Evidence for virus-mediated oncogenesis in bladder cancers arising in solid organ transplant recipients
Figures
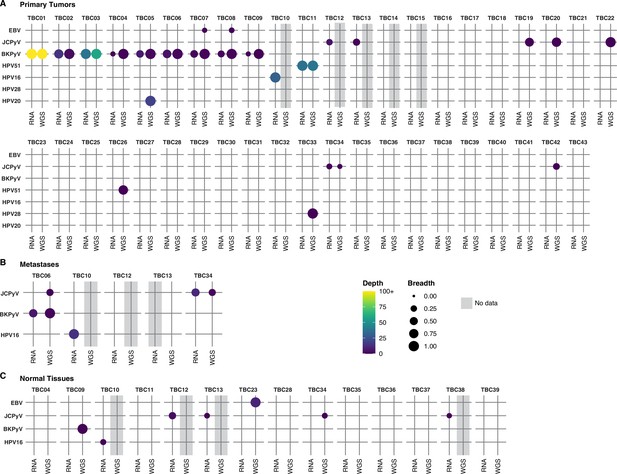
Detection of viral sequences.
(A) Primary tumors. (B) Metastatic tumors. (C) Normal tissues. Viral species are shown on the rows, and each case in the cohort (represented with a TBC number) is a column. TBC numbers represent a single case and are consistent across primary, metastatic, and normal tissues. Circle size represents the breadth or fraction of the viral genome covered, and color represents the average depth of coverage of the viral k-mers with all coverages over 100 binned together. Specimens without sequencing data have a gray background.
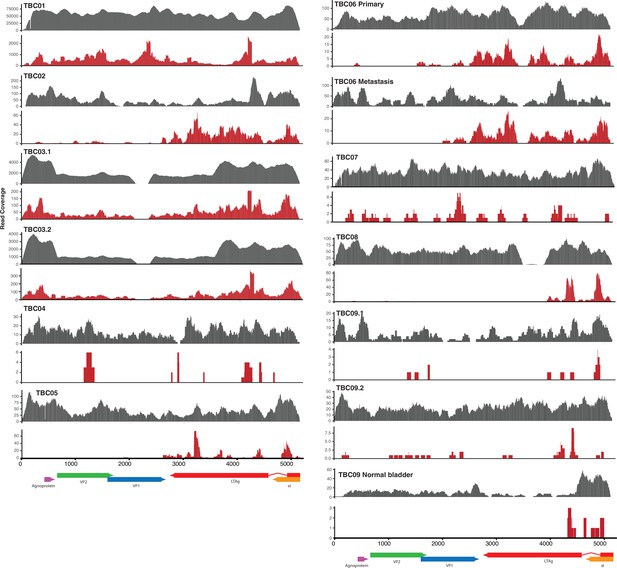
BK polyomavirus (BKPyV) DNA/RNA coverage plots.
Coverage plots for BKPyV DNA (gray) and RNA (red) in BKPyV-positive tumors.
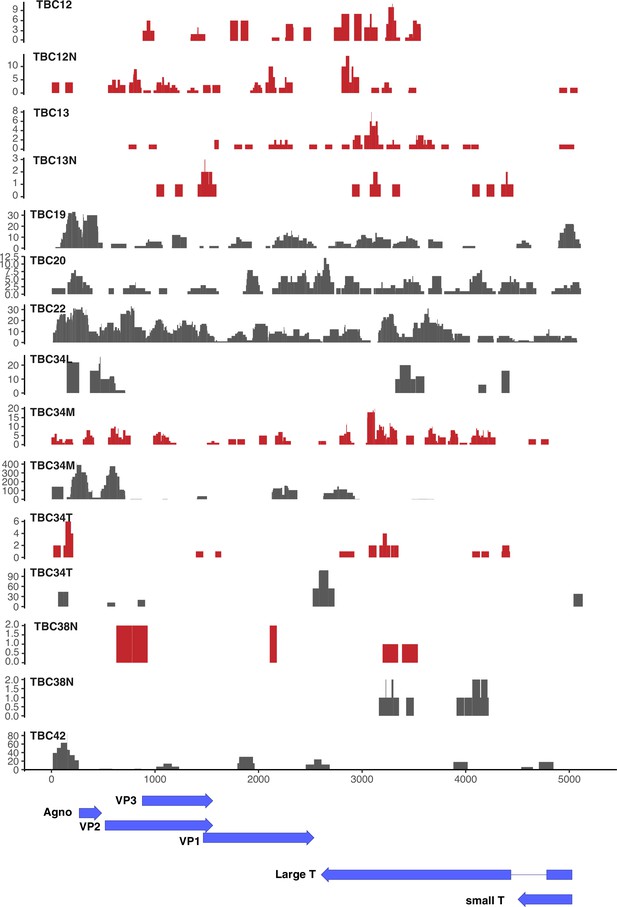
JC polyomavirus (JCPyV) DNA/RNA coverage plots.
