Network-based multi-omics integration reveals metabolic at-risk profile within treated HIV-infection
Figures
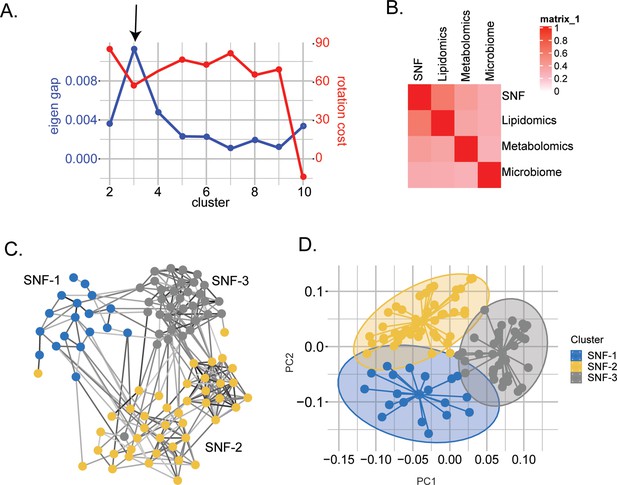
Similarity network fusion-based PWH stratification using lipidomics, metabolomics, and microbiome integration.
(A) Scatter plot showing the maximization of Eigen gap and the minimization of rotation cost for optimizing the number of clusters. (B) Concordance matrix between the combined network (SNF) and each omics network based on NMI calculation (0=no mutual information, 1=perfect correlation). (C) SNF-combined similarity network colored by clusters (SNF-1/HC-like=blue, SNF-2/severe at-risk=yellow, SNF-3/mild at-risk=grey) obtained after spectral clustering. Edges' color indicates the strength of the similarity (black = strong, grey = weak). (D) PCA plot of samples based on fused network. Samples are colored by condition.
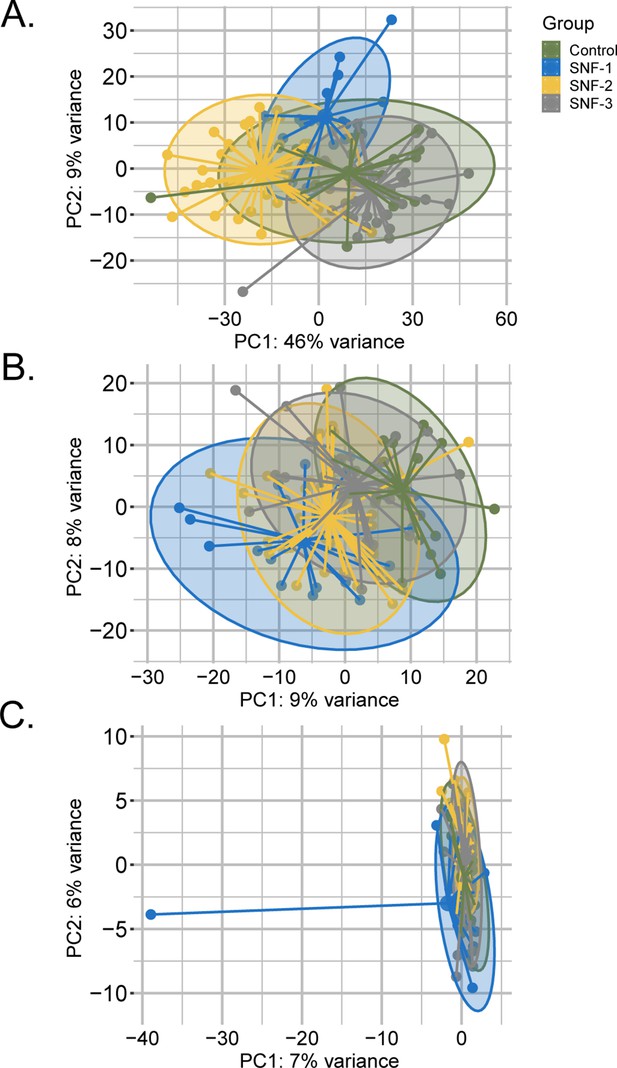
PCA plot of samples after prior standardization based on (a) Lipidomics (b) Metabolomics (c) Microbiome.
Variance proportions are written on each component axis. Samples are colored by condition (Ctrl = green, SNF-1=blue, SNF-2=yellow, SNF-3=grey).
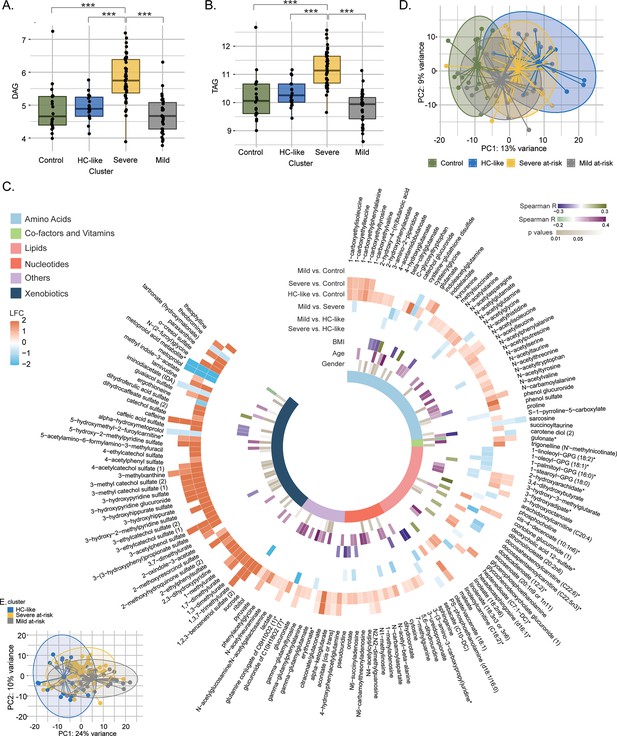
Lipidomics and metabolomics, characterization of the PWH clusters.
