diffusion model (solid lines) and then averaged to obtain the τD and α for each individual cell.
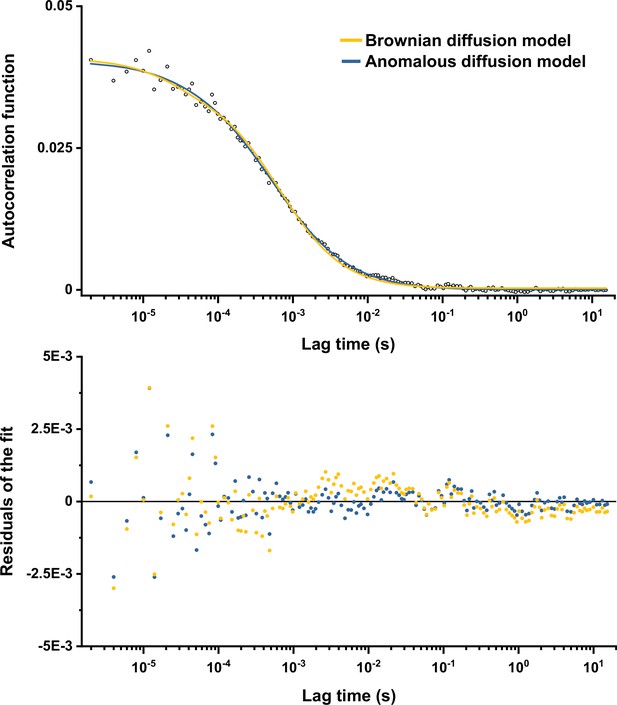
Comparison between fits of the experimental data with Brownian and anomalous diffusion models.
The experimental data (here the example for the R3 measurement from Figure 1—figure supplement 3) were fitted by the model of free Brownian diffusion and by the anomalous diffusion model as indicated by different colors (upper panel), with the corresponding values of residuals (lower panel).
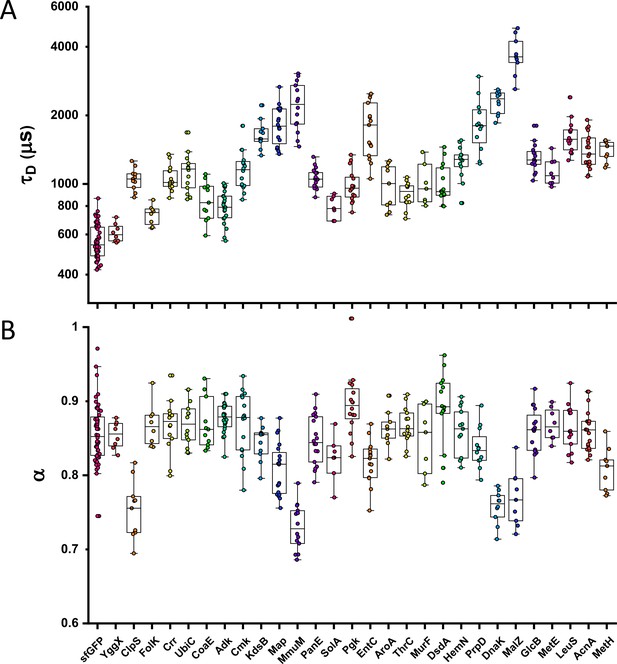
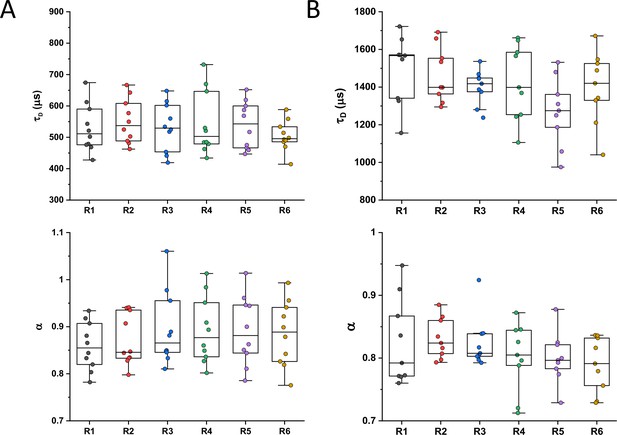
Individual measurements of τD and α for all Escherichia coli protein constructs included in the analysis of mass dependence.
The numbers of measured cells for each construct are shown in Appendix 6. Each dot in the box plot represents the values of τD (A) and α (B) for one individual cell. The averages values calculated from these datasets are shown in Figure 1.
-
Figure 1—figure supplement 5—source data 1
Individual values of τD from Figure 1—figure supplement 5A.
Individual values of α from Figure 1—figure supplement 5B.
- https://cdn.elifesciences.org/articles/82654/elife-82654-fig1-figsupp5-data1-v3.xlsx
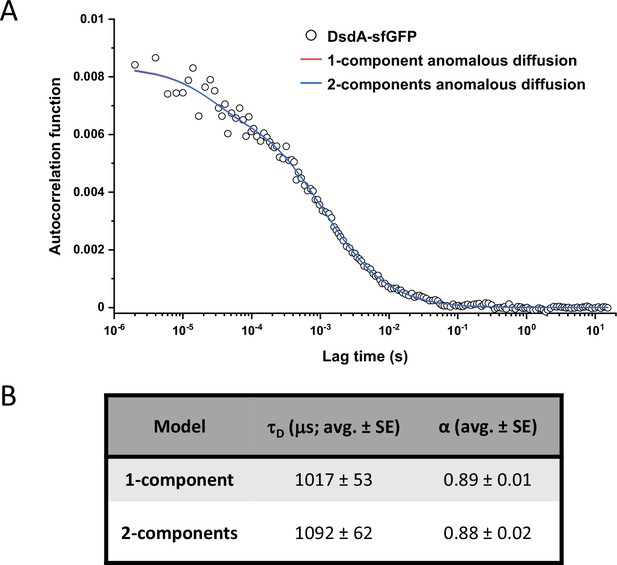
Comparison between one-component and two-components anomalous diffusion fit for DsdA-sfGFP.
(A) Fitting the experimental data for DsdA-sfGFP with a two-component model for anomalous diffusion, where the fast component corresponding to free sfGFP was fixed to 15% (corresponding to the measured degree of fusion protein degradation) with τD = 561 μs and α=0.86, that are, the average values obtained for sfGFP. For comparison, standard fitting with a one-component anomalous diffusion model is also shown. (B) The fitted values of τD with the two-component and one-component model.
-
Figure 1—figure supplement 6—source data 1
Individual values used to calculate mean and standard error of the mean values of τD and α values from Figure 1—figure supplement 6B.
- https://cdn.elifesciences.org/articles/82654/elife-82654-fig1-figsupp6-data1-v3.xlsx
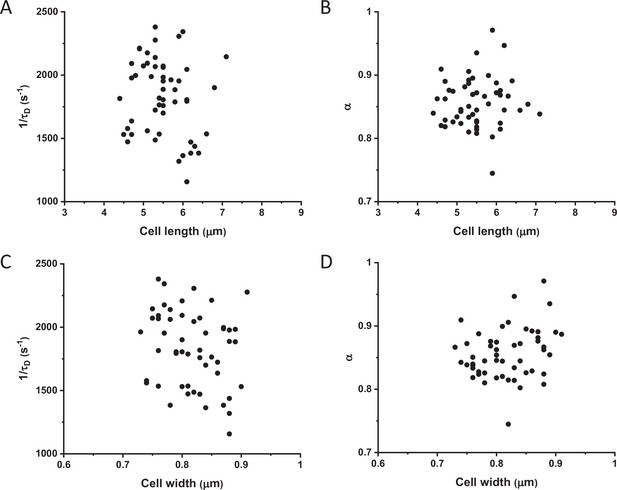
Mobility (1/τD) and anomaly of diffusion (α) of sfGFP in individual cells with different width and length.
Values of 1/τD (A, C) and α (B, D) for single cells expressing sfGFP plotted against the length (A, B) or width (C, D) of respective cell. The numbers of measured cells are shown in Appendix 6.
-
Figure 1—figure supplement 7—source data 1
Individual values of 1/τD and measurements of cell length from Figure 1—figure supplement 7A.
Individual values of α and measurements of cell length from Figure 1—figure supplement 7B. Individual values of 1/τD and measurements of cell width from Figure 1—figure supplement 7C. Individual values of α and measurements of cell width from Figure 1—figure supplement 7D.
- https://cdn.elifesciences.org/articles/82654/elife-82654-fig1-figsupp7-data1-v3.xlsx
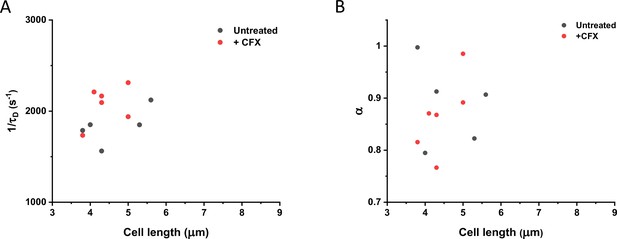
Comparison of protein mobility in cephalexin-treated and untreated cells.
Mobility of sfGFP measured in single cells treated with cephalexin (red dots) or in control untreated cells (black dots). Only cells of similar length were chosen for this comparison. No significant difference in protein mobility (1/τD; p=0.08) or anomaly of diffusion (α; p=0.67) is observed. The numbers of cells measured for each conditions are shown in Appendix 6.
-
Figure 1—figure supplement 8—source data 1
Individual values of 1/τD and measurements of cell length from Figure 1—figure supplement 8A.
Individual values of α and measurements of cell length from Figure 1—figure supplement 8B.
- https://cdn.elifesciences.org/articles/82654/elife-82654-fig1-figsupp8-data1-v3.xlsx
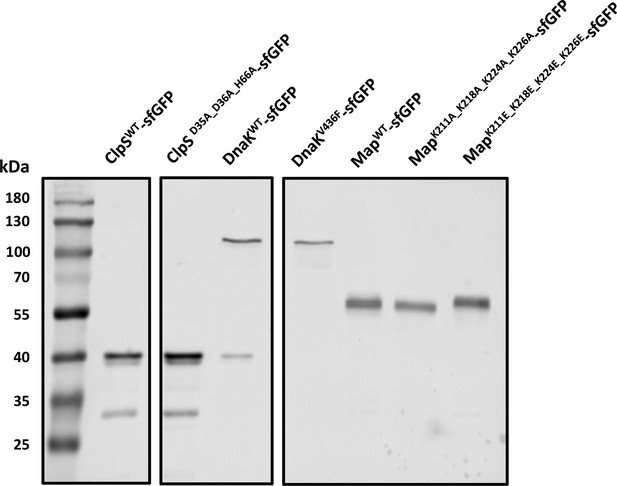
Expression analysis for the mutants with impaired interactions.
The expression of indicated point mutants of ClpS, DnaK, and Map was analyzed by SDS-PAGE and immunoblotting using a primary antibody specific for GFP. All mutants displayed a dominant band corresponding to the expected molecular mass of the full-length fusion, and comparable to that of the wild-type counterpart. ClpSD35A_D36A_H66A-sfGFP was measured in the same ΔclpA background as subsequently used for the FCS experiments.
-
Figure 1—figure supplement 9—source data 1
Uncropped western blot image for Figure 1—figure supplement 9.
- https://cdn.elifesciences.org/articles/82654/elife-82654-fig1-figsupp9-data1-v3.zip
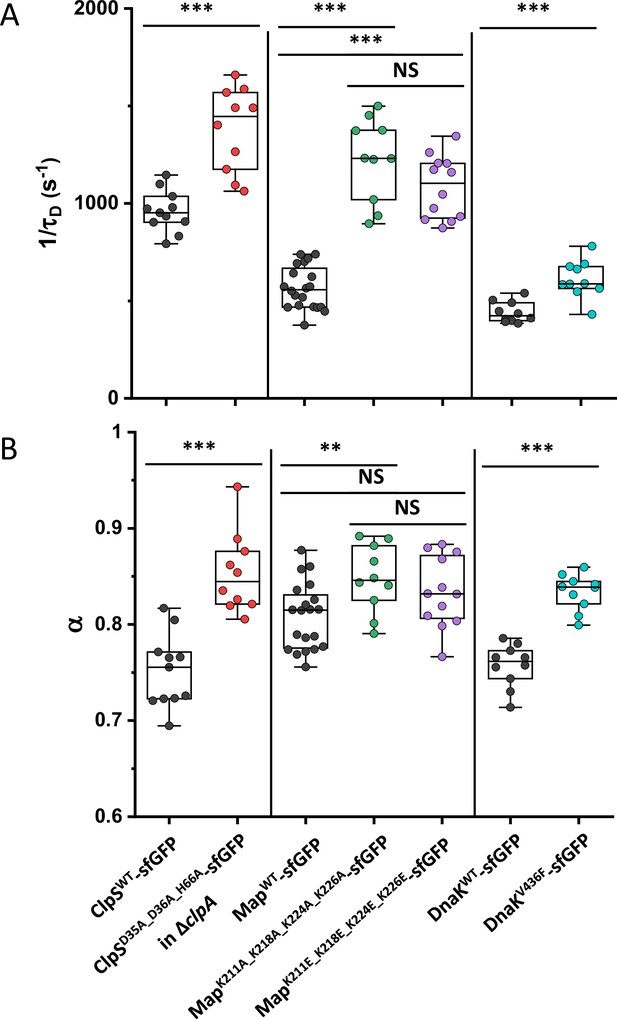
Mobility (1/τD) and anomaly of diffusion (α) of ClpS, Map and DnaK and of indicated mutants with disrupted protein interactions.
Each dot in the box plot represents the values of 1/τD (A) and α (B) for one individual cell. The numbers of cells measured for each construct are shown in Appendix 6. ClpS mutant was measured in ΔclpA background. *** p<0.0001; * p<0.05; NS: no statistically significant difference in a two-tailed heteroscedastistic t-test. Exact p-values can be found in Appendix 5.
-
Figure 1—figure supplement 10—source data 1
Individual values of 1/τD from Figure 1—figure supplement 10A.
Individual values of α from Figure 1—figure supplement 10B.