Coverage plots for JCPyV DNA (gray) and RNA (red) in JCPyV-positive tumors.
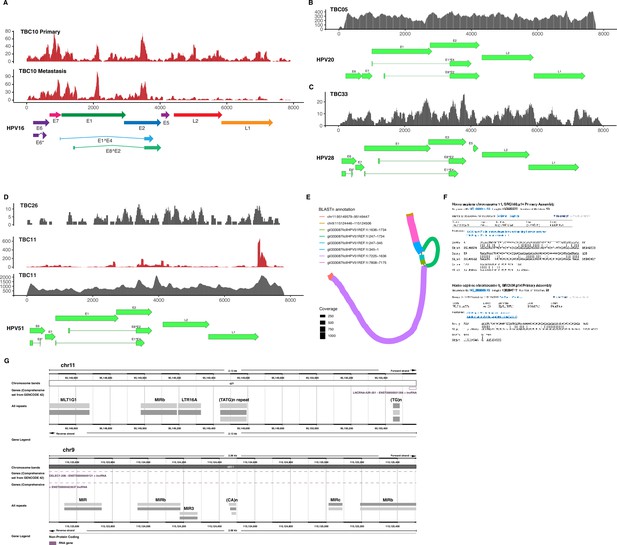
All human papillomavirus (HPV) DNA/RNA coverage plots.
(A-D) Coverage plots for HPV DNA (gray) and RNA (red) in HPV16 (A), HPV20 (B), HPV28 (C), and HPV51 (D) -positive tumors. Diagrams of open reading frames for each respective type are below the coverage plots. (E) Annotated assembly graph of TBC11 HPV51. (F) Details of the top human BLASTn hits from the assembly in (E). (G) 1 kb upstream and downstream of TBC11 HPV51 predicted human integration junctions with genes and repeats annotated.
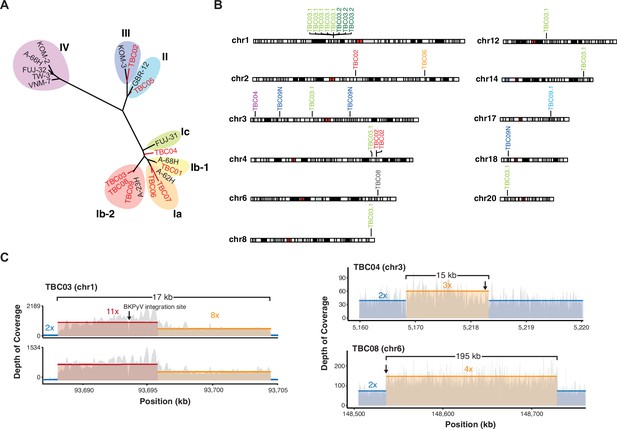
Virus diversity and integration.
(A) Phylogenetic tree of BK polyomavirus (BKPyV) large T antigen (LTag) sequences detected in tumors (red) and reference genotypes with representative strain names. (B) Sites of BKPyV integration into host chromosomes are indicated with case numbers. Two separate sections from separate formal-fixed paraffin-embedded (FFPE) blocks of the primary tumor were sequenced for case TBC03 (samples TBC03.1 and TBC03.2). Two separate sections were also sequenced for case TBC09, but an integration site was only detected in sample TBC09.1. Integration sites were also detected in normal tissue sample TBC09N. Black and gray bars indicate cytogenetic bands; red bars indicate centromeres. (C) Coverage plot of focal amplifications adjacent to BKPyV integration sites in cases TBC03.1, TBC03.2, TBC04, and TBC08. BKPyV integration junctions are indicated by a black arrow. Colored numbers in the body of the graph indicate coverage depth.