(A) Boxplots of DAG from untargeted lipid classes separated by groups. Significant stars are displayed for each comparison with *FDR <0.05, **FDR <0.01, ***FDR <0.001 (limma). (B) Boxplots of TAG from untargeted lipid classes separated by groups. (C) PCA plot of samples after prior standardization based on significant metabolites between at least one pairwise comparison (limma, FDR <0.05). Variance proportions are written on each component axis. Samples are colored by condition. (D) Circular heatmap of the top 159 metabolites (FDR <0.005). Metabolites are represented as slices and labeled around the plot. LogFold Change from significant metabolites between groups is displayed in the first six outer layers. The 7th to 9th layers represents the coefficient of correlation between metabolites and BMI, metabolites and age (Spearman, p <0.1, absolute R>0.15) and the p-value from significant associations between metabolites and gender (χ2, p<0.1). The inner layer represents the pathway of each metabolite. (E) PCA plot based on metabolites differing clusters adjusted for transmission mode and CD4 count.
-
Figure 2—source data 1
Table of differential lipid abundance analysis SNFs by lipids classes by clusters and corrected for transmission mode and CD4 count.
- https://cdn.elifesciences.org/articles/82785/elife-82785-fig2-data1-v3.xlsx
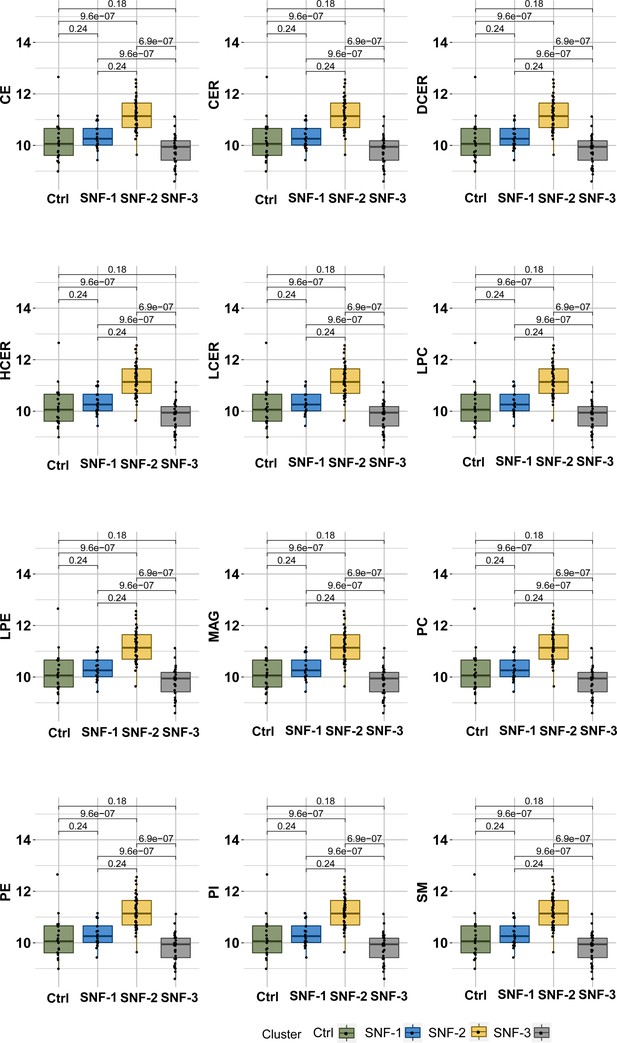
Boxplots of untargeted lipid classes separated by groups.
Color is based on groups (Ctrl = green, SNF-1=blue, SNF-2=yellow, SNF-3=grey). p Values are displayed for each comparison (Mann Withney U Test).
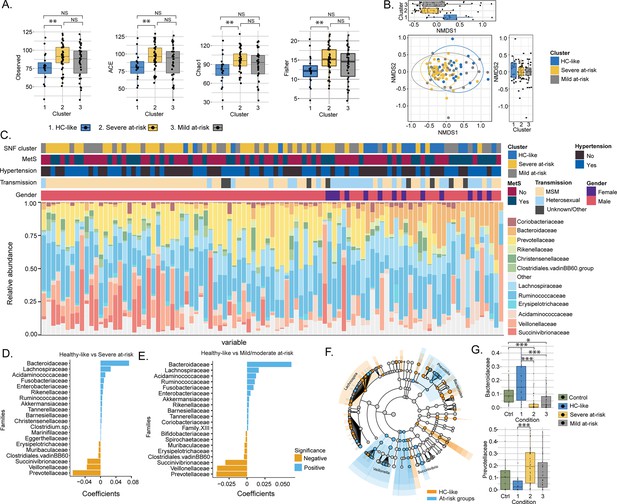
Transmission mode drove cluster differences in microbiome data.
