Dysregulation of mTOR signaling mediates common neurite and migration defects in both idiopathic and 16p11.2 deletion autism neural precursor cells
Figures
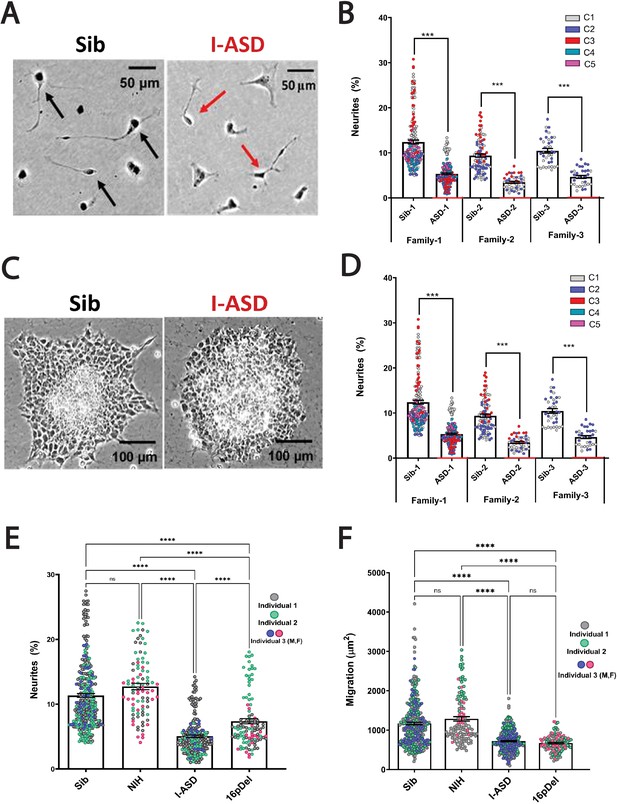
Reduced neurite outgrowth and cell migration in both I-ASD and 16pDel neural precursor cells (NPCs).
(A) Representative image: neurite outgrowth at 48 hr: Sib NPCs have more cells with neurites (black arrows) than I-ASD NPCs (red arrows). (B) Quantification of % neurites in three pairs of I-ASD and Sibs: I-ASD NPCs have fewer neurites (%) than Sib NPCs in all families. For neurite graphs, each dot represents the % of cells with neurites in a single dish. Colors represent different clones. (C) Representative image: Sib neurospheres have migrating carpet of cells (dark) that move further than that of I-ASD neurosphere. (D) Cell migration in three pairs of I-ASD and Sibs: I-ASD NPCs migrate less than Sib NPCs in all families. For migration graphs, each dot represents an individual neurosphere. Student’s t-test: all Sib vs I-ASD comparisons. (E) Neurite % in Sibs, NIH, I-ASD, and 16pDel ASD. Each dot represents data from a single dish with different colors denoting different individuals. Both 16pDel and I-ASD have reduced neurites compared to Sib and NIH NPCs. (F) NPC migration in Sibs, NIH, I-ASD, and 16pDel ASD. Both 16pDel and I-ASD NPCs have reduced migration compared to both Sib and NIH. One-way ANOVA for all 16pDel experiments. For all graphs: NS = p>0.05, p≤0.05 = *, p≤0.01 = **, p≤0.001 = ***, p≤0.0001 = ****. Error bars represent standard error of means (SEM). For detailed N values please see Supplementary file 1.
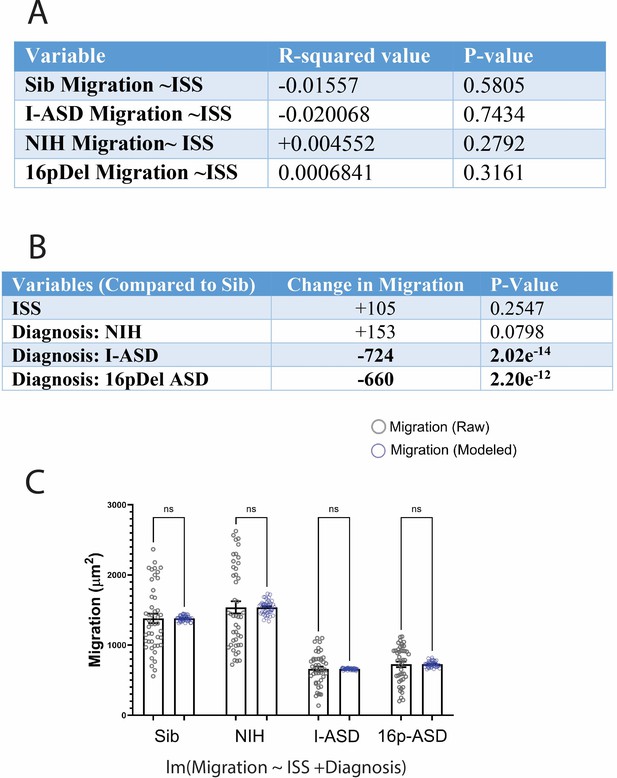
Evaluation of the relationship between initial sphere size (ISS) and neurosphere migration.