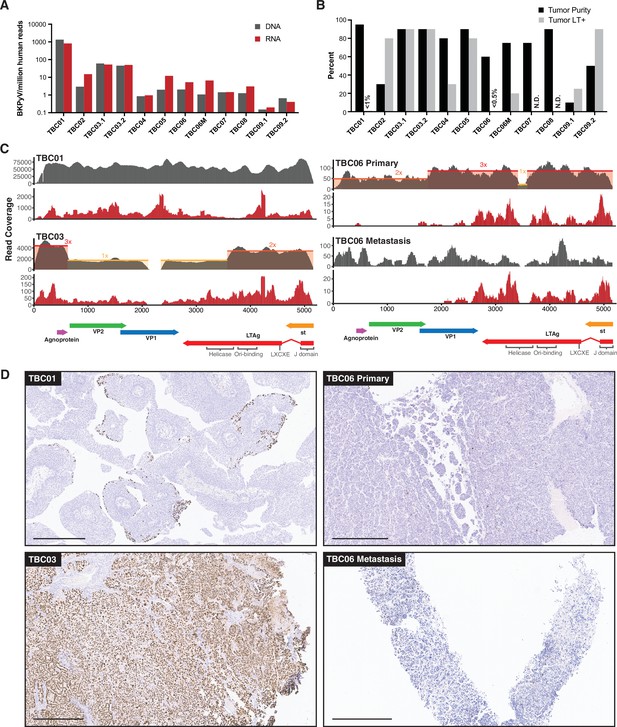
BK polyomavirus (BKPyV) DNA, RNA, and large T antigen (LTag) detection in tumors.
(A) Barplots showing the abundance of BKPyV DNA and RNA reads standardized to human reads (B) Barplots of histologically estimated percent tumor purity and Immunohistochemistry (IHC)-positivity for polyomavirus LTag expression. N.D. indicates no IHC image data were generated. (C) Representative coverage plots for BKPyV DNA (gray) and RNA (red) in BKPyV-positive tumors. Relative copy numbers are indicated by colored boxes and highlight the borders of duplications and deletions in the viral genome. (D) Selected images for LTag IHC highlighting positive staining for BKPyV-positive tumors with scale bars representing 500 microns.
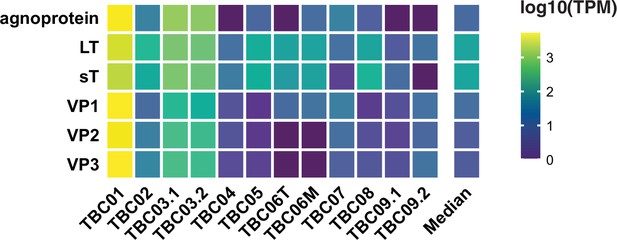
BK polyomavirus (BKPyV) gene expression.
Heatmap of normalized expression (transcripts per million, TPM) of BKPyV genes per tumor.
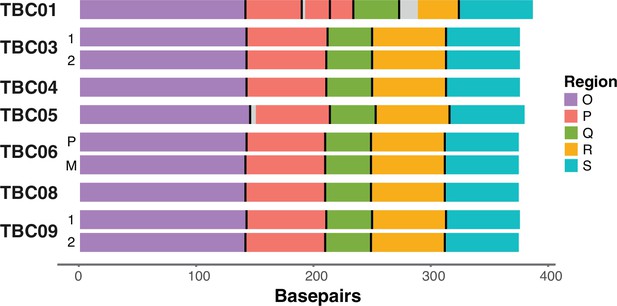
Diagrams of the assembled BK polyomavirus (BKPyV) NCCR structures and rearrangements in tumors.
P=primary tumor, M=metastatic tumor.
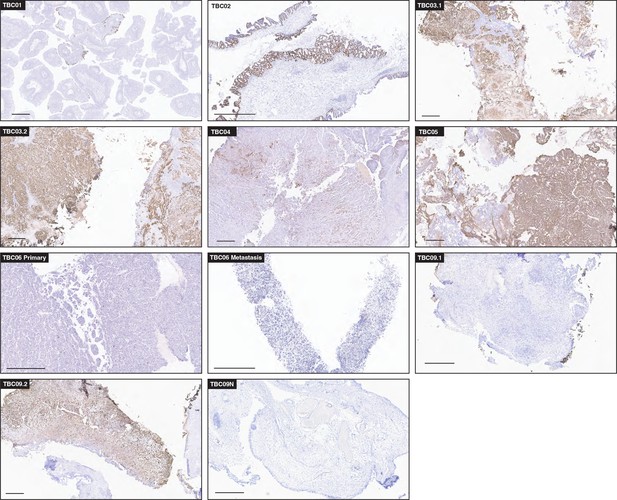
T antigen immunohistochemistry (IHC) in BK polyomavirus (BKPyV)-positive tumors.