(A) Boxplots of alpha diversity indices (Observed, ACE, Chao1, Fisher) separated by HIV cluster. Significant stars are shown for each comparison (Mann-Whitney U test). (B) Non-metric multidimensional scaling (NMDS) plot of Bray-Curtis distances. Samples are colored by clusters. Boxplots based on NMDS1 and NMDS2 are represented. (C) Barplot represents the relative abundance of bacteria at the family level for each patient. Patient information is displayed above the barplot, including cluster, metabolic syndrome (MetS: yes/no), hypertension (yes/no), transmission mode, and gender. (D) Barplot showing the top microbial families by representing their coefficient from PERMANOVA between SNF-1 and SNF-2. (E) Barplot showing the top microbial families between SNF-1 and SNF-3. (F) LEfSe cladogram representing cluster-specific microbial communities to HC-like and to at-risk groups (SNF-2/SNF-3). Top families from PERMANOVA are labeled. (G) Boxplot of relative abundance at family level for Bacteroides (top) and Prevotella (bottom). Significant stars are shown for significant comparisons (Mann-Whitney U test).
-
Figure 3—source data 1
Alpha diversity indices statistics.
- https://cdn.elifesciences.org/articles/82785/elife-82785-fig3-data1-v3.xlsx
-
Figure 3—source data 2
Permutational multivariate analysis of variance at the family level.
- https://cdn.elifesciences.org/articles/82785/elife-82785-fig3-data2-v3.xlsx
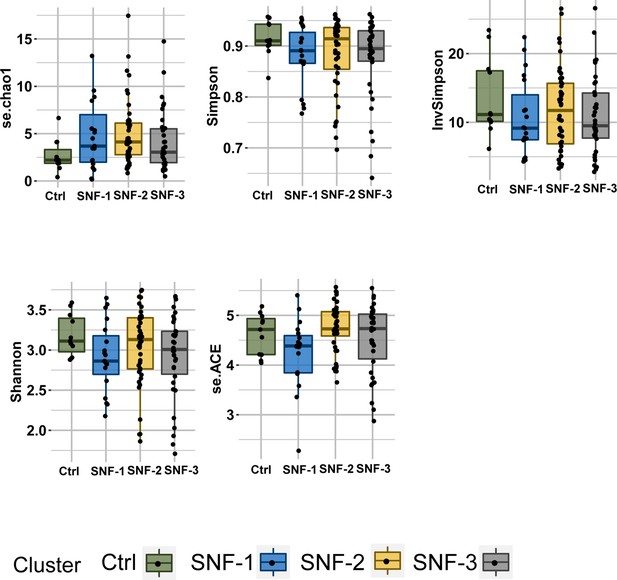
Boxplots of alpha diversity indices (se.chao1,Simpson, Shannon, se.ACE, InvSimpson) separated by HIV-cluster.
Color is based on groups (Ctrl = green, SNF-1=blue, SNF-2=yellow, SNF-3=grey).
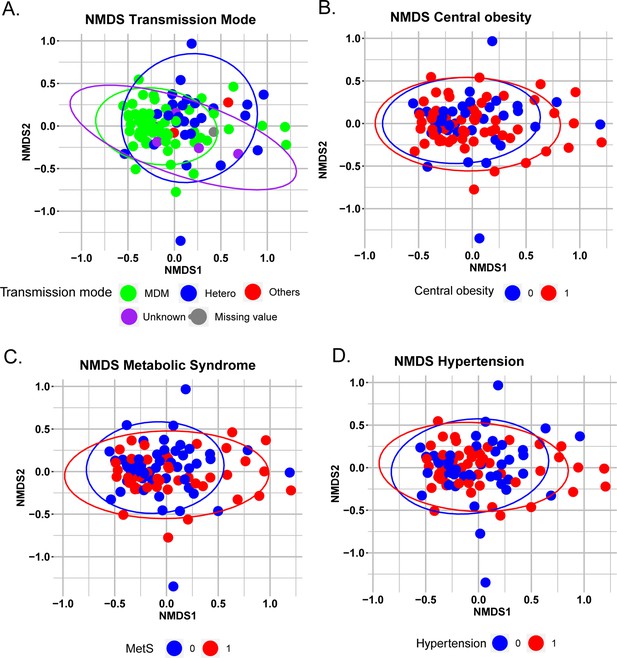
Non-metric multidimensional scaling (NMDS) plot of Bray-Curtis distances.
Samples are colored by (A) Transmission mode (B) Central obesity (C) Metabolic Syndrome (D) Hypertension.
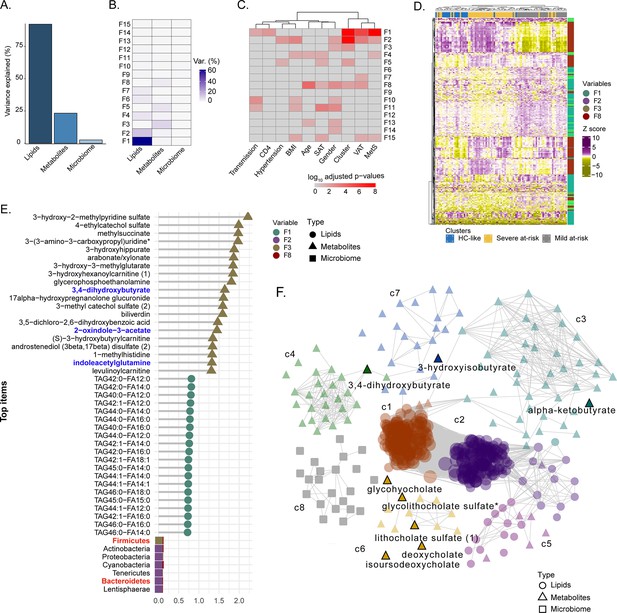
Factor analysis highlights the essential features for cluster separation and potential microbiome-derived metabolites importance (A) Barplot of total variance explained by MOFA model per view.