- https://cdn.elifesciences.org/articles/82654/elife-82654-fig1-figsupp10-data1-v3.xlsx
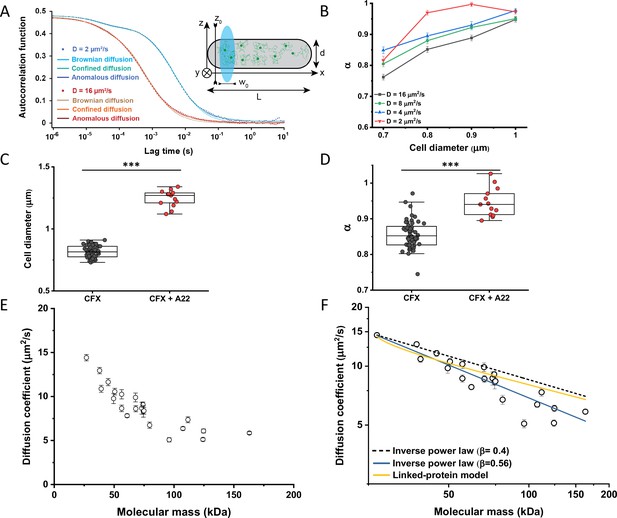
Protein diffusion in bacterial cytoplasm corrected for confinement.
(A) Representative ACFs of simulated fluorescence intensity fluctuations. Simulations were performed in a confined geometry of a cell with indicated length L and diameter d, and dimensions of the measurement volume ω0 and z0, representing an experimental FCS measurement (Inset; see Materials and methods) for two different values of the ansatz diffusion coefficient. Solid lines are fits by the models of unconfined Brownian diffusion, anomalous diffusion and by the Ornstein-Uhlenbeck (OU) model of Brownian diffusion under confinement, as indicated. (B) The exponent α extracted from the fit of the anomalous diffusion model to the ACFs data that were simulated at different values of the cell diameter. Corresponding values of the diffusion coefficient are shown in Figure 2—figure supplement 7. (C, D) Escherichia coli cells treated with cephalexin alone or with cephalexin and 1 µg/ml of A22 (see Materials and methods), show A22-dependent increase in the measured cell diameter (C) and higher values of the exponent α extracted from the fit to the ACF measurements (D). The numbers of cells measured for each condition are shown in Appendix 6. ***p<0.0001 in a two-tailed heteroscedastistic t-test. Exact p-values can be found in Appendix 5. (E) Dependence of the diffusion coefficient calculated from fitting the experimental ACFs with the OU model of confined diffusion. Only the subset of apparently freely diffusing constructs from Figure 1D has been analyzed with the OU model (see also Table 1). Each circle represents the average value for all individual cells that have been measured for that particular construct (Appendix 6), and the error bars represent the standard error of the mean. Error bars that are not visible are smaller than the symbol size. (F) Fit of the mass dependence with an inverse power law (solid blue line, exponent β=0.56±0.05), and predictions of the Stokes-Einstein relation (black dashed line) and of the model describing diffusion of two linked globular proteins (solid yellow line), both with exponent β=0.4. ACF, autocorrelation function; FCS, fluorescence correlation spectroscopy.
-
Figure 2—source data 1
Average and error from each simulation in Figure 2B.
Individual measurements of cell diameters from Figure 2C. Individual measurements of α from Figure 2D. Individual mean and standard error of the mean of diffusion coefficient values from Figure 2E and F.
- https://cdn.elifesciences.org/articles/82654/elife-82654-fig2-data1-v3.xlsx
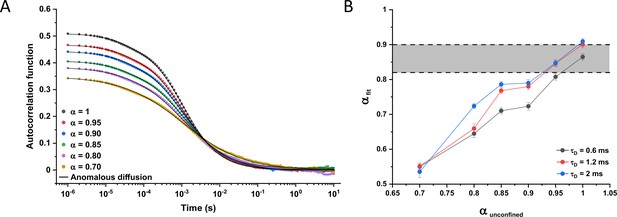
Simulations of particles undergoing fractional Brownian motion.
(A) Examples of simulated autocorrelation functions (ACFs) obtained for particles diffusing under fractional Brownian motion in the confined geometry of a cell of 0.85 μm diameter, with different degrees of anomaly of diffusion (values of α indicated by different colors) and with a fixed diffusion time (τD=[w02/2Γα] 1/α) of 1.2 ms. Data were fitted with the anomalous diffusion model. Similar results were obtained for other values of τD (0.6 and 2 ms). (B) Corresponding anomalous diffusion exponents from the fitting of the simulated ACFs in (A) (αfit, mean values of 5 replicates) compared with the ansatz values of anomalous diffusion exponent for unconfined diffusion (αunconfined) used in simulations for the indicated values of τD. The gray shaded area represents the typical values of α obtained from the fit of experimental ACFs (0.82–0.9). The fitted value of τD was 16%, 10%, and 5% lower than expected from the ansatz τD 0.6, 1.2, and 2 ms, respectively, irrespective of the value of α.
-
Figure 2—figure supplement 1—source data 1
Mean values and standard errors of the mean from Figure 2—figure supplement 1B.
- https://cdn.elifesciences.org/articles/82654/elife-82654-fig2-figsupp1-data1-v3.xlsx
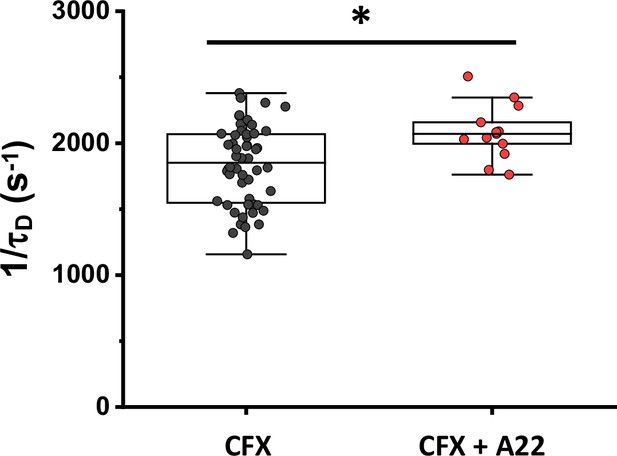
Mobility of sfGFP in cells treated with cephalexin (CFX) or the combination of cephalexin and A22.
Each dot in the box plot represents the values of 1/τD for one individual cell. The numbers of cells measured for each condition are shown in Appendix 6. Cultures are grown for ~3.5 hr in absence or presence of A22 before being both treated for 45 min with cephalexin. *p<0.05 in a two-tailed heteroscedastistic t-test. Exact p-values can be found in Appendix 5.
-
Figure 2—figure supplement 2—source data 1
Individual values of 1/τD from Figure 2—figure supplement 2.
- https://cdn.elifesciences.org/articles/82654/elife-82654-fig2-figsupp2-data1-v3.xlsx
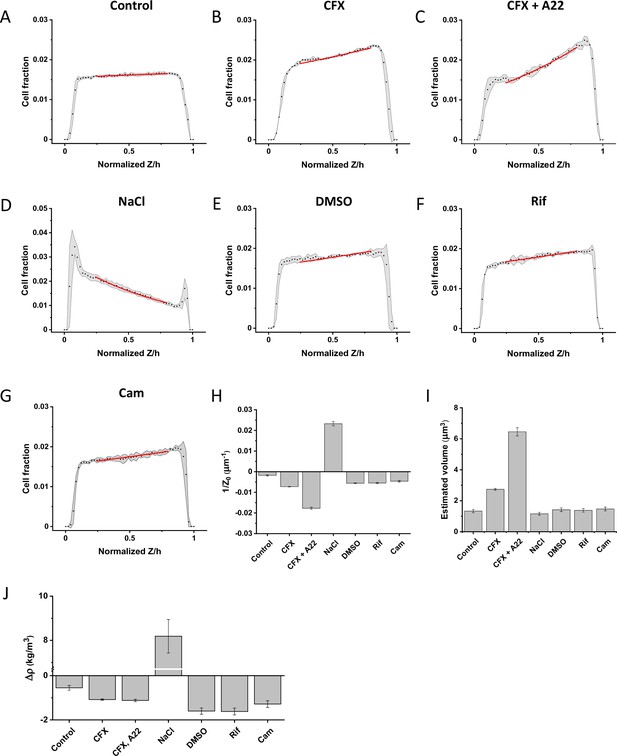
Sedimentation assay of cellular density for indicated treatments.
Sedimentation assays were performed using non-motile and non-aggregating (ΔfliC Δflu) variant of the Escherichia coli strain used in the other experiments. Cells were grown and treated as described in the correspondent sections and assayed for their density in motility buffer (MB) containing 20% iodixanol to match the density of control untreated cells (A). Treatments with cephalexin (CFX; B), cephalexin and A22 (C), 100 mM NaCl (D), DMSO (E), rifampicin (Rif; F), and chloramphenicol (Cam; G) are shown. Dots represent the cell fraction at each given Z position normalized on the total height of the microfluidic channel (50 μm). The gray shadings indicate the standard deviation of the three technical replicates and the red lines represent the exponential fit to the data used to determine the decay length Z0. (H) Average values of 1/Z0 calculated from three technical replicates for each condition. Error bars represent the standard error of the mean. (I) Estimated cell volume for cells in each of the tested conditions. Error bars represent the standard error of the mean. (J) Calculated values of cellular density mismatch for MB with 20% iodixanol, calculated from the values values of 1/Z0 and cell volume. Error bars represent the standard error of the mean. See Materials and methods for details of calculations and cell volume estimation.
-
Figure 2—figure supplement 3—source data 1
Mean values and standard errors of the mean from Figure 2—figure supplement 3H.
Mean values and standard errors of the mean from Figure 2—figure supplement 3I. Mean values and standard errors of the mean from Figure 2—figure supplement 3J.
- https://cdn.elifesciences.org/articles/82654/elife-82654-fig2-figsupp3-data1-v3.xlsx
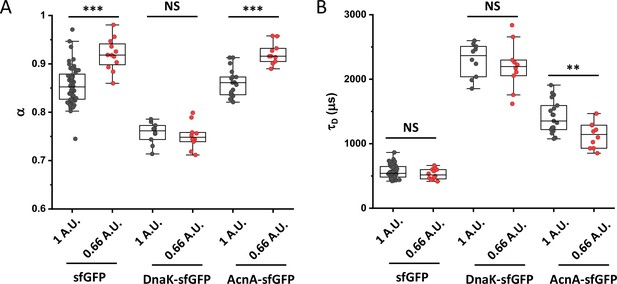
Apparent anomaly of diffusion and residence time for different pinhole sizes.
FCS measurements for indicated protein constructs were performed at the suboptimal pinhole size of 0.66 Airy units (A.U.) to extract values of the anomaly of diffusion (α; A) and residence time (τD; B). The numbers of cells measured for each condition are shown in Appendix 6. ***p<0.0001; **p<0.001; NS: no statistically significant difference in a two-tailed heteroscedastistic t-test.Exact p-values can be found in Appendix 5.
-
Figure 2—figure supplement 4—source data 1
Individual values of α from Figure 2—figure supplement 4A.
Individual values of τD from Figure 2—figure supplement 4B.
- https://cdn.elifesciences.org/articles/82654/elife-82654-fig2-figsupp4-data1-v3.xlsx
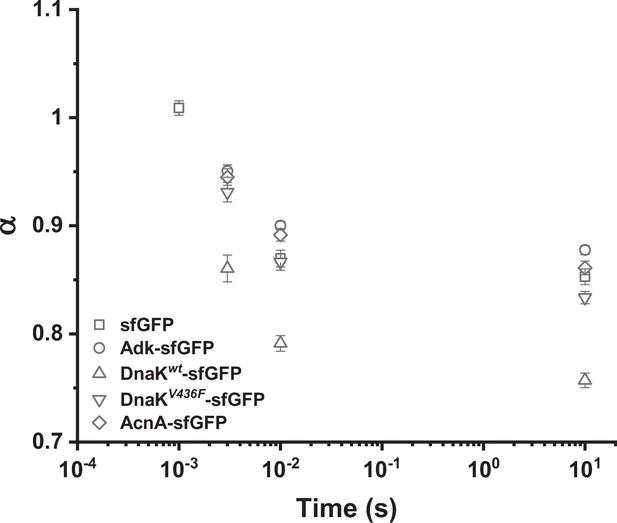
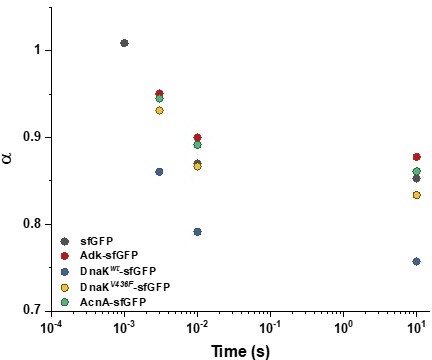
Apparent anomaly of diffusion (α) extracted from analysis of ACFs at shorter time scales.
ACFs measured for indicated proteins were truncated to shorter time scales and fitted with the anomalous diffusion model. The extracted anomaly of diffusion α is plotted as a function of time to which the analysis was limited. The shortest time lag (10–3 s) was analyzed only for sfGFP because for the slower-diffusing constructs this value was comparable to the inflection point of the ACF curve where α is extrapolated. The numbers of cells measured for each construct are shown in Appendix 6.
-
Figure 2—figure supplement 5—source data 1
Average values and standard errors of the mean of α from Figure 2—figure supplement 5.
- https://cdn.elifesciences.org/articles/82654/elife-82654-fig2-figsupp5-data1-v3.xlsx
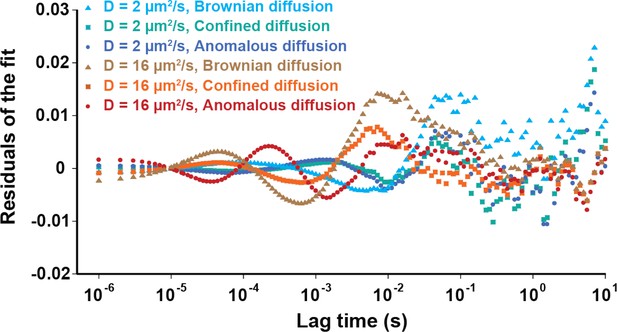
Residuals of fitting the simulated ACFs with different models.
Simulated fluorescence intensity ACF data were fitted by the models of anomalous and Brownian diffusion, as well as by the OU model of Brownian diffusion under confinement, as indicated.
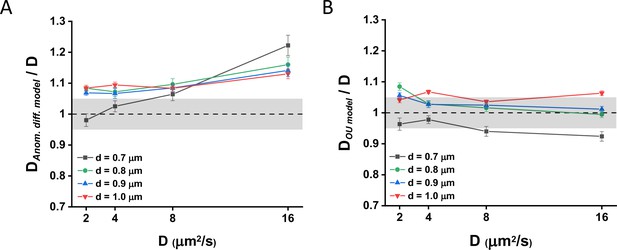
Diffusion coefficients fitted from simulation data.