Selected images for T antigen IHC highlighting positive staining for BKPyV-positive tumors with a scale bar representing 500 microns.
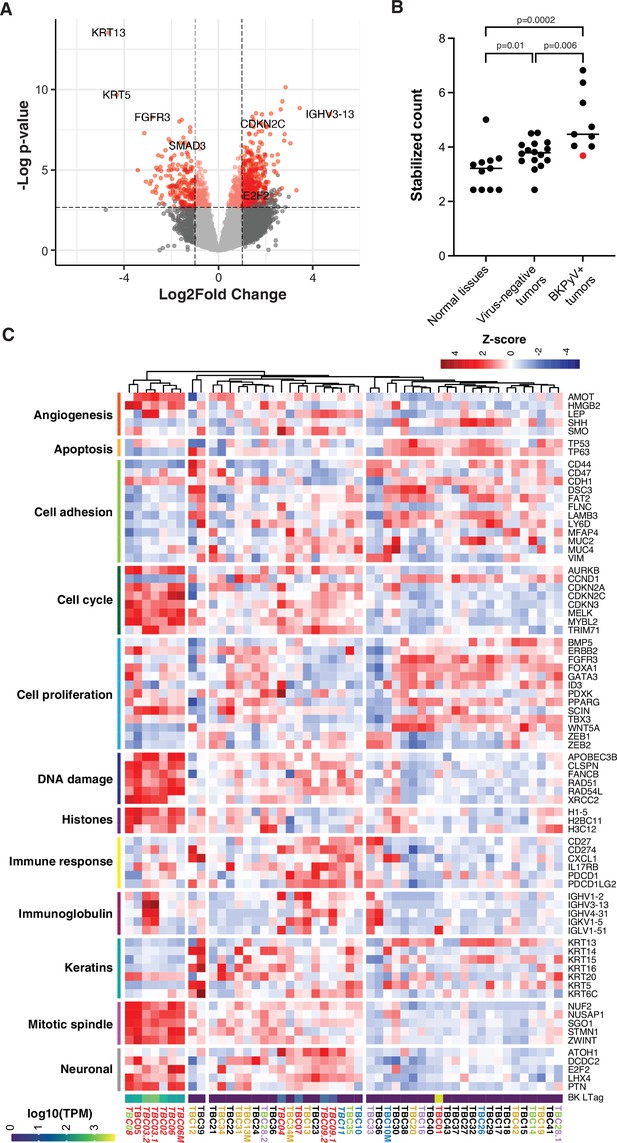
Differential gene expression in BK polyomavirus (BKPyV)-positive tumors.
(A) Volcano plot of differential gene expression between BKPyV-positive and virus-negative tumors. Significantly differentially expressed genes (q-value <0.05, DESeq2) with a fold change greater than two are in red, and genes with a fold change less than two are in pink. Non-significant genes are in gray. (B) Variance stabilized counts for APOBEC3B expression from DESeq2 grouped by normal tissues, virus-negative tumors, and BKPyV-positive tumors showing significantly increased expression in BKPyV-positive tumors (Mann-Whitney U test). TBC01 is indicated by a red dot. (C) Heatmap of Z-scores of significantly differentially expressed genes and genes relevant to bladder cancer grouped by gene ontology. High expression is red, low expression is blue. Tumors names are colored by likely etiology: BKPyV-positive, red; JC polyomavirus (JCPyV)-positive, goldenrod; HR-HPV-positive, blue; torque teno virus (TTV)-positive, green; aristolochic acid, purple; undetermined, black; multiple colors reflect multiple detected viruses or etiologies. Tumors with evidence of integration are in italics. BKPyV LTag expression is shown as log10(transcripts per million [TPM]).