(B) Variance decomposition plot. The percentage of variance is explained by each factor for each view. (C) External covariate association with factors plot. Association is represented with log10 adjusted p-values from Pearson correlation. (D) Heatmap representing levels of microbial communities, metabolites, and lipids with the higher absolute weight in MOFA factors associated with cluster (F1, F2, F3, F5, F8). Samples are labeled according to the study groups. Data were Z-score transformed. The type of data (lipid, metabolite, microbe) is displayed on the right. (E) Top 20 features with higher absolute weight in MOFA factors associated with cluster (F1, F2, F3, F5, F8) from lipidome, metabolome, and microbiome. Microbiome-derived metabolites and bacterial phylum of interest are colored in blue and red, respectively. (F) MOFA features differing clusters and interactions extracted from the three-layers consensus co-expression network. Microbiome-derived metabolites are labeled.
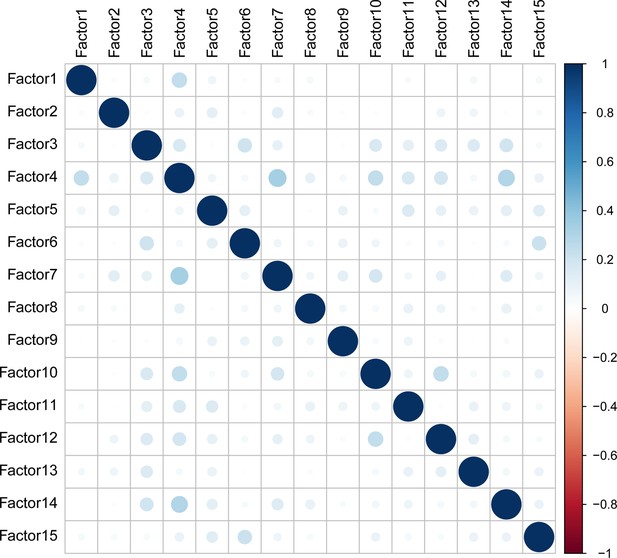
Correlation matrix of MOFA factors.
Size and transparency are proportional to the absolute coefficient of correlation. Color is displayed as a gradient-based coefficient of correlation from –1 (red) to 1 (blue).
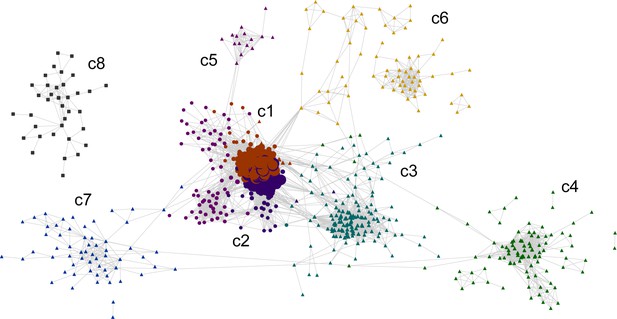
Cytoscape consensus co-expression network.
Color and label are based on communities.
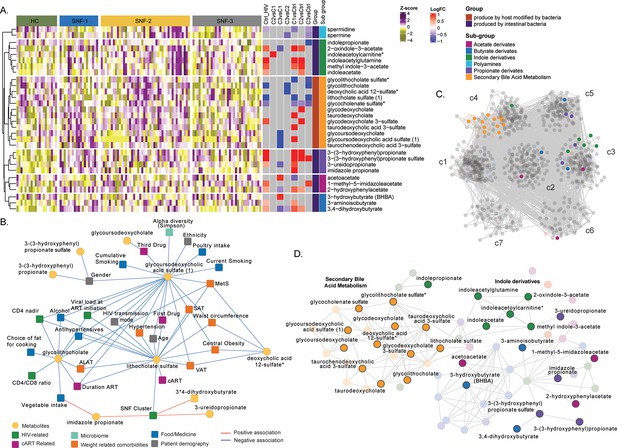
Microbiome-associated metabolites are affected in HIV clusters (A) Heatmap representing abundances of microbiome-derived metabolites differing in at least one comparison.
Data were Z-score transformed. Significant logFC (limma, FDR <0.05) of pairwise comparisons between conditions, groups, and under groups of microbiome-derived metabolites are displayed on the right. (B) Cytoscape network showing significant positive and negative associations between clinical parameters and microbiome-derived metabolites (univariate linear regression, FDR <0.05). Clinical parameters are colored based on categories. (C) Co-expression network of metabolomics data in PWH. Metabolites are grouped by communities, and microbiome-derived metabolites are labeled and colored based on the subgroup. (D) The subset of microbiome-derived metabolites from the co-expression network. Non-significant metabolites in all comparisons are displayed with transparency. Significant microbiome-derived metabolites between at least two conditions are labeled.