R studio used for all analyses and models presented. (A) Table showing the relationship between ISS and neurosphere migration for Sibs, I-ASD, NIH, and 16pDel neurospheres (Migration~ISS, separate linear regression model for each group). As shown in the table, R2 values for all groups are very low indicating minimal relationship between ISS and migration. Moreover, there are no statistically significant relationships between ISS and migration in any diagnosis group as indicated by all p>0.05. Table showing the relationship between neurosphere migration, ISS, and diagnosis using model lm(Migration~ISS+Diagnosis) allowing us to understand how ISS and ‘Diagnosis’ influence migration when compared to control conditions. Once again, ISS as a whole (regardless of diagnosis) has minimal impact on neurosphere migration (increase in 105 μm2 p=0.255). On the other hand, I-ASD and 16pDel ‘Diagnosis’ has a significant impact (both in terms of effect size and statistical significance) on neurosphere migration. (C) Graph depicting actual neurosphere migration data in Sib, NIH, I-ASD, 16pDel ASD plotted against predicted data generated by removing the effect of ISS on neurosphere migration using linear regression outputs in R studio. By applying Student’s t-test, we find that there is no significant difference between the actual neurosphere migration data and the predicted migration data in which the impacts of ISS were controlled for with statistical linear modeling. This indicates that ISS has minimal impact on migration and consideration of ISS in our data is not necessary to study neurosphere migration. For all graphs: p≤0.05 = *, p≤0.01 = **, p≤0.001 = ***, p≤0.0001 = ****. Error bars represent SEM. N=3 individuals, 2 clones, and 2 neural inductions for each subfigure.
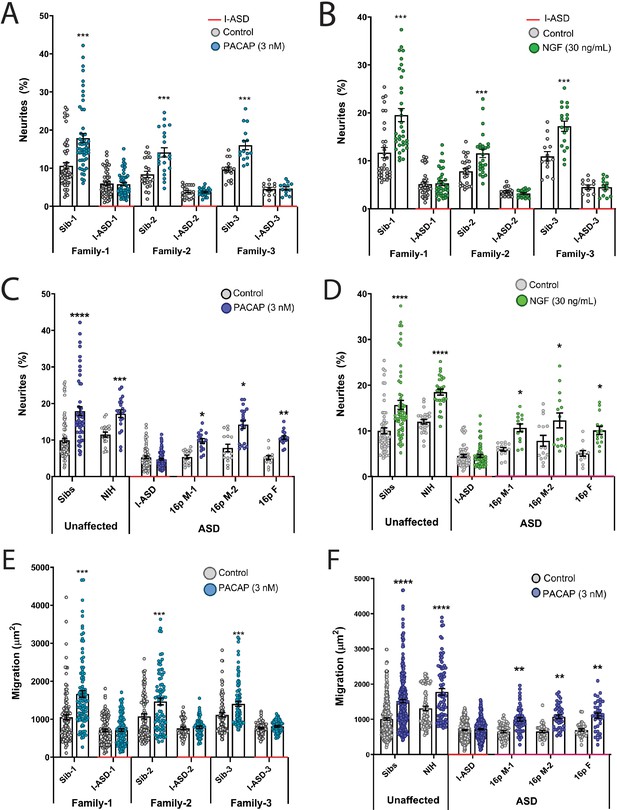
Autism spectrum disorder (ASD) subtype-specific extracellular factor (EF) responses in I-ASD and 16pDel neural precursor cells (NPCs).
(A and B) 3 nM PACAP (A) and 30 ng/mL nerve growth factor (NGF) (B) increases neurite outgrowth in all Sibs but fails to stimulate neurite outgrowth in all I-ASD NPCs. (C and D) 3 nM PACAP (C) and 30 ng/mL NGF (D) stimulate neurite outgrowth in Sibs, NIH, and all three 16pDel NPC but not in I-ASD NPCs. (E) 3 nM PACAP increases cell migration in all Sib NPCs but fails to stimulate migration in all I-ASD NPCs. (F) 3 nM PACAP stimulates migration in Sibs, NIH, and all three 16pDel patient NPCs but does not change migration in I-ASD NPCs. Two-way ANOVA for all comparisons, p<0.01 for all unaffected vs affected comparisons. For all graphs: p≤0.05 = *, p≤0.01 = **, p≤0.001 = ***, p≤0.0001 = ****. Error bars represent SEM. For detailed N values please see Supplementary file 1.