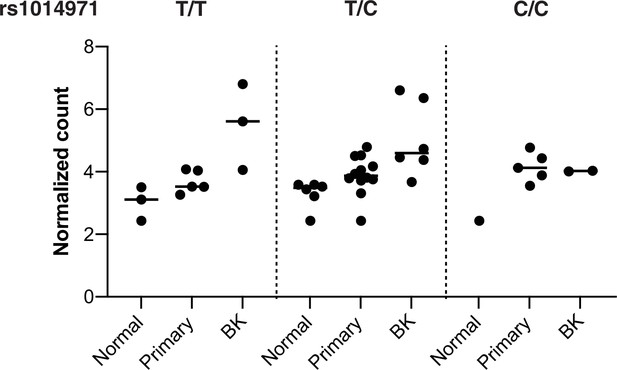
APOBEC3B germline variant and expression by BK polyomavirus (BKPyV) status.
Stabilized counts of APOBEC3B expression divided by tissue type (primary tumor, normal tissue), BKPyV status (BK), and germline variant rs1014971 status.
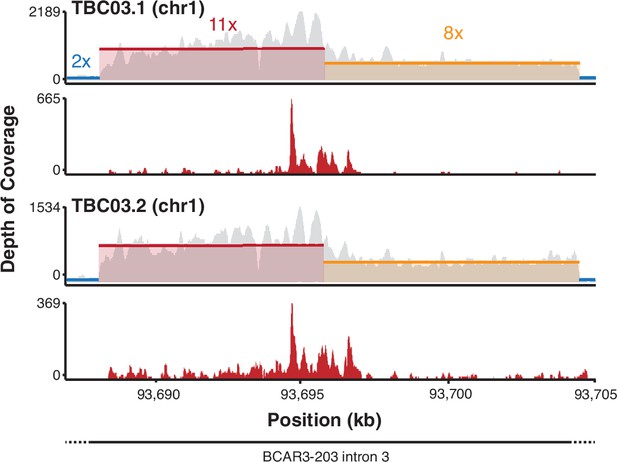
Host transcripts from the BK polyomavirus (BKPyV) integration site at BCAR3.
Coverage plots of DNA (gray) and RNA (red) from TBC03.1 and TBC03.2 on chromosome 1 at the BCAR3 locus.
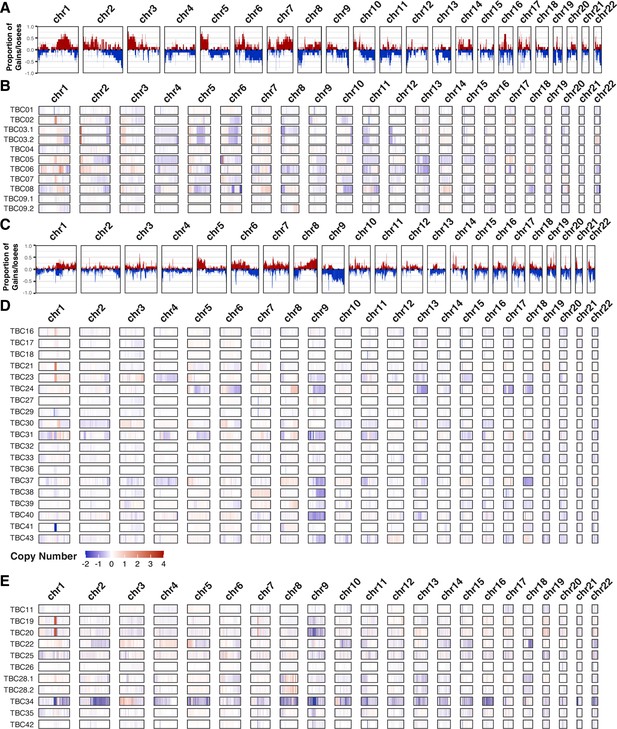
Copy number variants.
Frequency plots for large copy number variants in BK polyomavirus (BKPyV)-positive tumors (panel A) and virus-negative tumors (panel C). Frequency of gains/amplifications is shown in red; losses/deletions are shown in blue. Sample level copy number variant spectra for BKPyV-positive tumors (panel B), virus-negative tumors (panel D), and all other tumors (panel E). Complete deletions are in dark blue and high copy amplifications are in red.