-
Figure 5—source data 1
Univariate linear regression between clinical parameters and microbiome-derived metabolites differing groups.
- https://cdn.elifesciences.org/articles/82785/elife-82785-fig5-data1-v3.xlsx
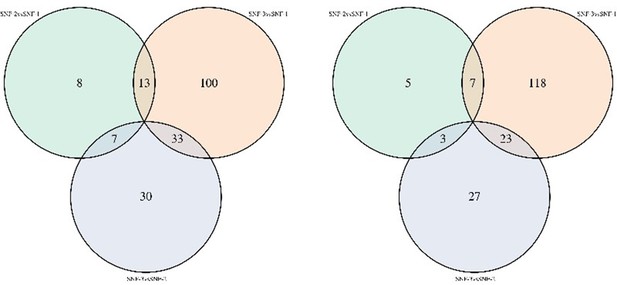
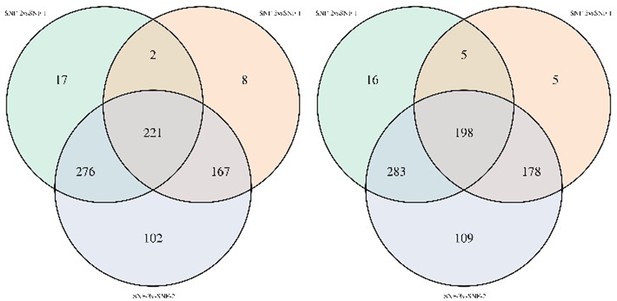
Venn diagram of metabolites significantly different between clusters in noncorrected limma model (left) and model corrected for transmission mode and CD4 count (right).
Tables
Patient characteristics.
| Complete Cohort | SNF-1 | SNF- 2 | SNF-3 | P values | |
|---|---|---|---|---|---|
| At-risk Classification | HC-like | Severe at risk | Mild | ||
| N | 97 | 19 | 44 | 34 | |
| Age in years, Median (IQR) | 54 (48–63) | 60 (48–68) | 54 (48–62) | 54 (51–60) | 0.75 |
| Gender, Male, N (%) | 84 (87) | 15 (79) | 40 (91) | 29 (85) | 0.36 |
| Ethnicity Caucasian, N (%) | 79 (81) | 15 (79) | 38 (87) | 26 (77) | 0.49 |
| Mode of transmission, N (%) Homosexual/bisexual Heterosexual Other/unknown | 63 (65) 26 (27) 8 (8) | 9 (47) 7 (37) 3 (16) | 36 (81) 6 (14) 2 (5) | 18 (53) 13 (38) 3 (9) | 0.017 |
| CD4 Nadir, cells/mL, Median (IQR) | 235 (123–320) | 240 (127–330) | 240 (145–365) | 223 (42–290) | 0.49 |
| CD4 at ART Initiation, cells/mL, Median (IQR) | 287 (155–410) | 270 (120–360) | 318 (192–463) | 240 (108–320) | 0.11 |
| Viral Load at ART initiation, log copies/mL, Median (IQR) | 5.02 (4.34–5.61) | 4.87 (4.32–5.5) | 5.11 (4.74–5.61) | 4.94 (4.2–5.55) | 0.35 |
| CD4 at sampling, cells/mL, Median(IQR) | 713 (570–900) | 680 (540–958) | 762 (689–923) | 610 (475–819) | 0.015 |
| CD8 at sampling, cells/mL, Median (IQR) | 775 (600–1100) | 780 (630–879) | 894 (638–1300) | 700 (530–870) | 0.054 |
| Viral load (<50 copies/mL), N (%) | 97 (100) | 19 (100) | 44 (100) | 34 (100) | 1 |
| Duration of treatment in years, median (IQR) | 15 (9–18) | 15 (13–18) | 15 (8–18) | 14 (7–17) | 0.73 |
| Current Treatment, 1st drug, N (%) ABC TDF/TAF Other | 31 (32) 42 (43) 24 (25) | 8 (42) 8 (42) 3 (16) | 13 (30) 19 (43) 12 (27) | 10 (29) 15 (44) 9 (27) | 0.84 |
| Current Treatment, 3rd drug, N (%) NNRTI PI/r INSTI Other | 38 (39) 18 (19) 15 (15) 26 (27) | 8 (42) 4 (21) 4 (21) 3 (16) | 14 (32) 11 (25) 6 (14) 13 (29) | 16 (47) 3 (9) 5 (15) 10 (29) | 0.45 |
| BMI, Mean (SD) | 24 (22–27) | 22 (19–25) | 26 (23–28) | 24 (22–27) | 0.003 |
| VAT, Median (IQR) | 89 (36–142) | 41 (19–106) | 127 (79–196) | 69 (26–100) | 0.0001 |
| SAT, Median (IQR) | 111 (70–167) | 69 (33–115) | 117 (82–174) | 119 (83–190) | 0.02 |
| MetS, N (%) | 43 (44) | 6 (32) | 31 (70) | 6 (17) | 0.000009 |
| Central obesity, N(%) | 57 (59) | 8 (42) | 32 (73) | 17 (50) | 0.033 |
| Waist circumference (cm) | 94 (87–101) | 90 (84–95) | 100 (91–105) | 90 (87–97) | 0.0007 |
| Hypertension, N (%) | 49 (51) | 5 (26) | 23 (52) | 21 (62) | 0.04 |
-
Table 1—source data 1
Comparative characteristics of the HC and PWH.