Diffusion coefficients computed () from the diffusion times extracted from the fit of the Brownian simulation data with (A) the anomalous diffusion model or (B) the Ornstein-Uhlenbeck (OU) model of Brownian diffusion under confinement at various value of the ansatz D and of the cell diameter d, normalized by the ansatz D. The gray areas represent ±5% accuracy.
-
Figure 2—figure supplement 7—source data 1
Average and error from simulation in Figure 2—figure supplement 7A.
Average and error from simulation in Figure 2—figure supplement 7B.
- https://cdn.elifesciences.org/articles/82654/elife-82654-fig2-figsupp7-data1-v3.xlsx
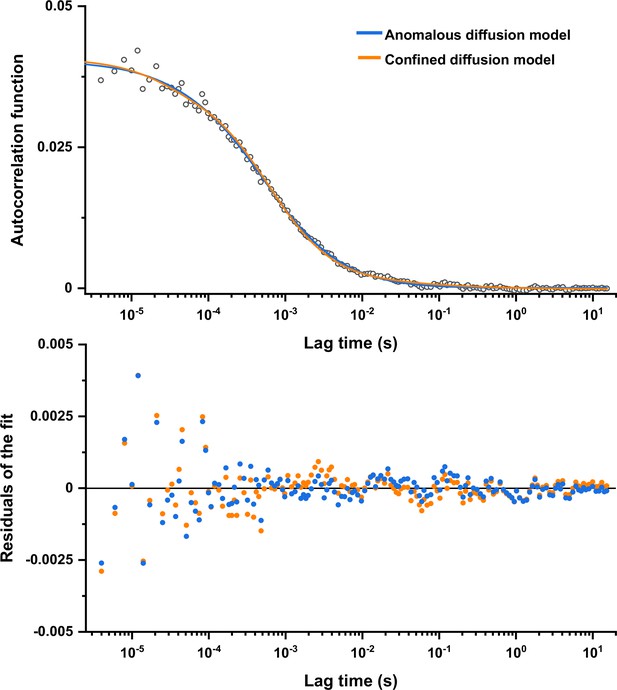
Comparison between fits of the experimental data with confined diffusion and anomalous diffusion models.
The experimental data (here the example for the R3 measurement from Figure 1—figure supplement 2) were fitted by the model of confined diffusion and by the anomalous diffusion model as indicated by different colors (upper panel), yielding comparable values of residuals (lower panel).
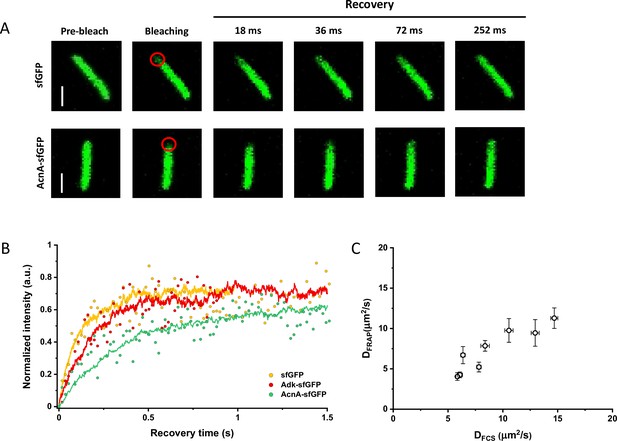
Comparison between protein diffusion coefficients measured by FCS and FRAP.
(A) Examples of FRAP measurements for two different constructs, sfGFP and AcnA-sfGFP. A 3×3 pixels area close to one cell pole (red circle) was photobleached with a high-intensity laser illumination for 48 ms and the recovery of fluorescence in the bleached area was monitored for 11 s with the time resolution of 18 ms. The scales bars are 2 μm. (B) Representative curves of fluorescence recovery in FRAP experiments and their fitting using simFRAP. The experimental data (colored dots) are used by the simFRAP algorithm to simulate the underlying diffusional process (colored lines). The simulation is then used to compute the diffusion coefficient. The simulation proceeds until the recovery curve reaches a plateau, therefore it is interrupted at a different time for each curve. (C) Correlation between the diffusion coefficients measured in FCS experiments (DFCS, fitting with the OU model; data from Figure 2E) and in FRAP experiment (DFRAP, fitting with simFRAP). The numbers of cells measured for each construct with each technique are shown in Appendix 6. Error bars represent the standard error of the mean. Error bars that are not visible are smaller than the symbol size. FCS, fluorescence correlation spectroscopy; FRAP, fluorescence recovery after photobleaching; OU, Ornstein-Uhlenbeck.
-
Figure 3—source data 1
Individual mean and standard error of the mean of diffusion coefficient values from Figure 3C.
- https://cdn.elifesciences.org/articles/82654/elife-82654-fig3-data1-v3.xlsx
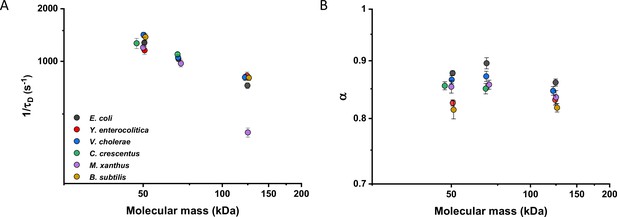
Mobility of homologous proteins from other bacterial species in Escherichia coli.
Mass dependence of protein mobility (1/τD; A) and anomaly of diffusion (α; B) of sfGFP fusions to homologues of Adk, Pgk, and AcnA from indicated bacterial species (E.c. = Escherichia coli; Y.e. = Yersinia enterocolitica; V.c. = Vibrio cholerae; C.c. = Caulobacter crescentus; M.x. = Myxococcus xanthus; B.s. = Bacillus subtilis) compared with that of their counterpart from E. coli. Each symbol represents the average value for all individual cells that have been measured for that construct and the error bars represent the standard error of the mean. Error bars that are not visible are smaller than the symbol size. The numbers of cells measured for each construct are shown in Appendix 6.
-
Figure 4—source data 1
Individual mean and standard error of the mean of 1/τD values from Figure 4A.
Individual mean and standard error of the mean of α values from Figure 4B.
- https://cdn.elifesciences.org/articles/82654/elife-82654-fig4-data1-v3.xlsx
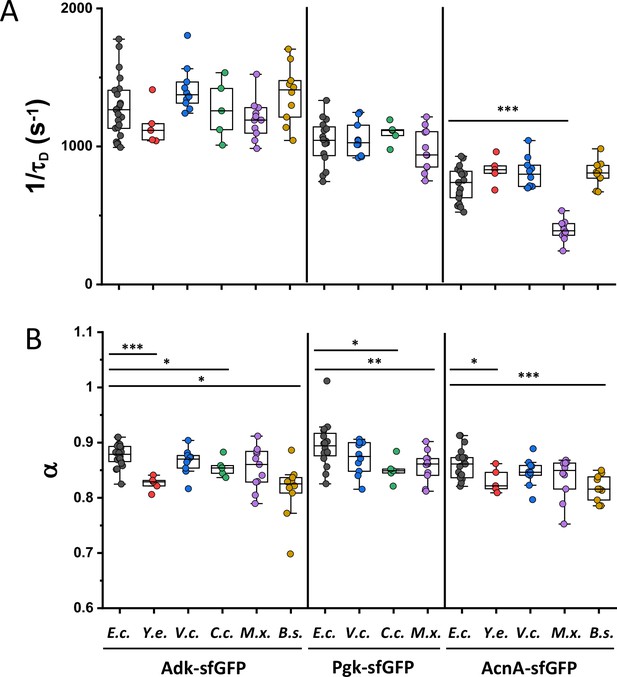
Mobility of homologous proteins from other bacterial species in Escherichia coli.
Each dot represents the protein mobility (1/τD; A) or the anomalous diffusion exponent (α; B) from one individual cell expressing the indicated sfGFP fusions to homologues of Adk, Pgk, and AcnA from indicated bacterial species (E.c. = Escherichia coli; Y.e. = Yersinia enterocolitica; V.c. = Vibrio cholerae; C.c. = Caulobacter crescentus; M.x. = Myxococcus xanthus; B.s. = Bacillus subtilis) compared with that of their counterpart from E. coli. The numbers of cells measured for each construct are shown in Appendix 6. ***p<0.0001; **p<0.001; *p<0.05 in a two-tailed heteroscedastistic t-test. When not indicated, no statistically significant difference is observed. Exact p-values can be found in Appendix 5.
-
Figure 4—figure supplement 1—source data 1
Individual values of 1/τD from Figure 4—figure supplement 1A.
Individual values of α from Figure 4—figure supplement 1B.
- https://cdn.elifesciences.org/articles/82654/elife-82654-fig4-figsupp1-data1-v3.xlsx
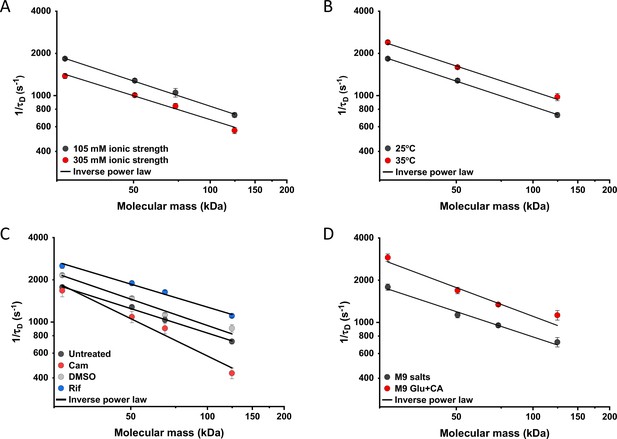
Effects of physicochemical perturbations and cell growth on mobility of differently sized proteins.
Each dot represents the average value of protein mobility (1/τD) of all the cells measured for the construct of the indicated molecular mass . The numbers of cells measured for each construct in each condition are shown in Appendix 6. Error bars represent the standard error. Error bars that are not visible are smaller than the symbol size. The solid black lines are the fit with an inverse power law to extract the size dependence of protein mobility (β) in that condition. (A) Protein mobility measured in cells that were resuspended in either tethering buffer (ionic strength of 105 mM; β=0.60±0.01) or in the same buffer but supplemented with additional 100 mM NaCl (total ionic strength of 305 mM; β=0.57±0.05). The measurements were performed in agarose pads prepared at the same ionic strength. (B) Protein mobility at different environmental temperatures. As for the other experiments, Escherichia coli cultures were grown at 37°C and bacterial cells during the measurements were incubated at 25°C (β=0.60±0.01) or at 35°C (β=0.60±0.05), as indicated. (C) Protein mobility in control cells (β=0.58±0.02) and after treatment with chloramphenicol (Cam; 200 µg/ml; β=0.88±0.11), rifampicin (Rif; 200 µg/ml, in 0.1% v/v DMSO; β=0.54±0.04), or DMSO control (0.1% v/v; β=0.62±0.07) as indicated. Antibiotics were added to growing E. coli culture 60 min prior to harvesting. (D) Protein mobility in non-growing cells incubated at 35°C on agarose pads containing only M9 salts (β=0.60±0.05) in comparison with growing cell incubated on pads with M9 salts supplemented with 20 mM glucose and 0.2% casamino acids (Glu+CA; β=0.68± 0.10).
-
Figure 5—source data 1
Individual mean and standard error of the mean of 1/τD values from Figure 5A.
- https://cdn.elifesciences.org/articles/82654/elife-82654-fig5-data1-v3.xlsx
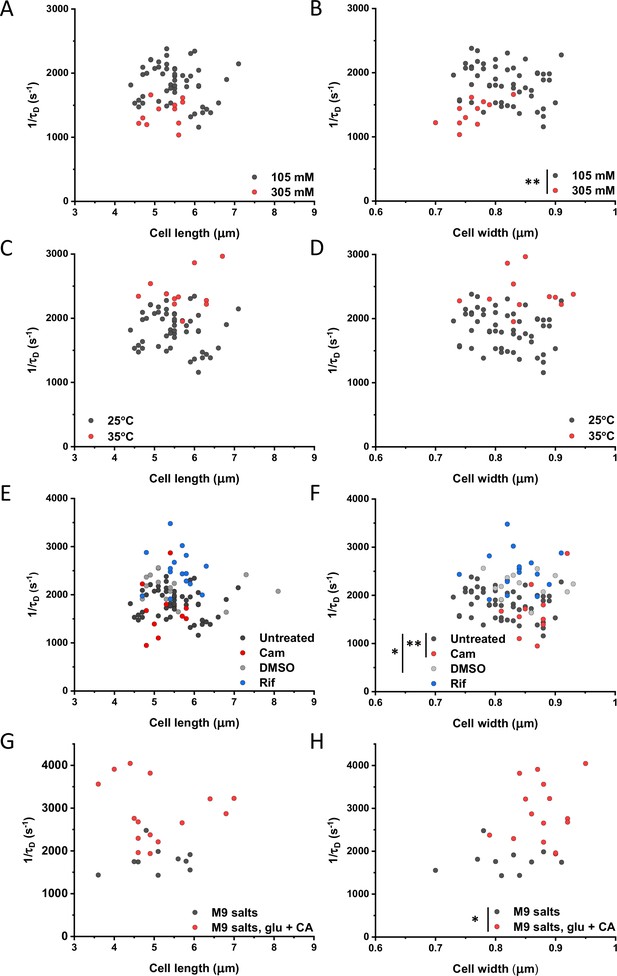
Mobility of sfGFP as a function of length and width of individual cells upon indicated perturbations.
The mobility (1/τD) of sfGFP in single cells plotted as a function of the respective cell length (A, C, E, G) or cell width (B, D, F, H) at different ionic strengths (A, B), environmental temperatures (C, D), after treatments with different antibiotics (E, F) and cell growth (G, H). The numbers of measured cells are shown in Appendix 6. Significance analysis was performed for the respective cell dimension. When not indicated, no significant difference is observed. ** p<0.001; * p<0.05 in a two-tailed heteroscedastistic t-test. Exact p-values can be found in Appendix 5.
-
Figure 5—figure supplement 1—source data 1
Individual measurements of cell length from Figure 5—figure supplement 1A.