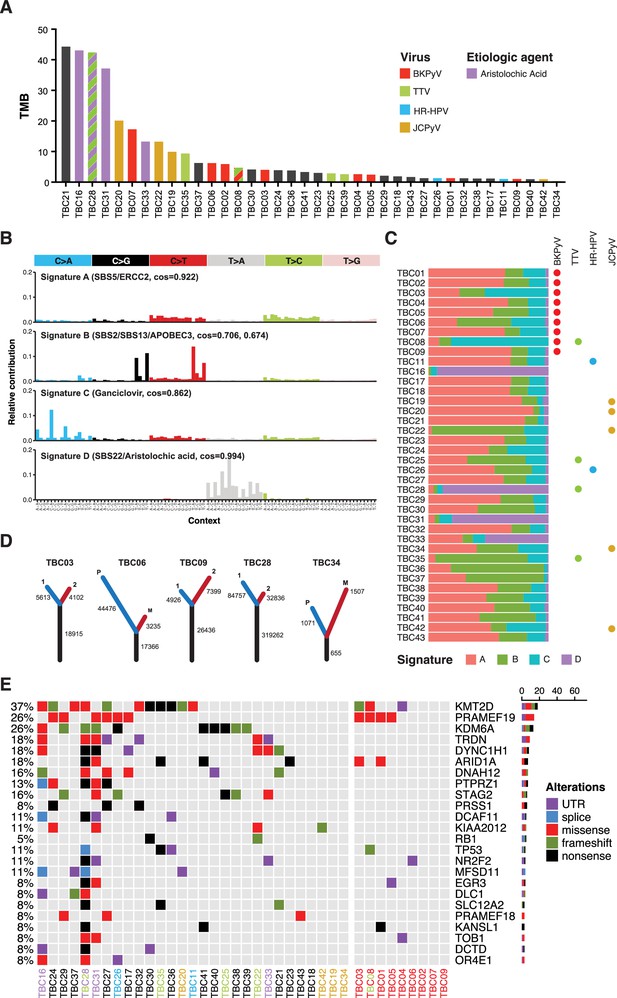
Somatic point mutations and mutation signature analysis.
(A) Tumor mutation burden (TMB, non-synonymous mutations per million bases) for each tumor in this study. Bars are colored by viral positivity (red, BK polyomavirus (BKPyV); green, TTV; blue, HR-HPV; goldenrod, JC polyomavirus (JCPyV)) or etiologic agent (aristolochic acid, purple; black, undetermined). Multiple colors reflect multiple detected viruses or etiologies. (B) Barplots of the contribution of each trinucleotide substitution for the four deconvoluted signatures with the likely mutation process indicated. (C) Proportion of each deconvoluted signature that contributes to each sample with virus status indicated by colored circles (red, BKPyV; green, TTV; blue, HR-HPV; goldenrod, JCPyV). (D) Number of unique and common trunk mutations in primary-metastatic tumor pairs and tumors with multi-region sequencing. For TBC03, TBC09, and TBC28, branches one and two refer to two separate areas of the same tumor. For TBC06 and TBC34, branches P and M refer to the primary tumor and metastasis, respectively. (E) Oncoprint for the top mutated genes in bladder cancers of transplant patients. Tumors IDs are colored by likely etiology: BKPyV-positive, red; JCPyV-positive, goldenrod; HR-HPV-positive, blue; TTV-positive, green; aristolochic acid, purple; undetermined, black. The percent of modified tumors is shown on the left and the count of the variants in each gene is represented by the barplot on the right.
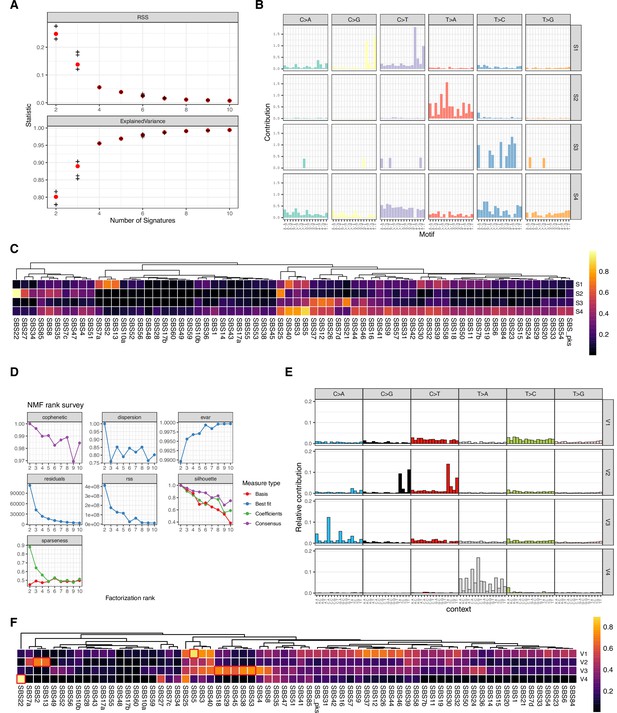
Mutations signature deconvolution.