- https://cdn.elifesciences.org/articles/82785/elife-82785-table1-data1-v3.xlsx
-
Table 1—source data 2
Non-significant characteristics between the cohorts.
- https://cdn.elifesciences.org/articles/82785/elife-82785-table1-data2-v3.xlsx
List of parameters used in the study.
| Description | Type | Definition |
|---|---|---|
| Ethnicity defined as in the Danish HIV cohort | logical | 1: caucasian, 2: asian, 3: black, 4: other 5: inuit |
| Origin defined as in the Danish HIV cohort | logical | 1: Denmark, 2: Other Scandinavian country 3: Other European country, 4: Turkia, 5: Pakistan, India and Sri Lanka, 6: Arabian countries / Iran, 7: Other (Africa, Asia, Greenland) |
| Mode of HIV transmission | logical | 1.Homo/biseksuel, 2. IV drug use, 3.homoseksuel+misbrug4.hæmofili 5.blodtransfusion 6.heteroseksuel, 7. andet 8. unknown 9.perinatal |
| Gender | logical | 0: female, 1:male |
| Computed age | numeric | Unit: years |
| Body mass index | numeric | Unit: kg/m2 |
| Physical activity in spare time | logical | 1: Inactive 2: Slightly active 3: Moderately active 4: Very active |
| Meat intake (Beef) | numeric | Times per week in average |
| Meat intake (Poultry) | numeric | Times per week in average |
| Choice of fat for preparing warm dishes | logical | 0: None, 1: Butter, 2: Spreadable (Kærgården), 3: Shortening (Stegemargarine), 4: Vegetable margarine (Plantemargarine), 5: Olive oil, 6: Other oil, 7: Other |
| Vegetable intake | logical | 0: Never 1: 1–3/month, 2: 1–2/week, 3: 3–4/week, 4: 5–6/week, 5: 1/day, 6: 2–3/day, 7:>3 /day |
| Fruit intake (whole fruit/portion of fruit) | logical | 0: Never, 1: 1–3/month, 2: 1–2/week, 3: 3–4/week, 4: 5–6/week, 5: 1/day, 6: 2–3/day, 7:>3 /day |
| *Weekly alcohol consumption | numeric | Unit:gram |
| Cumulative smoking | numeric | Unit: Pack years in current and previous smokers |
| Current Smoking | logical | 0: No, 1: Yes |
| Current CD4 count (closest to date of inclusion) | numeric | cell / ul |
| Current CD8 count | numeric | cell / ul |
| Ratio CD4/CD8 | numeric | x |
| CD4_nadir | numeric | cell / ul |
| CD4 at ART initiation | numeric | cell / ul |
| Current viral load | numeric | copies / ul |
| VL at ART initiation | numeric | copies / ul |
| Log10 of VL at ART initiation | numeric | x |
| SNF clusters | logical | 1:SNF-1, 2: SNF-2, 3: SNF-3 |
| Duration of current cART | numeric | Unit: months |
| Duration of current cART | numeric | Unit: years |
| Duration of previous cART | numeric | Unit: months |
| 3rd Drug | logical | NNRTI, PI/r,INSTI, Other/unknow |
| 1st drug | logical | ABC, TDF/TAF, Other/unknown |
| Subcutaneous adipose tissue (SAT) | numeric | squared centimeters |
| Visceral adipose tissue (VAT) | numeric | squared centimeters |
| Waist circumference | numeric | Unit: cm |
| Systolic blood pressure, right arm | numeric | Unit: mm mercury |
| Diastolic blood pressure, right arm | numeric | Unit: mm mercury |
| Metabolic syndrome (MetS)* | logical | 0: No, 1: Yes |
| Central obesity | logical | 0: No,1: Yes |
| hypertension (*JNC7 definition) | logical | 0: No, 1: Yes |
| Diabetes | logical | 0: No, 1: Yes |
| Estimated glomerular function rate (eGFR) | numeric | Unit: reads mL/min/1.73 m2 |
| Alanine aminotransferase (ALAT) | numeric | Unit:Units per liter |
| Antihypertensives | logical | 0: No, 1: Yes |
List of microbiome-derived metabolites.