Individual measurements of cell width from Figure 5—figure supplement 1B. Individual measurements of cell length from Figure 5—figure supplement 1C. Individual measurements of cell width from Figure 5—figure supplement 1D. Individual measurements of cell length from Figure 5—figure supplement 1E. Individual measurements of cell width from Figure 5—figure supplement 1F. Individual measurements of cell length from Figure 5—figure supplement 1G. Individual measurements of cell width from Figure 5—figure supplement 1H.
- https://cdn.elifesciences.org/articles/82654/elife-82654-fig5-figsupp1-data1-v3.xlsx
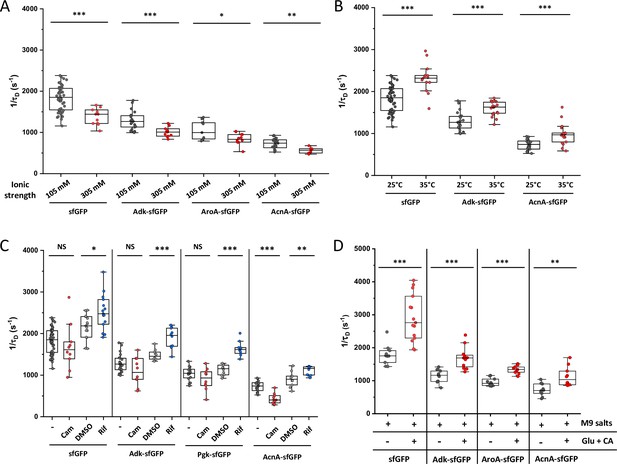
Effect of different perturbations on protein mobility (1/τD) in individual cells.
Experiments for ionic strength (A), environmental temperature (B), antibiotics treatment (C), and growth rate (D) are shown for indicated proteins. Individual measurements and significance analysis of data from Figure 5. Each dot in the box plot represents the value of 1/τD for one individual cell measured in the indicated condition. The numbers of cells measured for each construct in each condition are shown in Appendix 6. *** p<0.0001; ** p<0.001; * p<0.05; NS: no statistically significant difference in a two-tailed heteroscedastistic t-test. Exact p-values can be found in Appendix 5.
-
Figure 5—figure supplement 2—source data 1
Individual values of 1/τD from Figure 5—figure supplement 2A.
Individual values of 1/τD from Figure 5—figure supplement 2B. Individual values of 1/τD from Figure 5—figure supplement 2C. Individual values of 1/τD from Figure 5—figure supplement 2D.
- https://cdn.elifesciences.org/articles/82654/elife-82654-fig5-figsupp2-data1-v3.xlsx
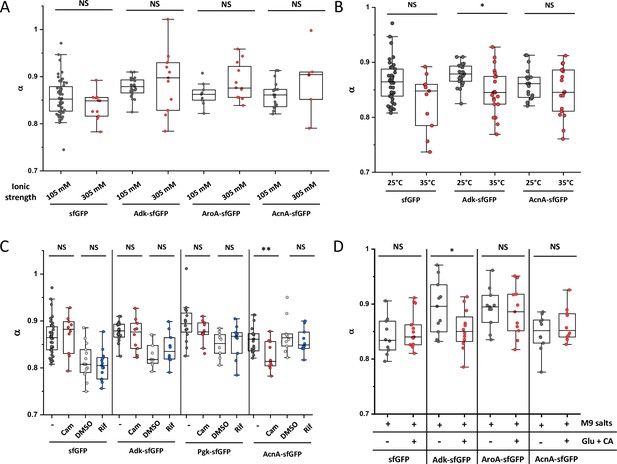
Effect of different perturbations on the anomaly of protein diffusion (α) in individual cells.
Experiments for ionic strength (A), environmental temperature (B), antibiotics treatment (C), and growth rate (D) are shown for indicated proteins. Each dot in the box plot represents the value of α for one individual cell measured in the indicated condition. The numbers of cells measured for each construct in each condition are shown in Appendix 6. ** p<0.001; * p<0.05; NS: no statistically significant difference in a two-tailed heteroscedastistic t-test. Exact p-values can be found in Appendix 5.
-
Figure 5—figure supplement 3—source data 1
Individual values of α from Figure 5—figure supplement 3A.
Individual values of α from Figure 5—figure supplement 3B. Individual values of α from Figure 5—figure supplement 3C. Individual values of α from Figure 5—figure supplement 3D.
- https://cdn.elifesciences.org/articles/82654/elife-82654-fig5-figsupp3-data1-v3.xlsx
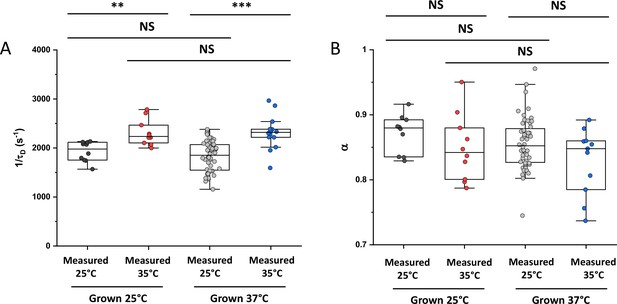
Effect of growth and measurement temperature on protein diffusion.
Mobility (1/τD; A) and anomaly of diffusion (α; B) for sfGFP in cells grown either at 25 °C or at 37 °C and measured either at 25 °C or at 35 °C, as in Figure 4A. The numbers of cells measured for each construct in each condition are shown in Appendix 6. *** p<0.0001; ** p<0.001; NS: no statistically significant difference in a two-tailed heteroscedastistic t-test. Exact p-values can be found in Appendix 5.
-
Figure 5—figure supplement 4—source data 1
Individual values of 1/τD from Figure 5—figure supplement 4A.
Individual values of α from Figure 5—figure supplement 4B.
- https://cdn.elifesciences.org/articles/82654/elife-82654-fig5-figsupp4-data1-v3.xlsx
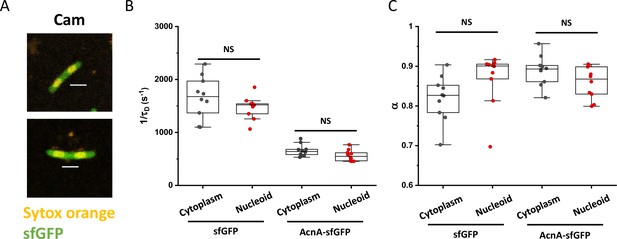
Influence of nucleoid on protein mobility.
(A) Bacterial cells were treated with chloramphenicol to achieve nucleoid compaction and stained with the DNA-binding dye SYTOX Orange. Scale bars are 2 μm. The mobility (1/τD; B) and anomaly of diffusion (α; C) of sfGFP and of one larger construct (AcnA-sfGFP) was measured in both the cytoplasm and in the nucleoid of chloramphenicol treated cells. The numbers of cells measured for each construct in each condition are shown in Appendix 6. NS: no statistically significant difference in a two-tailed heteroscedastistic t-test. Exact p-values can be found in Appendix 5.
-
Figure 5—figure supplement 5—source data 1
Individual values of 1/τD from Figure 5—figure supplement 5B.
Individual values of α from Figure 5—figure supplement 5C.
- https://cdn.elifesciences.org/articles/82654/elife-82654-fig5-figsupp5-data1-v3.xlsx
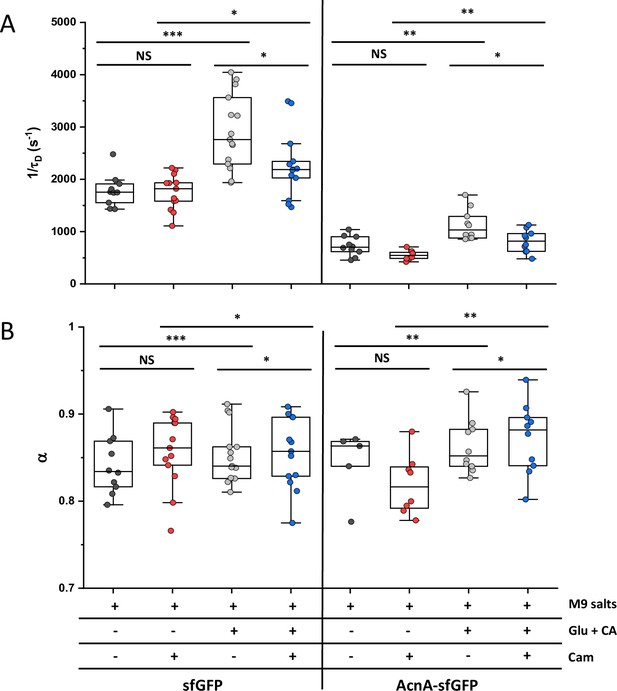
Effect of nutrient availability and growth on protein mobility.
The mobility (1/τD; A) and anomaly of diffusion (α; B) of indicated protein constructs was measured in cells incubated at 35 °C on agarose pads containing either only M9 salts or M9 salts together with 20 mM glucose and 0.2% casamino acids (Glu +CA). Where indicated, chloramphenicol (Cam) was also added to the agarose pads. The numbers of cells measured for each construct in each condition are shown in Appendix 6. *** p<0.0001; ** p<0.001; * p<0.05; NS: no statistically significant difference in a two-tailed heteroscedastistic t-test. Exact p-values can be found in Appendix 5.
-
Figure 5—figure supplement 6—source data 1
Individual values of 1/τD from Figure 5—figure supplement 6A.
Individual values of α from Figure 5—figure supplement 6B.
- https://cdn.elifesciences.org/articles/82654/elife-82654-fig5-figsupp6-data1-v3.xlsx
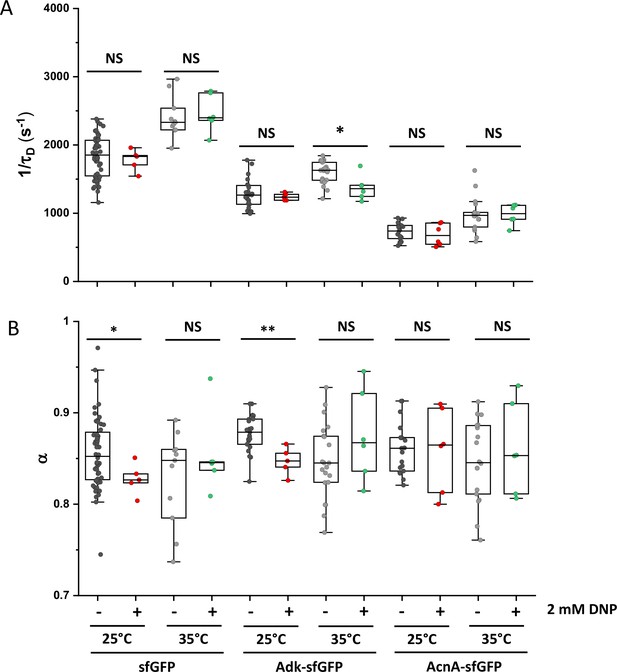
Effect of DNP on protein mobility.
The mobility (1/τD; A) and the anomaly of diffusion (α; B) of sfGFP and of two constructs with higher molecular mass was measured in bacterial cells treated in batch for 60 minutes with 2 mM DNP and compared with the respective untreated control. The numbers of cells measured for each construct in each condition are shown in Appendix 6. Measurements were performed on 1% agarose pads prepared in tethering buffer and supplemented with 2 mM DNP at the indicated incubation temperature. ** p<0.001; * p<0.05; NS: no statistically significant difference in a two-tailed heteroscedastistic t-test. Exact p-values can be found in Appendix 5.
-
Figure 5—figure supplement 7—source data 1
Individual values of 1/τD from Figure 5—figure supplement 7A.
Individual values of α from Figure 5—figure supplement 7B.
- https://cdn.elifesciences.org/articles/82654/elife-82654-fig5-figsupp7-data1-v3.xlsx
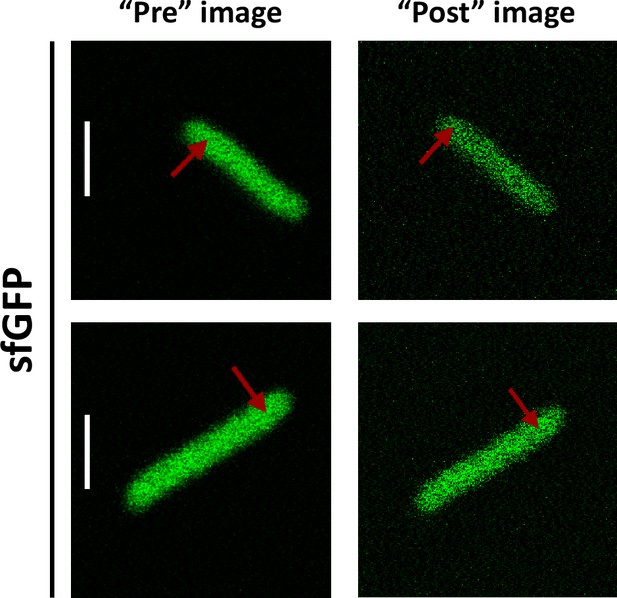
Typical examples of presence or absence of lateral focal drift during FCS measurements.
Substantial lateral drift could be observed for <10% of experiments (upper images), whereas most measurement showed no perceptible lateral drift (lower images). FCS, fluorescence correlation spectroscopy. Scale bars are 2 μm.
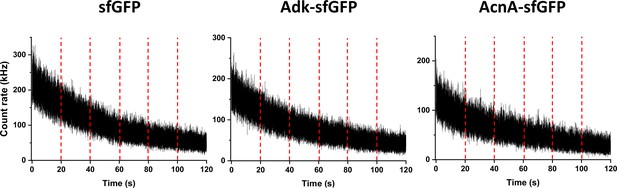
Typical traces of fluorescence intensity during FCS measurements.
Examples of fluorescence intensity traces for indicated protein fusions. The vertical red dashed lines separate sequential fluorescence intensity acquisitions on the same cell. FCS, fluorescence correlation spectroscopy.
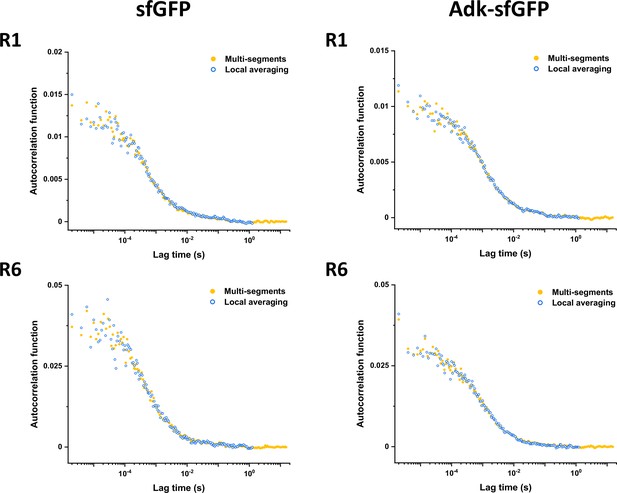
Results of detrending with multi-segments and local averaging approaches.