(A) Residual sum of squares and explained variance for 2–10 signatures deconvoluted by SomaticSignatures. (B) Barplot of base substitution contributions to each of the four deconvoluted signatures from SomaticSignatures. (C) Heatmap of cosine similarities of four signatures deconvoluted by Somatic Signatures versus known Single Base Substitution Signatures (SBS). (D) NMF rank survey results for 2–10 signature deconvolution by MutationalPatterns. (E) Barplot of base substitution contributions to each of the four deconvoluted signatures from MutationalPatterns. (F) Heatmap of cosine similarities of four signatures deconvoluted by MutationalPatterns versus known SBS with closest matches highlighted in red.
Tables
Characteristics of post-transplant bladder cancer cases (N=43).
| Characteristic | Statistic | |
|---|---|---|
| Median | IQR | |
| Age in years at diagnosis | 65 | 60, 71 |
| Years from transplant to diagnosis | 5.8 | 3, 7 |
| N | % | |
| Sex | ||
| Female | 13 | 30 |
| Male | 30 | 70 |
| Transplanted organ | ||
| Kidney | 24 | 56 |
| Liver | 4 | 9 |
| Heart and/or lung | 14 | 33 |
| Pancreas | 1 | 2 |
| Race | ||
| Non-Hispanic White | 30 | 70 |
| Asian/Pacific Islander | 8 | 19 |
| Hispanic | 5 | 12 |
| Summary stage | ||
| In situ | 12 | 28 |
| Localized | 19 | 46 |
| Regional | 7 | 14 |
| Distant | 5 | 12 |
| Grade | ||
| Low | 20 | 47 |
| High | 22 | 51 |
| Papillary urothelial neoplasm of low malignant potential | 1 | 2 |
-
IQR: interquartile range.
BK polyomavirus (BKPyV) integrations sites and microhomology.
| ID | Human sequence match | Virus sequence match | Maximum MH length | MH sequence | Chromosome | Position | Nearest gene (Symbol) | Nearest gene (Ensembl ID) | Distance to Nearest Gene | Nearest RE | Distance to nearest RE |
|---|---|---|---|---|---|---|---|---|---|---|---|
| TBC02 | CATCATGATGATGGG | GATGGGCAGCCTA | 5 | ATGGG | chr2 | 120378301 | INHBB | ENSG00000163083 | –26499 | MIRb | –45 |
| TBC02 | CTCCTGCTCATGAA | CATGAAGGT TAAGCATGCTA | 5 | ATGAA | chr4 | 145732354 | C4orf51 | ENSG00000237136 | 0 | AluSq2 | –474 |
| TBC02 | ACCATTTAATTCCCAA | AGTGGAAATTAC | 2 | AC | chr4 | 145732375 | C4orf51 | ENSG00000237136 | 0 | AluSq2 | –495 |
| TBC03.1 | GCCTTTCTTG TGGACTGGGT | ATTTTCATTTCT ACTGGGGTCAGGA | 0 | No overlap | chr1 | 93693546 | BCAR3 | ENSG00000137936 | 0 | MIRb | 377 |
| TBC03.1 | TCTGTTTCT TATTTCAGAA | GGGTTCTCCTG TTTATAAGGTC | 2 | TC | chr1 | 93693570 | BCAR3 | ENSG00000137936 | 0 | MIRb | 353 |
| TBC03.1 | AGAGCCTTG GTGGTGG | GGTGGCAAA CAGTGCAG | 5 | GGTGG | chr1 | 93693890 | BCAR3 | ENSG00000137936 | 0 | MIRb | 33 |
| TBC03.1 | GATACTTTTT AGACATGC | AACCATGACC TCAGGAAGGA | 4 | CATG | chr1 | 93694075 | BCAR3 | ENSG00000137936 | 0 | MIRb | 0 |