| BIOCHEMICAL | SUPER.PATHWAY | SUB.PATHWAY | group | under_group |
|---|---|---|---|---|
| butyrate/isobutyrate (4:0) | Lipid | Short Chain Fatty Acid | produced_by_intestinal_bacteria | Short Chain Fatty Acid |
| N-acetylputrescine | Amino Acid | Polyamine Metabolism | produced_by_intestinal_bacteria | Polyamines |
| spermidine | Amino Acid | Polyamine Metabolism | produced_by_intestinal_bacteria | Polyamines |
| spermine | Amino Acid | Polyamine Metabolism | produced_by_intestinal_bacteria | Polyamines |
| 1-methyl-4-imidazoleacetate | Amino Acid | Histidine Metabolism | produced_by_intestinal_bacteria | Acetate derivates |
| 1-methyl-5-imidazoleacetate | Amino Acid | Histidine Metabolism | produced_by_intestinal_bacteria | Acetate derivates |
| 1-ribosyl-imidazoleacetate* | Amino Acid | Histidine Metabolism | produced_by_intestinal_bacteria | Acetate derivates |
| 1H-indole-7-acetic acid | Xenobiotics | Bacterial/Fungal | produced_by_intestinal_bacteria | Indole derivatives |
| 2,3-dihydroxy-2-methylbutyrate | Amino Acid | Leucine, Isoleucine and Valine Metabolism | produced_by_intestinal_bacteria | Butyrate derivates |
| 2-aminobutyrate | Amino Acid | Glutathione Metabolism | produced_by_intestinal_bacteria | Butyrate derivates |
| 2-hydroxybutyrate/2-hydroxyisobutyrate | Amino Acid | Glutathione Metabolism | produced_by_intestinal_bacteria | Butyrate derivates |
| 2-hydroxyphenylacetate | Amino Acid | Phenylalanine Metabolism | produced_by_intestinal_bacteria | Acetate derivates |
| 2-oxindole-3-acetate | Xenobiotics | Food Component/Plant | produced_by_intestinal_bacteria | Indole derivatives |
| 2 R,3R-dihydroxybutyrate | Lipid | Fatty Acid, Dihydroxy | produced_by_intestinal_bacteria | Butyrate derivates |
| 2 S,3R-dihydroxybutyrate | Lipid | Fatty Acid, Dihydroxy | produced_by_intestinal_bacteria | Butyrate derivates |
| 3,4-dihydroxybutyrate | Lipid | Fatty Acid, Dihydroxy | produced_by_intestinal_bacteria | Butyrate derivates |
| 3-(3-hydroxyphenyl)propionate | Xenobiotics | Benzoate Metabolism | produced_by_intestinal_bacteria | Propionate derivates |
| 3-(3-hydroxyphenyl)propionate sulfate | Xenobiotics | Benzoate Metabolism | produced_by_intestinal_bacteria | Propionate derivates |
| 3-aminoisobutyrate | Nucleotide | Pyrimidine Metabolism, Thymine containing | produced_by_intestinal_bacteria | Butyrate derivates |
| 3-carboxy-4-methyl-5-pentyl-2-furanpropionate (3-CMPFP) | Lipid | Fatty Acid, Dicarboxylate | produced_by_intestinal_bacteria | Propionate derivates |
| 3-formylindole | Xenobiotics | Food Component/Plant | produced_by_intestinal_bacteria | Indole derivatives |
| 3-hydroxy-2-ethylpropionate | Amino Acid | Leucine, Isoleucine and Valine Metabolism | produced_by_intestinal_bacteria | Propionate derivates |
| 3-hydroxybutyrate (BHBA) | Lipid | Ketone Bodies | produced_by_intestinal_bacteria | Butyrate derivates |
| 3-hydroxyisobutyrate | Amino Acid | Leucine, Isoleucine and Valine Metabolism | produced_by_intestinal_bacteria | Butyrate derivates |
| 3-indoleglyoxylic acid | Xenobiotics | Food Component/Plant | produced_by_intestinal_bacteria | Indole derivatives |
| 3-methyl-2-oxobutyrate | Amino Acid | Leucine, Isoleucine and Valine Metabolism | produced_by_intestinal_bacteria | Butyrate derivates |
| 3-phenylpropionate (hydrocinnamate) | Xenobiotics | Benzoate Metabolism | produced_by_intestinal_bacteria | Propionate derivates |
| 3-ureidopropionate | Nucleotide | Pyrimidine Metabolism, Uracil containing | produced_by_intestinal_bacteria | Propionate derivates |
| 4-imidazoleacetate | Amino Acid | Histidine Metabolism | produced_by_intestinal_bacteria | Acetate derivates |
| 6-hydroxyindole sulfate | Xenobiotics | Chemical | produced_by_intestinal_bacteria | Indole derivatives |
| 7-hydroxyindole sulfate | Amino Acid | Tryptophan Metabolism | produced_by_intestinal_bacteria | Indole derivatives |
| acetoacetate | Lipid | Ketone Bodies | produced_by_intestinal_bacteria | Acetate derivates |
| alpha-ketobutyrate | Amino Acid | Methionine, Cysteine, SAM and Taurine Metabolism | produced_by_intestinal_bacteria | Butyrate derivates |
| citalopram propionate* | Xenobiotics | Drug - Psychoactive | produced_by_intestinal_bacteria | Propionate derivates |
| gamma-glutamyl-2-aminobutyrate | Peptide | Gamma-glutamyl Amino Acid | produced_by_intestinal_bacteria | Butyrate derivates |
| guanidinoacetate | Amino Acid | Creatine Metabolism | produced_by_intestinal_bacteria | Acetate derivates |
| hydantoin-5-propionate | Amino Acid | Histidine Metabolism | produced_by_intestinal_bacteria | Propionate derivates |
| imidazole propionate | Amino Acid | Histidine Metabolism | produced_by_intestinal_bacteria | Propionate derivates |
| iminodiacetate (IDA) | Xenobiotics | Chemical | produced_by_intestinal_bacteria | Acetate derivates |