Comparison of experimental ACFs corrected using either multi-segments or local averaging approaches (as indicated) for sfGFP and Adk-sfGFP and different data acquisition segments (R1 vs. R6). ACF, autocorrelation function.
Values of τD or α for the six sequential ACFs.
Values were determined by fitting the anomalous diffusion model to experimental ACFs for the six sequential time segments per individual cell expressing sfGFP (A) or MetH-sfGFP (B). ACF, autocorrelation function.
Tables
Molecular mass, biological function, and measured parameters for all studied sfGFP fusion constructs.
The concentration of expression inducer and the number of cells measured with each technique is also indicated.
| Protein name | Molecular mass of sfGFP fusion construct | Biological function in E. coli | IPTG concentration used for FCS (FRAP) | Number of cells analyzed by FCS | τD (µs; mean ± SEM) | α (mean ± SEM) | Diffusion coefficient, FCS (μm2/s, mean ± SEM) | Number of cells analyzed by FRAP | Diffusion coefficient, FRAP (μm2/s, mean ± SEM) |
|---|---|---|---|---|---|---|---|---|---|
| sfGFP | 26.9 | – | 5 µM (15 µM) | 52 | 561±14 | 0.86±0.01 | 14.7±0.3 | 11 | 11.3±1.3 |
| YggX | 39.2 | Probable Fe (2+)-trafficking protein | 5 µM (5 µM) | 8 | 611±19 | 0.85±0.01 | 12.9±0.4 | 10 | 9.4±1.6 |
| ClpS | 39.2 | ATP-dependent Clp protease adapter protein | 0 µM | 11 | 1054±33 | 0.75±0.01 | |||
| FolK | 45.1 | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase | 0 µM | 8 | 734±24 | 0.87±0.01 | 11.6±0.4 | ||
| Crr | 45.2 | Component of glucose-specific phosphotransferase enzyme IIA | 0 µM | 14 | 1065±36 | 0.87±0.01 | |||
| UbiC | 45.7 | Chorismate pyruvate-lyase | 15 µM | 14 | 1140±58 | 0.87±0.01 | |||
| ThpR | 46.9 | RNA 2′,3′-cyclic phosphodiesterase | Discarded due to instability of sfGFP fusion construct | ||||||
| CoaE | 49.6 | Dephospho-CoA kinase | 0 µM | 11 | 854±47 | 0.87±0.01 | 9.8±0.6 | ||
| Adk | 50.6 | Adenylate kinase | 5 µM (15 µM) | 23 | 802±26 | 0.88±0.00 | 10.6±0.4 | 16 | 9.8±1.5 |
| Cmk | 51.7 | Cytidylate kinase | 5 µM | 16 | 1163±58 | 0.87±0.01 | |||
| NagD | 54.1 | Ribonucleotide monophosphatase | Discarded due to non-uniform protein localization | ||||||
| KdsB | 54.6 | 3-deoxy-manno-octulosonate cytidylyltransferase | 0 µM | 11 | 1659±70 | 0.84±0.01 | |||
| Map | 56.3 | Methionine aminopeptidase | 0 µM | 20 | 1830±78 | 0.81±0.01 | |||
| MmuM | 60.4 | Homocysteine S-methyltransferase | 5 µM | 14 | 2241±138 | 0.73±0.01 | |||
| RihA | 60.8 | Pyrimidine-specific ribonucleoside hydrolase | Discarded due to non-uniform protein localization | ||||||
| PanE | 60.8 | 2-dehydropantoate 2-reductase | 0 µM (5 µM) | 18 | 1059±26 | 0.85±0.01 | 7.8±0.2 | 11 | 5.2±0.6 |
| SolA | 67.9 | N-methyl-L-tryptophan oxidase | 0 µM | 7 | 795±31 | 0.82±0.01 | 9.9±0.5 | ||
| Pgk | 68.1 | Phosphoglycerate kinase | 0 µM | 16 | 991±41 | 0.90±0.01 | 8.6±.0.3 | ||
| EntC | 69.9 | Isochorismate synthase | 15 µM | 15 | 1777±119 | 0.82±0.01 | |||
| AroA | 73.1 | 3-phosphoshikimate 1-carboxyvinyltransferase | 5 µM | 9 | 995±69 | 0.86±0.01 | 8.7±0.7 | ||
| ThrC | 74.1 | Threonine synthase | 0 µM | 14 | 908±28 | 0.87±0.01 | 9.1±0.3 | ||
| MurF | 74.4 | UDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-alanine ligase | 0 µM | 7 | 1008±76 | 0.85±0.02 | 8.3±0.7 | ||
| DsdA | 74.9 | D-serine dehydratase | 0 µM | 14 | 1017±53 | 0.89±0.01 | 8.4±0.4 | 10 | 7.8±0.7 |
| HemN | 79.7 | Oxygen-independent coproporphyrinogen III oxidase | 0 µM | 13 | 1262±54 | 0.86±0.01 | 6.7±0.4 | ||
| PrpD | 80.9 | 2-methylcitrate dehydratase | 0 µM | 12 | 1866±140 | 0.84±0.01 | |||
| DnaK | 96.0 | Molecular chaperone | 5 µM | 10 | 2296±78 | 0.76±0.01 | |||
| MalZ | 96.0 | Maltodextrin glucosidase | 0 µM | 9 | 3725±229 | 0.77±0.01 | |||
| GlcB | 107.5 | Malate synthase G | 5 µM (15 µM) | 16 | 1315±45 | 0.86±0.01 | 6.4±0.2 | 10 | 6.7±1.1 |
| MetE | 111.7 | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase | 5 µM | 8 | 1137±53 | 0.87±0.01 | 7.4±0.3 | ||
| LeuS | 124.2 | Leucine--tRNA ligase | 0 µM | 14 | 1637±75 | 0.86±0.01 | 5.1±0.2 | ||
| AcnA | 124.7 | Aconitate hydratase A | 5 µM (15 µM) | 19 | 1415±56 | 0.86±0.01 | 6.1±0.2 | 10 | 4.3±0.4 |
| MetH | 163.0 | Methionine synthase | 0 µM (5 µM) | 9 | 1402±45 | 0.81±0.01 | 5.8±0.1 | 15 | 4.0±0.5 |
List of primers used in this study.
| Primer name | Sense | Nucleotide sequence | Description |
|---|---|---|---|
| NBp1 | RW | ACCCATGGCACACTCCTTCACTAG | Amplify pTrc99A |
| NBp2 | RW | CTTGGACATGCTACCTCCGCCCCCTTAGTACAACGGTGACGCCGG | Amplify ubiC gene of E. coli MG1655 and fuse it to linker-sfgfp |
| NBp3 | FW | GGGGGCGGAGGTAGCATGTCCAAGGGTGAAGAGCTATTTAC | Amplify pTrc99A |
| NBp4 | FW | GTACTAGTGAAGGAGTGTGCCATGGGTATGTCACACCCCGCGTTAAC | Amplify ubiC gene of E. coli MG1655 and fuse it to trc promoter |
| NBp5 | FW | TTGACAATTAATCATCCGGCTCG | Sequence pTrc99A |
| NBp7 | RW | CTTGGACATGCTACCTCCGCCCCCGTACAACGGTGACGCCGG | Amplify ubiC gene from K12 and fuse it to linker-sfGFP |
| NBp8 | FW | GTACTAGTGAAGGAGTGTGCCATGGGTATGCGTATCATTCTGCTTGGCG | Amplify adk gene of E. coli MG1655 and fuse it to trc promoter |
| NBp9 | RW | CTTGGACATGCTACCTCCGCCCCCGCCGAGGATTTTTTCCAGATCAG | Amplify adk gene of E. coli MG1655 and fuse it to linker-sfGFP |
| NBp10 | FW | GTACTAGTGAAGGAGTGTGCCATGGGTATGTCGCAGAATAATCCGTT | Amplify mmuM gene of E. coli MG1655 and fuse it to trc promoter |
| NBp11 | RW | CTTGGACATGCTACCTCCGCCCCCGCTTCGCGCTTTTAACG | Amplify mmuM gene of E. coli MG1655 and fuse it to linker-sfGFP |
| NBp12 | FW | GTACTAGTGAAGGAGTGTGCCATGGGTATGGAAAACGCTAAAATGAACTCG | Amplify dsdA gene of E. coli MG1655 and fuse it to trc promoter |
| NBp13 | RW | CTTGGACATGCTACCTCCGCCCCCACGGCCTTTTGCCAGATATTG | Amplify dsdA gene of E. coli MG1655 and fuse it to linker-sfGFP |
| NBp14 | FW | GTACTAGTGAAGGAGTGTGCCATGGGTATGTCTGTACAGCAAATCGACTGGG | Amplify hemN gene from K12 genome and fuse it to trc promoter |
| NBp15 | RW | CTTGGACATGCTACCTCCGCCCCCAATCACCCGAGAGAACTGCTGC | Amplify hemN gene of E. coli MG1655 and fuse it to linker-sfGFP |
| NBp16 | FW | GTACTAGTGAAGGAGTGTGCCATGGGTATGAGTCAAACCATAACCCAGAG | Amplify glcB gene of E. coli MG1655 and fuse it to trc promoter |
| NBp17 | RW | CTTGGACATGCTACCTCCGCCCCCATGACTTTCTTTTTCGCGTAAAC | Amplify glcB gene of E. coli MG1655 and fuse it to linker-sfGFP |
| NBp18 | RW | GATTTAATCTGTATCAGG | Sequence pTrc99A |
| NBp19 | FW | GTACTAGTGAAGGAGTGTGCCATGGGTATGACAGTGGCGTATATTGC | Amplify folK gene of E. coli MG1655 and fuse it to trc promoter |
| NBp20 | RW | CTTGGACATGCTACCTCCGCCCCCCCATTTGTTTAATTTGTCAA | Amplify folK gene of E. coli MG1655 and fuse it to linker-sfGFP |
| NBp21 | FW | GTACTAGTGAAGGAGTGTGCCATGGGTATGGCTATCTCAATCAAGACCCC | Amplify map gene of E. coli MG1655 and fuse it to trc promoter |
| NBp22 | RW | CTTGGACATGCTACCTCCGCCCCCTTCGTCGTGCGAGATTATCG | Amplify map gene of E. coli MG1655 and fuse it to linker-sfGFP |
| NBp23 | FW | GTACTAGTGAAGGAGTGTGCCATGGGTATGAAACTCTACAATCTGAAAG | Amplify thrC gene of E. coli MG1655 and fuse it to trc promoter |
| NBp24 | RW | CTTGGACATGCTACCTCCGCCCCCCTGATGATTCATCATCAATTTAC | Amplify thrC gene of E. coli MG1655 and fuse it to linker-sfGFP |
| NBp25 | FW | GTACTAGTGAAGGAGTGTGCCATGGGTATGTCAGCTCAAATCAACAACATCCG | Amplify prpD gene of E. coli MG1655 and fuse it to trc promoter |
| NBp26 | RW | CTTGGACATGCTACCTCCGCCCCCAATGACGTACAGGTCGAGATACTC | Amplify prpD gene of E. coli MG1655 and fuse it to linker-sfGFP |
| NBp27 | FW | GTACTAGTGAAGGAGTGTGCCATGGGTATGTTAAATGCATGGCACCTGC | Amplify malZ gene of E. coli MG1655 and fuse it to trc promoter |
| NBp28 | RW | CTTGGACATGCTACCTCCGCCCCCGTTCATCCATACCGTAGCCGAAATG | Amplify malZ gene of E. coli MG1655 and fuse it to linker-sfGFP |
| NBp29 | FW | GTACTAGTGAAGGAGTGTGCCATGGGTATGTCTGAACCGCAACGTCTG | Amplify thrP gene of E. coli MG1655 and fuse it to trc promoter |
| NBp30 | RW | CTTGGACATGCTACCTCCGCCCCCTTGCGTTAGCGCCCAGC | Amplify thrP gene of E. coli MG1655 and fuse it to linker-sfGFP |
| NBp31 | FW | GTACTAGTGAAGGAGTGTGCCATGGGTATGTCTGTAATTAAGATGACCGATC | Amplify pgk gene of E. coli MG1655 and fuse it to trc promoter |
| NBp32 | RW | CTTGGACATGCTACCTCCGCCCCCCTTCTTAGCGCGCTCTTCG | Amplify pgk gene of E. coli MG1655 and fuse it to linker-sfGFP |
| NBp33 | FW | GTACTAGTGAAGGAGTGTGCCATGGGTATGAGGTATATAGTTGCCTTAACGG | Amplify coaE gene of E. coli MG1655 and fuse it to trc promoter |
| NBp35 | FW | GTACTAGTGAAGGAGTGTGCCATGGGTATGACGGCAATTGCCCC | Amplify cmk gene of E. coli MG1655 and fuse it to trc promoter |
| NBp37 | FW | GTACTAGTGAAGGAGTGTGCCATGGGTATGGATACGTCACTGGCTGAG | Amplify entC gene of E. coli MG1655 and fuse it to trc promoter |
| NBp39 | FW | GTACTAGTGAAGGAGTGTGCCATGGGTATGATTAGCGTAACCCTTAGCC | Amplify murF gene of E. coli MG1655 and fuse it to trc promoter |
| NBp41 | FW | GTACTAGTGAAGGAGTGTGCCATGGGTATGAAAATTACCGTATTGGGATGCG | Amplify panE gene of E. coli MG1655 and fuse it to trc promoter |
| NBp53 | FW | TCCAAGGGTGAAGAGCTATTTACTGGG | Deletion of ATG from sfgfp in dsdA-sfgfp, ubiC-sfgfp, thrC-sfgfp, malZ-sfgfp * |
| NBp54 | RW | GCTACCTCCGCCCCCACG | Deletion of ATG from sfgfp in dsdA-sfgfp * |
| NBp55 | FW | TCCAAGGGTGAAGAGCTATTTACTGGGGTTG | Deletion of ATG from sfgfp in adk-sfgfp * |
| NBp56 | RW | GCTACCTCCGCCCCCGCC | Deletion of ATG from sfgfp in adk-sfgfp * |
| NBp57 | FW | TCCAAGGGTGAAGAGCTATTTACTGGGG | Deletion of ATG from sfgfp in mmuM-sfgfp and folK-sfgfp * |
| NBp58 | RW | GCTACCTCCGCCCCCGCT | Deletion of ATG from sfgfp in mmuM-sfgfp * |