| TBC03.1 | CCTCAAAGC CACCCACTCC | TTTCCATGA GCCCCAAA | 5 | CCAAA | chr1 | 93694843 | BCAR3 | ENSG00000137936 | 0 | MER5A | –92 |
| TBC03.1 | CAATTTTTTTTTTTT | TTTTTTTATT TGTAAGGGTG | 7 | TTTTTTT | chr12 | 50449935 | LARP4 | ENSG00000161813 | 0 | AluSc | 0 |
| TBC03.1 | TGCAAGGTG CTTCATGTAT | AGGGGGCTTA AAGGATGCA | 4 | TGCA | chr14 | 95764390 | ENSG00000257275 | –6735 | MIRb | 0 | |
| TBC03.1 | TAGCCAAAA AAAAAAAGG | AAAAAAAAA GGCCACAG | 11 | AAAAAAAAAGG | chr20 | 8525269 | PLCB1 | ENSG00000182621 | 0 | MamSINE1 | 154 |
| TBC03.1 | CAATTTGGA AAACAAT | ATGCAAGGG CAGTGCACA | 2 | AT | chr3 | 73059264 | PPP4R2 | ENSG00000163605 | 0 | MER103C | 69 |
| TBC03.1 | TAAAAAGTGTCA | AAGTGTCAA TAGAGAAAAA | 8 | AAGTGTCA | chr4 | 142307350 | INPP4B | ENSG00000109452 | 0 | L2a | 0 |
| TBC03.1 | TCACACAAT TT-TACTCCTCT | ACACTTTTTAC ACTCCTCTA | 8 | ACTCCTCT | chr8 | 140923993 | PTK2 | ENSG00000169398 | 0 | L2a | 0 |
| TBC03.2 | GTTGAGTT GGAGCA | CATCTAAATAA TCTCTCAAACT | 2 | CA | chr1 | 93693160 | BCAR3 | ENSG00000137936 | 0 | MER5A1 | –10 |
| TBC03.2 | ACCCAGTCCA CAAGAAAGGC | CCAGTAGAA ATGAAAAT | 0 | No overlap | chr1 | 93693546 | BCAR3 | ENSG00000137936 | 0 | MIRb | 377 |
| TBC03.2 | TCTGTTTCT TATTTCAG | GTTCTCCTGT TTATAAGGTC | 2 | TC | chr1 | 93693570 | BCAR3 | ENSG00000137936 | 0 | MIRb | 353 |
| TBC04 | GAGTGAGT TCATAG | CAACACTGTG GTGAG-TGAGTT | 4 | GAGT | chr3 | 5202593 | EDEM1 | ENSG00000134109 | 0 | L2b | –466 |
| TBC06 | CAGACATT -AGGA | TGAGGACC TAACCTGT | 4 | AGGA | chr2 | 201676427 | MPP4 | ENSG00000082126 | 0 | MIR1_Amn | 0 |
| TBC08 | TCCACTTT CAGTACTT | TGCAAAA AATCAAAT | 1 | T | chr6 | 148535326 | SASH1 | ENSG00000111961 | 0 | AluSq | 995 |
| TBC09.1 | GGGGCGG TAACTAGAAG | ACTAGAAG CTTGTCGT | 8 | ACTAGAAG | chr17 | 61340185 | BCAS3 | ENSG00000141376 | 0 | L2-3_Crp | 0 |
| TBC09N | GAGAAAAT AGGACTCGG | AAGATTCGC CTGAGAAAA | 7 | GAGAAAA | chr18 | 8169205 | PTPRM | ENSG00000173482 | 0 | MER127 | –648 |
| TBC09N | TCCATCC TCCTCTAC | CTCCTCT ACATTGT | 9 | CTCCTCTAC | chr3 | 34028749 | LINC01811 | ENSG00000226320 | 130585 | L2b | 0 |
| TBC09N | ATGTAAT ATAAAACT | CATGATT TTAACCCAG | 0 | No overlap | chr3 | 117678477 | ENSG00000239268 | 0 | L2c | 0 |
-
MH: microhomology; RE: Repeat element.
Additional files
-
Supplementary file 1
Supporting information.
(a) Checkerboard table of samples used in this study. (b) Sequencing metrics. (c) Reference sequences used in this study. (d) Tumor torque teno virus similarities. (e) BKPyV-positive tumor vs virus-free tumor significantly differentially expressed genes. (f) Non-synonymous point mutations. (g) Copy number variants.
- https://cdn.elifesciences.org/articles/82690/elife-82690-supp1-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/82690/elife-82690-mdarchecklist1-v2.docx