| indole-3-carboxylate | Amino Acid | Tryptophan Metabolism | produced_by_intestinal_bacteria | Indole derivatives |
| indoleacetate | Amino Acid | Tryptophan Metabolism | produced_by_intestinal_bacteria | Indole derivatives |
| indoleacetoylcarnitine* | Amino Acid | Tryptophan Metabolism | produced_by_intestinal_bacteria | Indole derivatives |
| indoleacetylglutamine | Amino Acid | Tryptophan Metabolism | produced_by_intestinal_bacteria | Indole derivatives |
| indolelactate | Amino Acid | Tryptophan Metabolism | produced_by_intestinal_bacteria | Indole derivatives |
| indolepropionate | Amino Acid | Tryptophan Metabolism | produced_by_intestinal_bacteria | Indole derivatives |
| methyl indole-3-acetate | Xenobiotics | Food Component/Plant | produced_by_intestinal_bacteria | Indole derivatives |
| phenylacetate | Amino Acid | Phenylalanine Metabolism | produced_by_intestinal_bacteria | Acetate derivates |
| taurine | Amino Acid | Methionine, Cysteine, SAM and Taurine Metabolism | produced_by_host_modified_by_bacteria | Taurine |
| deoxycholate | Lipid | Secondary Bile Acid Metabolism | produced_by_host_modified_by_bacteria | Secondary Bile Acid Metabolism |
| deoxycholic acid 12-sulfate* | Lipid | Secondary Bile Acid Metabolism | produced_by_host_modified_by_bacteria | Secondary Bile Acid Metabolism |
| deoxycholic acid glucuronide | Lipid | Secondary Bile Acid Metabolism | produced_by_host_modified_by_bacteria | Secondary Bile Acid Metabolism |
| glycocholenate sulfate* | Lipid | Secondary Bile Acid Metabolism | produced_by_host_modified_by_bacteria | Secondary Bile Acid Metabolism |
| glycodeoxycholate | Lipid | Secondary Bile Acid Metabolism | produced_by_host_modified_by_bacteria | Secondary Bile Acid Metabolism |
| glycodeoxycholate 3-sulfate | Lipid | Secondary Bile Acid Metabolism | produced_by_host_modified_by_bacteria | Secondary Bile Acid Metabolism |
| glycohyocholate | Lipid | Secondary Bile Acid Metabolism | produced_by_host_modified_by_bacteria | Secondary Bile Acid Metabolism |
| glycolithocholate | Lipid | Secondary Bile Acid Metabolism | produced_by_host_modified_by_bacteria | Secondary Bile Acid Metabolism |
| glycolithocholate sulfate* | Lipid | Secondary Bile Acid Metabolism | produced_by_host_modified_by_bacteria | Secondary Bile Acid Metabolism |
| glycoursodeoxycholate | Lipid | Secondary Bile Acid Metabolism | produced_by_host_modified_by_bacteria | Secondary Bile Acid Metabolism |
| glycoursodeoxycholic acid sulfate (1) | Lipid | Secondary Bile Acid Metabolism | produced_by_host_modified_by_bacteria | Secondary Bile Acid Metabolism |
| hyocholate | Lipid | Secondary Bile Acid Metabolism | produced_by_host_modified_by_bacteria | Secondary Bile Acid Metabolism |
| isoursodeoxycholate | Lipid | Secondary Bile Acid Metabolism | produced_by_host_modified_by_bacteria | Secondary Bile Acid Metabolism |
| lithocholate sulfate (1) | Lipid | Secondary Bile Acid Metabolism | produced_by_host_modified_by_bacteria | Secondary Bile Acid Metabolism |
| taurochenodeoxycholic acid 3-sulfate | Lipid | Secondary Bile Acid Metabolism | produced_by_host_modified_by_bacteria | Secondary Bile Acid Metabolism |
| taurocholenate sulfate* | Lipid | Secondary Bile Acid Metabolism | produced_by_host_modified_by_bacteria | Secondary Bile Acid Metabolism |
| taurodeoxycholate | Lipid | Secondary Bile Acid Metabolism | produced_by_host_modified_by_bacteria | Secondary Bile Acid Metabolism |
| taurodeoxycholic acid 3-sulfate | Lipid | Secondary Bile Acid Metabolism | produced_by_host_modified_by_bacteria | Secondary Bile Acid Metabolism |
| taurohyocholate* | Lipid | Secondary Bile Acid Metabolism | produced_by_host_modified_by_bacteria | Secondary Bile Acid Metabolism |
| taurolithocholate 3-sulfate | Lipid | Secondary Bile Acid Metabolism | produced_by_host_modified_by_bacteria | Secondary Bile Acid Metabolism |
| ursodeoxycholate | Lipid | Secondary Bile Acid Metabolism | produced_by_host_modified_by_bacteria | Secondary Bile Acid Metabolism |
Additional files
-
Supplementary file 1
Table of differential lipid abundance analysis on individual lipids abundances between clusters corrected for transmission mode and CD4 count.
- https://cdn.elifesciences.org/articles/82785/elife-82785-supp1-v3.xlsx
-
Supplementary file 2
False discovery rate after differential metabolite analysis between the groups.
- https://cdn.elifesciences.org/articles/82785/elife-82785-supp2-v3.xlsx
-
Supplementary file 3
Table of differential metabolite abundance analysis after adjustment of the CD4 count at sampling and route of transmission.
- https://cdn.elifesciences.org/articles/82785/elife-82785-supp3-v3.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/82785/elife-82785-mdarchecklist1-v3.docx