| NBp59 | RW | GCTACCTCCGCCCCCGTA | Deletion of ATG from sfgfp in ubiC-sfgfp * |
| NBp60 | FW | TCCAAGGGTGAAGAGCTATTTACTGG | Deletion of ATG from sfgfp in glcB-sfgfp * |
| NBp61 | RW | GCTACCTCCGCCCCCATG | Deletion of ATG from sfgfp in glcB-sfgfp * |
| NBp62 | FW | TCCAAGGGTGAAGAGCTATTTACTG | Deletion of ATG from sfgfp in hemN-sfgfp, map-sfgfp, prpD-sfgfp * |
| NBp63 | RW | GCTACCTCCGCCCCCAAT | Deletion of ATG from sfgfp in hemN-sfgfp and prpD-sfgfp * |
| NBp64 | RW | GCTACCTCCGCCCCCTTC | Deletion of ATG from sfgfp in map-sfgfp * |
| NBp65 | RW | GCTACCTCCGCCCCCCTG | Deletion of ATG from sfgfp in thrC-sfgfp * |
| NBp66 | RW | GCTACCTCCGCCCCCCCA | Deletion of ATG from sfgfp in folK –sfgfp * |
| NBp67 | RW | GCTACCTCCGCCCCCGTT | Deletion of ATG from sfgfp in malZ-sfgfp * |
| NBp68 | FW | GGGGGCGGAGGTAGCTCCAAGGGTGAAGAGCTATTTACTG | Amplification of backbone flexible linker-sfgfp without ATG |
| NBp81 | RW | GCTCTTCACCCTTGGAGCTACCTCCGCCCCCTGCGAGAGCCAATTTCTGG | Amplify cmk gene of E. coli MG1655 and fuse it to linker-sfgfp |
| NBp82 | RW | GCTCTTCACCCTTGGAGCTACCTCCGCCCCCCGGTTTTTCCTGTGAGACAAAC | Amplify coaE gene of E. coli MG1655 and fuse it to linker-sfgfp |
| NBp83 | RW | GCTCTTCACCCTTGGAGCTACCTCCGCCCCCATGCAATCCAAAAACGTTCAACAT | Amplify entC gene of E. coli MG1655 and fuse it to linker -sfgfp deleted STOP |
| NBp84 | RW | GCTCTTCACCCTTGGAGCTACCTCCGCCCCCACATGTCCCATTCTCCTGTAAAG | Amplify murF gene of E. coli MG1655 and fuse it to linker-sfgfp |
| NBp85 | RW | GCTCTTCACCCTTGGAGCTACCTCCGCCCCCTTGCGTTAGCGCCCAGC | Amplify thrP gene of E. coli MG1655 and fuse it to linker-sfgfp |
| NBp86 | RW | GCTCTTCACCCTTGGAGCTACCTCCGCCCCCCCAGGGGCGAGGCAAAC | Amplify panE gene of E. coli MG1655 and fuse it to linker-sfgfp |
| NBp87 | RW | GCTCTTCACCCTTGGAGCTACCTCCGCCCCCCTTCTTAGCGCGCTCTTCG | Amplify pgk gene gene of E. coli MG1655 and fuse it to linker-sfgfp |
| NBp88 | FW | GTACTAGTGAAGGAGTGTGCCATGGGTATGGGTTTGTTCGATAAACTG | Amplify crr gene of E. coli MG1655 and fuse it to trc promoter |
| NBp89 | RW | TCACCCTTGGAGCTACCTCCGCCCCCCTTCTTGATGCGGATAACC | Amplify crr gene of E. coli MG1655 and fuse it to linker-sfgfp |
| NBp90 | FW | GTACTAGTGAAGGAGTGTGCCATGGGTATGCAAGAGCAATACCGCC | Amplify leuS gene of E. coli MG1655 and fuse it to trc promoter |
| NBp91 | RW | TTCACCCTTGGAGCTACCTCCGCCCCCGCCAACGACCAGATTGAGG | Amplify leuS gene of E. coli MG1655 and fuse it to linker-sfgfp |
| NBp92 | FW | GTACTAGTGAAGGAGTGTGCCATGGGTATGGCACTGCCAATTCTGTTAG | Amplify rihA gene of E. coli MG1655 and fuse it to trc promoter |
| NBp93 | RW | TTCACCCTTGGAGCTACCTCCGCCCCCAGCGTAAAATTTCAGACGATCAG | Amplify rihA gene of E. coli MG1655 and fuse it to linker-sfgfp |
| NBp94 | FW | GTACTAGTGAAGGAGTGTGCCATGGGTATGACCATTAAAAATGTAATTTGCGATATCG | Amplify nagA gene of E. coli MG1655 and fuse it to trc promoter |
| NBp95 | RW | TTCACCCTTGGAGCTACCTCCGCCCCCGATAACGTCGATTTCAGCGACTG | Amplify nagA gene of E. coli MG1655 and fuse it to linker-sfgfp |
| NBp96 | FW | GTACTAGTGAAGGAGTGTGCCATGGGTATGGGTAAAACGAACGACTG | Amplify clpS gene of E. coli MG1655 and fuse it to trc promoter |
| NBp97 | RW | TTCACCCTTGGAGCTACCTCCGCCCCCGGCTTTTTCTAGCGTACACAG | Amplify clpS gene of E. coli MG1655 and fuse it to linker-sfgfp |
| NBp98 | FW | GTACTAGTGAAGGAGTGTGCCATGGGTATGGAATCCCTGACGTTACAACC | Amplify aroA gene of E. coli MG1655 and fuse it to trc promoter |
| NBp99 | RW | TTCACCCTTGGAGCTACCTCCGCCCCCGGCTGCCTGGCTAATCCG | Amplify aroA gene of E. coli MG1655 and fuse it to linker-sfgfp |
| NBp100 | FW | GTACTAGTGAAGGAGTGTGCCATGGGTATGAGTTTTGTGGTCATTATTCCCG | Amplify kdsB gene of E. coli MG1655 and fuse it to trc promoter |
| NBp101 | RW | TTCACCCTTGGAGCTACCTCCGCCCCCGCGCATTTCAGCGCGAAC | Amplify kdsB gene of E. coli MG1655 and fuse it to linker-sfgfp |
| NBp102 | FW | GTACTAGTGAAGGAGTGTGCCATGGGTATGAAATACGATCTCATCATTATTGGCAG | Amplify solA gene of E. coli MG1655 and fuse it to trc promoter |
| NBp103 | RW | TTCACCCTTGGAGCTACCTCCGCCCCCTTGGAAGCGGGAAAGCCTG | Amplify solA gene of E. coli MG1655 and fuse it to linker-sfgfp |
| NBp107 | RW | CACCCTTGGAGCTACCTCCGCCCCCCCATTTGTTTAATTTGTCAAATGCTC | Amplify folK gene of E. coli MG1655 and fuse it to linker-sfgfp |
| NBp122 | FW | ACTAGTGAAGGAGTGTGCCATGGGTGTGAGCAGCAAAGTGGAACAAC | Amplify metH gene of E. coli MG1655 and insert it into pTrc99A fused to sfgfp |
| NBp123 | RW | CACCCTTGGAGCTACCTCCGCCCCCGTCCGCGTCATACCCCAGATTC | |
| NBp124 | FW | ACTAGTGAAGGAGTGTGCCATGGGTATGTCGTCAACCCTACGAG | Amplify acnA gene of E. coli MG1655 and insert into pTrc99A fused to sfgfp |
| NBp125 | RW | CACCCTTGGAGCTACCTCCGCCCCCCTTCAACATATTACGAATGACATAATGC | |
| NBp126 | FW | ACTAGTGAAGGAGTGTGCCATGGGTATGACAATATTGAATCACACCCTC | Amplify metE gene of E. coli MG1655 and insert into pTrc99A fused to sfgfp |
| NBp127 | RW | CACCCTTGGAGCTACCTCCGCCCCCCCCCCGACGCAAGTTC | |
| NBp177 | FW | ACTAGTGAAGGAGTGTGCCATGGGTATGAGCAGAACGATTTTTTGTAC | Amplify yggX of E. coli MG1655 and insert into pTrc99A fused to sfgfp |
| NBp178 | RW | CACCCTTGGAGCTACCTCCGCCCCCTTTTTTATCTTCCGGCGTATAG | |
| NBp179 | FW | ACTAGTGAAGGAGTGTGCCATGGGTATGAATCTGATCCTGTTCGG | Amplify adk gene from Caulobacter crescentus and insert into pTrc99A fused to sfgfp |
| NBp180 | RW | CACCCTTGGAGCTACCTCCGCCCCCTCCTGCAGCGACG | |
| NBp181 | FW | CAGACCATGTACTAGTGAAGGAGTGTGCCATGGGTATGACCTTCCGCACCCTC | Amplify pgk gene from Caulobacter crescentus and insert into pTrc99A fused to sfgfp |
| NBp182 | RW | CACCCTTGGAGCTACCTCCGCCCCCGGATTCGAGCGCCGC | |
| NBp183 | FW | ACTAGTGAAGGAGTGTGCCATGGGTATGGCGTCTGTGGACAGC | Amplify acnA gene from Caulobacter crescentus and insert into pTrc99A fused to sfgfp |
| NBp184 | RW | CACCCTTGGAGCTACCTCCGCCCCCGTCGGCCTTGGCCAGG | |
| NBp185 | FW | ACTAGTGAAGGAGTGTGCCATGGGTATGAACTTAGTCTTAATGGGG | Amplify adk from Bacillus subtilis and insert into pTrc99A fused to sfgfp |
| NBp186 | RW | CACCCTTGGAGCTACCTCCGCCCCCTTTTTTTAATCCTCCAAGAAGATCC | |
| NBp187 | FW | ACTAGTGAAGGAGTGTGCCATGGGTATGAATAAAAAAACTCTCAAAGACATCG | Amplify pgk from Bacillus subtilis and insert into pTrc99A fused to sfgfp |
| NBp188 | RW | CACCCTTGGAGCTACCTCCGCCCCCTTTATCGTTCAGTGCAGCTAC | |
| NBp189 | FW | ACTAGTGAAGGAGTGTGCCATGGGTATGGCAAACGAGCAAAAAAC | Amplify acnA from Bacillus subtilis and insert into pTrc99A fused to sfgfp |
| NBp190 | RW | CACCCTTGGAGCTACCTCCGCCCCCGGACTGCTTCATTTTTTCACG | |
| NBp191 | FW | ACTAGTGAAGGAGTGTGCCATGGGTATGAACCTGATCCTGTTGGGG | Amplify adk from Myxococcus xanthus and insert into pTrc99A fused to sfgfp |
| NBp192 | RW | CACCCTTGGAGCTACCTCCGCCCCCGGCCTTGCCCGCAG | |
| NBp193 | FW | ACTAGTGAAGGAGTGTGCCATGGGTATGATCCGTTACATCGATGATCTGC | Amplify pgk from Myxococcus xanthus and insert into pTrc99A fused to sfgfp |
| NBp194 | RW | CACCCTTGGAGCTACCTCCGCCCCCCCGCGTCTCCAGCG | |
| NBp195 | FW | ACTAGTGAAGGAGTGTGCCATGGGTATGACCGACAGTTTCGGC | Amplify acnA from Myxococcus xanthus and insert into pTrc99A fused to sfgfp |
| NBp196 | RW | CACCCTTGGAGCTACCTCCGCCCCCGCCCTTGGCCAGTTG | |
| NBp197 | FW | ACTAGTGAAGGAGTGTGCCATGGGTATGCGCATCATTCTTCTCGG | Amplify adk from Vibrio cholerae and insert into pTrc99A fused to sfgfp |
| NBp198 | RW | CACCCTTGGAGCTACCTCCGCCCCCAGCCAACGCTTTAGCAATGTC | |
| NBp199 | FW | ACTAGTGAAGGAGTGTGCCATGGGTATGTCTGTAATCAAGATGATTGACCTGG | Amplify pgk from Vibrio cholerae and insert into pTrc99A fused to sfgfp |
| NBp200 | RW | CACCCTTGGAGCTACCTCCGCCCCCCGCTTTAGCGCGTGCTTC | |
| NBp201 | FW | ACTAGTGAAGGAGTGTGCCATGGGTATGAACAGTCTGTATCGTAAAGC | Amplify acnA from Vibrio cholerae and insert into pTrc99A fused to sfgfp |
| NBp202 | RW | CACCCTTGGAGCTACCTCCGCCCCCCTGCGCCAAAAAGTCTTG | |
| NBp216 | FW | ACTAGTGAAGGAGTGTGCCATGGGTATGCGTATCATTCTGCTGG | amplify adk from Yersinia enterocolitica and insert into pTrc99A fused to sfgfp |
| NBp217 | RW | CACCCTTGGAGCTACCTCCGCCCCCACCGAGAATAGTCGCCAG | |
| NBp218 | FW | CTAGTGAAGGAGTGTGCCATGGGTATGTCTGTAATTAAGATGACCGATCTGG | Amplify pgk from Yersinia enterocolitica and insert into pTrc99A fused to sfgfp |
| NBp219 | RW | CACCCTTGGAGCTACCTCCGCCCCCCTGCTTAGCGCGCTCTTC | |
| NBp220 | FW | ACTAGTGAAGGAGTGTGCCATGGGTATGTCGTTGGATTTGCGGAAAAC | Amplify acnA from Yersinia enterocolitica and insert into pTrc99A fused to sfgfp |
| NBp221 | RW | CACCCTTGGAGCTACCTCCGCCCCCCAACATTTTGCGGATCACATAATGC | |
| NBp227 | FW | GATGGCTGGACGGTAGAAACCGAAGATCGCAGCTTGTCTGCAC | Site-directed mutagenesis of Lys to Glu in map-sfgfp |
| NBp228 | RW | TTCCATGGTGCGGATCTCTTTTTCACCCGCGTTGACCATTGG | |
| NBp229 | FW | GATGGCTGGACGGTAGCAACCGCAGATCGCAGCTTGTCTGCAC | Site-directed mutagenesis of Lys to Ala in map-sfgfp |
| NBp230 | RW | TGCCATGGTGCGGATCTCTTTTGCACCCGCGTTGACCATTGG | |
| NBp231 | FW | ACTAGTGAAGGAGTGTGCCATGGGTATGGGTAAAATAATTGGTATCG | Amplify dnaK from E. coli MG1655 and insert into pTrc99A fused to sfgfp |
| NBp232 | RW | CACCCTTGGAGCTACCTCCGCCCCCTTTTTTGTCTTTGACTTCTTC | |
| NBp234 | FW | CCAGTCTGCGTTTACCATCCATG | Site-directed mutagenesis of V436F in dnaK-sfgfp |
| NBp235 | RW | TTGTCTTCAGCGGTAGAG | |
| NBp240 | FW | TTTTCTTATGATGTAGAACGTGCAACGCAATTGATGCTCGCTGTTGCGTACCAGGGGAAGGCCATT | Site-directed mutagenesis of D35A, D36A, H66A in clpS-sfgfp |
| NBp241 | RW | GAATTTTTGTAACACGTCAATAACAAACTCCATCGGAGTGTACGCCGCATTGACTAATATCACTTTATACATAGATGGC | |
| Eri121 | FW | CAGTCATAGCCG AATAGCCT | Checking insertion of KanR cassette |
| Eri122 | RW | CGGTGCCCTGAA TGAACTGC |
-
*
Indicated constructs were erroneously generated omitting deletion of ATG start codon of sfgfp gene and thus corrected with site-directed mutagenesis.
List of plasmids generated for this study.
| Plasmid | Relevant genotype | Reference or source |
|---|---|---|
| pTrc99A | Ampr; expression vector; pBR ori; trc promoter, IPTG inducible | Amann et al., 1988 |
| pCP20 | Ampr, Camr; flp | Cherepanov and Wackernagel, 1995 |
| pNB1 | Ampr; sfGFP in pTrc99A | This work |
| pNB3 | Ampr; Adk-sfGFP in pTrc99A | This work |
| pNB4 | Ampr; CoaE-sfGFP in pTrc99A | This work |
| pNB5 | Ampr; Cmk-sfGFP in pTrc99A | This work |
| pNB6 | Ampr; Pgk-sfGFP in pTrc99A | This work |
| pNB7 | Ampr; MmuM-sfGFP in pTrc99A | This work |
| pNB8 | Ampr; PrpD-sfGFP in pTrc99A | This work |
| pNB9 | Ampr; DsdA-sfGFP in pTrc99A | This work |
| pNB11 | Ampr; GlcB-sfGFP in pTrc99A | This work |
| pNB13 | Ampr; HemN-sfGFP in pTrc99A | This work |
| pNB14 | Ampr; MapWT-sfGFP in pTrc99A | This work |
| pNB15 | Ampr; ThrC-sfGFP in pTrc99A | This work |
| pNB16 | Ampr; MalZ-sfGFP in pTrc99A | This work |
| pNB17 | Ampr; EntC-sfGFP in pTrc99A | This work |
| pNB18 | Ampr; ThpR-sfGFP in pTrc99A | This work |
| pNB19 | Ampr; AroA-sfGFP in pTrc99A | This work |
| pNB20 | Ampr; ClpSWT-sfGFP in pTrc99A | This work |
| pNB21 | Ampr; Crr-sfGFP in pTrc99A | This work |
| pNB22 | Ampr; KdsB-sfGFP in pTrc99A | This work |
| pNB23 | Ampr; LeuS-sfGFP in pTrc99A | This work |
| pNB24 | Ampr; MurF-sfGFP in pTrc99A | This work |
| pNB25 | Ampr; NagD-sfGFP in pTrc99A | This work |
| pNB26 | Ampr; RihA-sfGFP in pTrc99A | This work |
| pNB27 | Ampr; SolA-sfGFP in pTrc99A | This work |
| pNB28 | Ampr; UbiC-sfGFP in pTrc99A | This work |
| pNB29 | Ampr; PanE-sfGFP in pTrc99A | This work |
| pNB30 | Ampr; FolK-sfGFP in pTrc99A | This work |
| pNB39 | Ampr; AcnA-sfGFP in pTrc99A | This work |
| pNB40 | Ampr; MetE-sfGFP in pTrc99A | This work |
| pNB42 | Ampr; MetH-sfGFP in pTrc99A | This work |
| pNB44 | Ampr; YggX-sfGFP in pTrc99A | This work |
| pNB45 | Ampr; AdkC.c.-sfGFP in pTrc99A | This work |
| pNB46 | Ampr; AdkV.c.-sfGFP in pTrc99A | This work |
| pNB47 | Ampr; AdkM.x.-sfGFP in pTrc99A | This work |
| pNB48 | Ampr; AcnAM.x.-sfGFP in pTrc99A | This work |
| pNB49 | Ampr; AcnAV.c.-sfGFP in pTrc99A | This work |
| pNB51 | Ampr; AdkB.s.-sfGFP in pTrc99A | This work |
| pNB52 | Ampr; AcnAB.s.-sfGFP in pTrc99A | This work |
| pNB54 | Ampr; AdkY.e.-sfGFP in pTrc99A | This work |
| pNB56 | Ampr; PgkC.c.-sfGFP in pTrc99A | This work |
| pNB58 | Ampr; PgkV.c.-sfGFP in pTrc99A | This work |
| pNB59 | Ampr; PgkM.x.sfGFP in pTrc99A | This work |
| pNB60 | Ampr; AcnAY.e.-sfGFP in pTrc99A | This work |
| pNB61 | Ampr; DnaKWT-sfGFP in pTrc99A | This work |
| pNB62 | Ampr; MapK211E_K218E_K224E_K226E-sfGFP in pTrc99A | This work |
| pNB63 | Ampr; MapK211A_K218A_K224A_K226A-sfGFP in pTrc99A | This work |
| pNB64 | Ampr; DnaKV436F -sfGFP in pTrc99A | This work |
| pNB66 | Ampr; ClpSD35A_D36A_H66A-sfGFP in pTrc99A | This work |
Figure 1C.
| Testing pair | P-value |
|---|---|
| sfGFP vs Adk-sfGFP | 0.000000010 |
| Adk-sfGFP vs AcnA-sfGFP | 0.00000000044 |
Figure 1—figure supplement 10A.
| Testing pair | P-value |
|---|---|
| ClpSWT-sfGFP versus ClpSD35A_D36A_H66A-sfGFP | 0.000094 |
| MapWT-sfGFP versus MapLys→Ala-sfGFP | 0.0000012 |
| MapWT-sfGFP versus MapLys→Glu-sfGFP | 0.000000016 |
| MapLys→Ala-sfGFP versus MapLys→Glu-sfGFP | 0.10 |
| DnaKWT-sfGFP versus DnaKV436F-sfGFP | 0.00023 |
Figure 1—figure supplement 10B.
| Testing pair | P-value |
|---|---|
| ClpSWT-sfGFP versus ClpSD35A_D36A_H66A-sfGFP | 0.000014 |
| MapWT-sfGFP versus MapLys→Ala-sfGFP | 0.0092 |
| MapWT-sfGFP versus MapLys→Glu-sfGFP | 0.065 |
| MapLys→Ala-sfGFP versus MapLys→Glu-sfGFP | 0.37 |
| DnaKWT-sfGFP versus DnaKV436F-sfGFP | 0.00000019 |
Figure 2C.
| Testing pair | P-value |
|---|---|
| Untreated versus A22 treatment | 0.0000000000007 |
Figure 2D.
| Testing pair | P-value |
|---|---|
| Untreated versus A22 treatment | 0.000001 |
Figure 2—figure supplement 2.
| Testing pair | P-value |
|---|---|
| Untreated versus A22 treatment | 0.002 |
Figure 2—figure supplement 4A.
| Testing pair | P-value |
|---|---|
| sfGFP, 1 A.U. versus 0.66 A.U. | 0.00001 |
| DnaK-sfGFP 1 A.U. versus 0.66 A.U. | 0.60 |
| AcnA-sfGFP, 1 A.U. versus 0.66 A.U. | 0.000002 |
Figure 2—figure supplement 4B.
| Testing pair | P-value |
|---|---|
| sfGFP, 1 A.U. versus 0.66 A.U. | 0.24 |
| DnaK-sfGFP 1 A.U. versus 0.66 A.U. | 0.50 |
| AcnA-sfGFP, 1 A.U. versus 0.66 A.U. | 0.002 |
Figure 4—figure supplement 1A.
| Testing pair | P-value |
|---|---|
| AdkE.c.-sfGFP versus AdkY.e.-sfGFP | 0.17 |
| AdkE.c.-sfGFP versus AdkV.c. -sfGFP | 0.056 |
| AdkE.c.-sfGFP versus AdkC.c.-sfGFP | 0.93 |
| AdkE.c.-sfGFP versus AdkM.x.-sfGFP | 0.21 |
| AdkE.c.-sfGFP versus AdkB.s.-sfGFP | 0.23 |
| PgkE.c.-sfGFP versus PgkV.c.-sfGFP | 0.75 |
| PgkE.c.-sfGFP versus PgkC.c.-sfGFP | 0.26 |
| PgkE.c.-sfGFP versus PgkM.x.-sfGFP | 0.33 |
| AcnAE.c.-sfGFP versus AcnAY.e.-sfGFP | 0.093 |
| AcnAE.c.-sfGFP versus AcnAV.c.-sfGFP | 0.084 |
| AcnAE.c.-sfGFP versus AcnAM.x.-sfGFP | 0.0000000023 |
| AcnAE.c.-sfGFP versus AcnAB.s.-sfGFP | 0.069 |
Figure 4—figure supplement 1B.
| Testing pair | P-value |
|---|---|
| AdkE.c.-sfGFP versus AdkY.e.-sfGFP | 0.000060 |
| AdkE.c.-sfGFP versus AdkV.c. -sfGFP | 0.18 |
| AdkE.c.-sfGFP versus AdkC.c.-sfGFP | 0.042 |
| AdkE.c.-sfGFP versus AdkM.x.-sfGFP | 0.070 |
| AdkE.c.-sfGFP versus AdkB.s.-sfGFP | 0.0029 |
| PgkE.c.-sfGFP versus PgkV.c.-sfGFP | 0.11 |
| PgkE.c.-sfGFP versus PgkC.c.-sfGFP | 0.0082 |
| PgkE.c.-sfGFP versus PgkM.x.-sfGFP | 0.0087 |
| AcnAE.c.-sfGFP versus AcnAY.e.-sfGFP | 0.035 |
| AcnAE.c.-sfGFP versus AcnAV.c.-sfGFP | 0.16 |
| AcnAE.c.-sfGFP versus AcnAM.x.-sfGFP | 0.083 |
| AcnAE.c.-sfGFP versus AcnAB.s.-sfGFP | 0.00024 |
Figure 5—figure supplement 2A.
| Testing pair | P-value |
|---|---|
| sfGFP ionic strength 105 mM versus 305 mM | 0.0000036 |
| Adk-sfGFP ionic strength 105 mM versus 305 mM | 0.000044 |
| AroA-sfGFP ionic strength 105 mM versus 305 mM | 0.035 |
| AcnA-sfGFP ionic strength 105 mM versus 305 mM | 0.0018 |
Figure 5—figure supplement 2B.
| Testing pair | P-value |
|---|---|
| sfGFP 25°C versus 35°C | 0.00015 |
| Adk-sfGFP 25°C versus 35°C | 0.0000025 |
| AcnA-sfGFP 25°C versus 35°C | 0.00077 |
Figure 5—figure supplement 2C.
| Testing pair | P-value |
|---|---|
| sfGFP Untreated versus Chloramphenicol | 0.40 |
| sfGFP DMSO versus Rifampicin | 0.012 |
| Adk-sfGFP Untreated versus Chloramphenicol | 0.12 |
| Adk-sfGFP DMSO versus Rifampicin | 0.00048 |
| Pgk-sfGFP Untreated versus Chloramphenicol | 0.17 |
| Pgk-sfGFP DMSO versus Rifampicin | 0.0000012 |
| AcnA-sfGFP Untreated versus Chloramphenicol | 0.000011 |
| AcnA-sfGFP DMSO versus Rifampicin | 0.0085 |
Figure 5—figure supplement 2D.
| Testing pair | P-value |
|---|---|
| sfGFP M9 salts versus M9 salts, Glu+CA | 0.000023 |
| Adk-sfGFP M9 salts versus M9 salts, Glu+CA | 0.000070 |
| AroA-sfGFP M9 salts versus M9 salts, Glu+CA | 0.00000015 |
| AcnA-sfGFP M9 salts versus M9 salts, Glu+CA | 0.0022 |
Figure 5—figure supplement 3A.
| Testing pair | P-value |
|---|---|
| sfGFP ionic strength 105 mM versus 305 mM | 0.097 |
| Adk-sfGFP ionic strength 105 mM versus 305 mM | 0.54 |
| AroA-sfGFP ionic strength 105 mM versus 305 mM | 0.077 |
| AcnA-sfGFP ionic strength 105 mM versus 305 mM | 0.31 |
Figure 5—figure supplement 3B.
| Testing pair | P-value |
|---|---|
| sfGFP 25°C versus 35°C | 0.12 |
| Adk-sfGFP 25°C versus 35°C | 0.005 |
| AcnA-sfGFP 25°C versus 35°C | 0.26 |
Figure 5—figure supplement 3C.
| Testing pair | P-value |
|---|---|
| sfGFP Untreated versus Chloramphenicol | 0.32 |
| sfGFP DMSO versus Rifampicin | 0.50 |
| Adk-sfGFP Untreated versus Chloramphenicol | 0.53 |
| Adk-sfGFP DMSO versus Rifampicin | 0.32 |
| Pgk-sfGFP Untreated versus Chloramphenicol | 0.17 |
| Pgk-sfGFP DMSO versus Rifampicin | 0.59 |
| AcnA-sfGFP Untreated versus Chloramphenicol | 0.008 |
| AcnA-sfGFP DMSO versus Rifampicin | 0.42 |
Figure 5—figure supplement 3D.
| Testing pair | P-value |
|---|---|
| sfGFP M9 salts versus M9 salts, Glu+CA | 0.44 |
| Adk-sfGFP M9 salts versus M9 salts, Glu+CA | 0.035 |
| AroA-sfGFP M9 salts versus M9 salts, Glu+CA | 0.69 |
| AcnA-sfGFP M9 salts versus M9 salts, Glu+CA | 0.31 |
Figure 5—figure supplement 4A.
| Testing pair | P-value |
|---|---|
| sfGFP Grown 25°C, measured 25°C versus 35°C | 0.0020 |
| sfGFP Measured 25°C, grown 25°C versus 37°C | 0.060 |
| sfGFP Measured 35°C, grown 25°C versus 37°C | 0.98 |
| sfGFP Grown 37°C, measured 25°C versus 35°C | 0.000040 |
Figure 5—figure supplement 4B.
| Testing pair | P-value |
|---|---|
| sfGFP Grown 25°C, measured 25°C versus 35°C | 0.26 |
| sfGFP Measured 25°C, grown 25°C versus 37°C | 0.45 |
| sfGFP Measured 35°C, grown 25°C versus 37°C | 0.44 |
| sfGFP Grown 37°C, measured 25°C versus 35°C | 0.12 |
Figure 5—figure supplement 5A.
| Testing pair | P-value |
|---|---|
| sfGFP cytoplasm versus nucleoid | 0.20 |
| AcnA-sfGFP cytoplasm versus nucleoid | 0.062 |
Figure 5—figure supplement 5B.
| Testing pair | P-value |
|---|---|
| sfGFP cytoplasm versus nucleoid | 0.09 |
| AcnA-sfGFP cytoplasm versus nucleoid | 0.09 |
Figure 5—figure supplement 6A.
| Testing pair | P-value |
|---|---|
| sfGFP M9 salts versus M9 salts, Cam | 0.82 |
| sfGFP M9 salts versus M9 salts, Glu+CA | 0.000023 |
| sfGFP M9 salts, Cam vs M9 salts, Cam, Glu +CA | 0.019 |
| sfGFP M9 salts, Glu +CA versus M9 salts, Cam, Glu +CA | 0.019 |
| AcnA-sfGFP M9 salts versus M9 salts, Cam | 0.37 |
| AcnA-sfGFP M9 salts versus M9 salts, Glu+CA | 0.0022 |
| AcnA-sfGFP M9 salts, Cam versus M9 salts, Cam, Glu +CA | 0.0035 |
| AcnA-sfGFP M9 salts, Glu +CA versus M9 salts, Cam, Glu +CA | 0.014 |
Figure 5—figure supplement 6B.
| Testing pair | P-value |
|---|---|
| sfGFP M9 salts versus M9 salts, Cam | 0.33 |
| sfGFP M9 salts versus M9 salts, Glu+CA | 0.44 |
| sfGFP M9 salts, Cam versus M9 salts, Cam, Glu+CA | 0.91 |
| sfGFP M9 salts, Glu+CA versus M9 salts, Cam, Glu+CA | 0.80 |
| AcnA-sfGFP M9 salts versus M9 salts, Cam | 0.099 |
| AcnA-sfGFP M9 salts versus M9 salts, Glu+CA | 0.31 |
| AcnA-sfGFP M9 salts, Cam versus M9 salts, Cam, Glu+CA | 0.0085 |
| AcnA-sfGFP M9 salts, Glu+CA versus M9 salts, Cam, Glu+CA | 0.56 |
Figure 5—figure supplement 7A.
| Testing pair | P-value |
|---|---|
| sfGFP untreated versus DNP treatment, 25°C | 0.52 |
| sfGFP untreated versus DNP treatment, 35°C | 0.66 |
| Adk-sfGFP untreated versus DNP treatment, 25°C | 0.46 |
| Adk-sfGFP untreated versus DNP treatment, 35°C | 0.03 |
| AcnA-sfGFP untreated versus DNP treatment, 25°C | 0.59 |
| AcnA-sfGFP untreated versus DNP treatment, 35°C | 0.98 |
Figure 5—figure supplement 7B.
| Testing pair | P-value |
|---|---|
| sfGFP untreated versus DNP treatment, 25°C | 0.013 |
| sfGFP untreated versus DNP treatment, 35°C | 0.32 |
| Adk-sfGFP untreated versus DNP treatment, 25°C | 0.006 |
| Adk-sfGFP untreated versus DNP treatment, 35°C | 0.25 |
| AcnA-sfGFP untreated versus DNP treatment, 25°C | 0.94 |
| AcnA-sfGFP untreated versus DNP treatment, 35°C | 0.57 |
Figure 1 and Figure 1—figure supplement 5.
| Construct | Numerosity (n) | Construct | Numerosity (n) |
|---|---|---|---|
| sfGFP | 52 | EntC-sfGFP | 15 |
| YggX-sfGFP | 8 | AroA-sfGFP | 9 |
| ClpSWT-sfGFP | 11 | ThrC-sfGFP | 14 |
| FolK-sfGFP | 8 | MurF-sfGFP | 7 |
| Crr-sfGFP | 14 | DsdA-sfGFP | 14 |
| UbiC-sfGFP | 14 | HemN-sfGFP | 13 |
| CoaE-sfGFP | 11 | PrpD-sfGFP | 12 |
| Adk-sfGFP | 23 | DnaKWT-sfGFP | 10 |
| Cmk-sfGFP | 16 | MalZ-sfGFP | 9 |
| KdsB-sfGFP | 22 | GlcB-sfGFP | 16 |
| MapWT-sfGFP | 20 | MetE-sfGFP | 8 |
| MmuM-sfGFP | 14 | LeuS-sfGFP | 14 |
| PanE-sfGFP | 18 | AcnA-sfGFP | 19 |
| SolA-sfGFP | 7 | MetH-sfGFP | 9 |
| Pgk-sfGFP | 16 | ||
Figure 1—figure supplement 7.
| Construct | Numerosity (n) |
|---|---|
| sfGFP | Same as Appendix 6—table 1 |
Figure 1—figure supplement 8.
| Condition | Numerosity (n) |
|---|---|
| Untreated | 5 |
| Cephalexin | 6 |
Figure 1—figure supplement 10.
| Construct | Numerosity (n) |
|---|---|
| ClpSWT-sfGFP | Same as Appendix 6—table 1 |
| ClpSD35A_D36A_H66A-sfGFP | 10 |
| MapWT-sfGFP | Same as Appendix 6—table 1 |
| MapLys→Glu-sfGFP | 10 |
| MapLys→Ala-sfGFP | 12 |
| DnaKWT-sfGFP | Same as Appendix 6—table 1 |
| DnaKV436F-sfGFP | 10 |
Figure 2C and D and Figure 2—figure supplement 2.
| Condition | Numerosity (n) |
|---|---|
| Untreated | Same as Appendix 6—table 1 |
| A22-treated | 12 |
Figure 2—figure supplement 4.
| Condition | Numerosity (n) |
|---|---|
| sfGFP 1 A.U. | Same as Appendix 6—table 1 |
| sfGFP 0.66 A. U. | 12 |
| DnaK-sfGFP 1 A.U. | Same as Appendix 6—table 1 |
| DnaK-sfGFP 0.66 A.U, | 10 |
| AcnA-sfGFP 1 A.U. | Same as Appendix 6—table 1 |
| AcnA-sfGFP 0.66 A. U. | 10 |
Figure 2—figure supplement 5.
| Construct | Numerosity (n) |
|---|---|
| sfGFP | Same as Appendix 6—table 1 |
| Adk-sfGFP | Same as Appendix 6—table 1 |
| DnaK-sfGFP | Same as Appendix 6—table 1 |
| DnaKV436-sfGFP | Same as Appendix 6—table 4 |
| AcnA-sfGFP | Same as Appendix 6—table 1 |
Figure 3C.
| Construct | Numerosity (n) | Construct | Numerosity (n) |
|---|---|---|---|
| sfGFP | 11 | DsdA-sfGFP | 10 |
| YggX-sfGFP | 10 | GlcB-sfGFP | 10 |
| Adk-sfGFP | 16 | AcnA-sfGFP | 10 |
| PanE-sfGFP | 11 | MetH-sfGFP | 15 |
Figure 4 and Figure 4—figure supplement 1.
| Construct | Numerosity (n) | Construct | Numerosity (n) |
|---|---|---|---|
| AdkE.c.-sfGFP | Same as Appendix 6—table 1 | PgkC.c.-sfGFP | 5 |
| AdkY.e.-sfGFP | 5 | PgkM.x.-sfGFP | 11 |
| AdkV.c.-sfGFP | 10 | AcnAE.c.-sfGFP | Same as Appendix 6—table 1 |
| AdkC.c.-sfGFP | 5 | AcnAY.e.-sfGFP | 5 |
| AdkM.x.-sfGFP | 11 | AcnAV.c.-sfGFP | 10 |
| AdkB.s.-sfGFP | 10 | AcnAM.x.-sfGFP | 10 |
| PgkE.c.-sfGFP | Same as Appendix 6—table 1 | AcnAB.s.-sfGFP | 10 |
| PgkV.c.-sfGFP | 10 | ||
Figure 5A, Figure 5—figure supplement 1A, B, Figure 5—figure supplement 2, and Figure 5—figure supplement 3A.
| Construct and condition | Numerosity (n) | Construct and condition | Numerosity (n) |
|---|---|---|---|
| sfGFP, 105 mM | Same as Appendix 6—table 1 | AroA-sfGFP, 105 mM | Same as Appendix 6—table 1 |
| sfGFP, 305 mM | 11 | AroA-sfGFP, 305 mM | 12 |
| Adk-sfGFP, 105 mM | Same as Appendix 6—table 1 | AcnA-sfGFP, 105 mM | Same as Appendix 6—table 1 |
| Adk-sfGFP, 305 mM | 11 | AcnA-sfGFP, 305 mM | 6 |
Figure 5B, Figure 5—figure supplement 1C, D, Figure 5—figure supplement 2, and Figure 5—figure supplement 3B.
| Construct and condition | Numerosity (n) | Construct and condition | Numerosity (n) |
|---|---|---|---|
| sfGFP, 25°C | Same as Appendix 6—table 1 | AcnA-sfGFP, 25°C | Same as Appendix 6—table 1 |
| sfGFP, 35°C | 14 | AcnA-sfGFP, 35°C | 18 |
| Adk-sfGFP, 25°C | Same as Appendix 6—table 1 | ||
| Adk-sfGFP, 35°C | 21 | ||
Figure 5C, Figure 5—figure supplement 1E, F, Figure 5—figure supplement 2C, and Figure 5—figure supplement 3C.
| Construct and condition | Numerosity (n) | Construct and condition | Numerosity (n) |
|---|---|---|---|
| sfGFP, untreated | Same as Appendix 6—table 1 | Pgk-sfGFP, untreated | Same as Appendix 6—table 1 |
| sfGFP, chloramphenicol | 10 | Pgk-sfGFP, chloramphenicol | 10 |
| sfGFP, DMSO | 15 | Pgk-sfGFP, DMSO | 10 |
| sfGFP, rifampicin | 15 | Pgk-sfGFP, rifampicin | 10 |
| Adk-sfGFP, untreated | Same as Appendix 6—table 1 | AcnA-sfGFP, untreated | Same as Appendix 6—table 1 |
| Adk-sfGFP, chloramphenicol | 10 | AcnA-sfGFP, chloramphenicol | 10 |
| Adk-sfGFP, DMSO | 10 | AcnA-sfGFP, DMSO | 10 |
| Adk-sfGFP, rifampicin | 10 | AcnA-sfGFP, rifampicin | 10 |
Figure 5D, Figure 5—figure supplement 1G, H, Figure 5—figure supplement 2D, and Figure 5—figure supplement 3D.
| Construct and condition | Numerosity (n) | Construct and condition | Numerosity (n) |
|---|---|---|---|
| sfGFP, M9 salts | 10 | AroA-sfGFP, M9 salts | 11 |
| sfGFP, M9 salts, Glu+CA | 15 | AroA-sfGFP, M9 salts, Glu+CA | 11 |
| Adk-sfGFP, M9 salts | 11 | AcnA-sfGFP, M9 salts | 10 |
| Adk-sfGFP, M9 salts, Glu+CA | 12 | AcnA-sfGFP, M9 salts, Glu+CA | 10 |
Figure 5—figure supplement 4.
| Construct and condition | Numerosity (n) |
|---|---|
| sfGFP, grown 25°C, measured 25°C | 10 |
| sfGFP, grown 25°C, measured 35°C | 10 |
| sfGFP, grown 37°C, measured 25°C | Same as Appendix 6—table 1 |
| sfGFP, grown 37°C, measured 35°C | Same as Appendix 6—table 11 |
Figure 5—figure supplement 5.
| Construct and condition | Numerosity (n) |
|---|---|
| sfGFP, cytoplasm | 10 |
| sfGFP, nucleoid | 10 |
| AcnA-sfGFP, cytoplasm | 10 |
| AcnA-sfGFP, nucleoid | 10 |
Figure 5—figure supplement 6.
| Construct and condition | Numerosity (n) | Construct and condition | Numerosity (n) |
|---|---|---|---|
| sfGFP, M9 salts | Same as Appendix 6—table 12 | AcnA-sfGFP, M9 salts | Same as Appendix 6—table 12 |
| sfGFP, M9 salts, Cam | 13 | AcnA-sfGFP, M9 salts, Cam | 8 |
| sfGFP, M9 salts, Glu+CA | Same as Appendix 6—table 12 | AcnA-sfGFP, M9 salts, Glu+CA | Same as Appendix 6—table 12 |
| sfGFP, M9 salts, Cam, Glu+CA | 13 | AcnA-sfGFP, M9 salts, Cam, Glu+CA | 10 |
Figure 5—figure supplement 7.
| Construct and condition | Numerosity (n) |
|---|---|
| sfGFP, untreated, 25°C | Same as Appendix 6—table 1 |
| sfGFP, 2 mM DNP, 25°C | 5 |
| sfGFP, untreated, 35°C | Same as Appendix 6—table 10 |
| sfGFP, 2 mM DNP, 35°C | 6 |
| Adk-sfGFP, untreated, 25°C | Same as Appendix 6—table 1 |
| Adk-sfGFP, 2 mM DNP, 25°C | 5 |
| Adk-sfGFP, untreated, 35°C | Same as Appendix 6—table 10 |
| Adk-sfGFP, 2 mM DNP, 35°C | 6 |
| AcnA-sfGFP, untreated, 25°C | Same as Appendix 6—table 1 |
| AcnA-sfGFP, 2 mM DNP, 25°C | 5 |
| AcnA-sfGFP, untreated, 35°C | Same as Appendix 6—table 10 |
| AcnA-sfGFP, 2 mM DNP, 35°C | 6 |