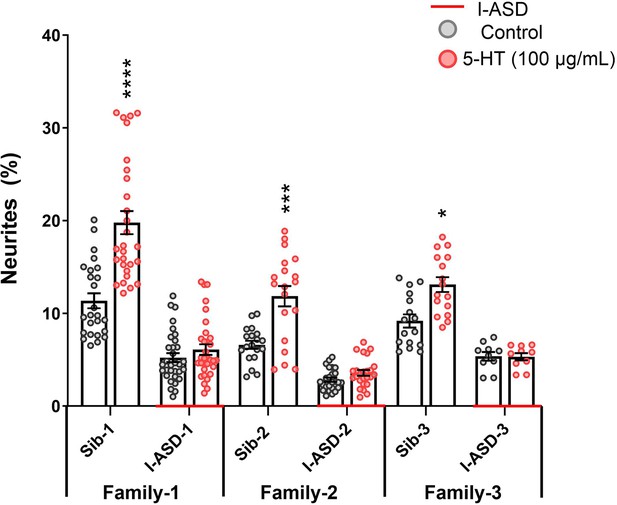
Effects of 5-HT on I-ASD and Sib neural precursor cells (NPCs).
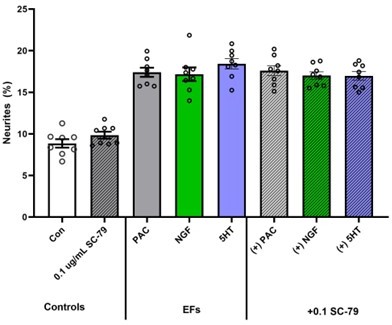
5-HT increases neurite outgrowth in Sib neural precursor cells (NPCs) but has no effect on I-ASD-affected NPCs (Sib vs ASD comparisons – Student’s t-test). Asterisk guide: p≤0.05 = *, p≤0.01 = **, p≤0.001 = ***, p≤0.0001 = ****. Error bars represent SEM. N Values: 2 clones per individual, 2 neural inductions per clone, 2-3 dishes per neural induction.
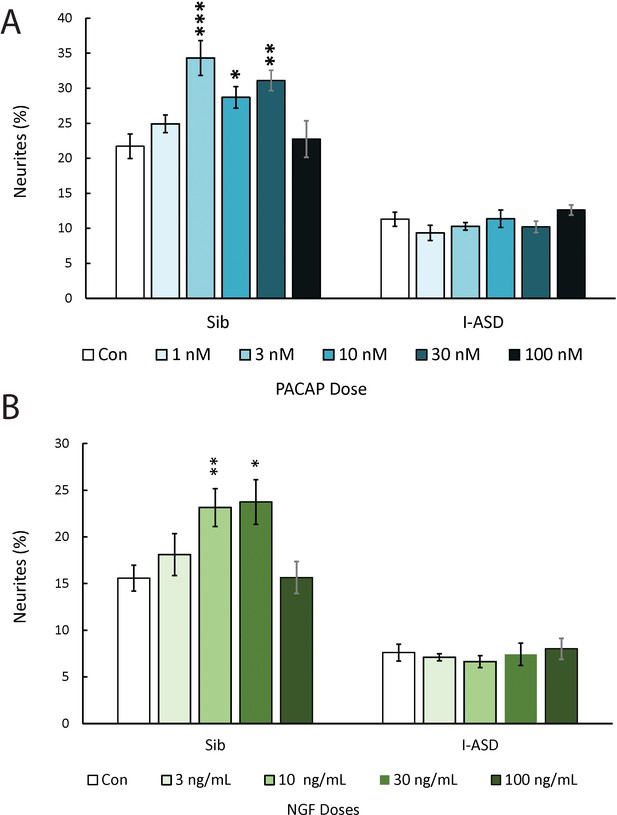
Dose-response graphs for extracellular factors (EFs).
(A) Dose-response experiments – PACAP: Sib is responsive to PACAP at numerous concentrations whereas I-ASD has no response at any concentration (one-way ANOVA for Sib vs I-ASD comparison). (B) Dose-response experiments – nerve growth factor (NGF): Sib is responsive to NGF at numerous concentrations whereas I-ASD has no response at any concentration (Sib vs I-ASD, one-way ANOVA). For all graphs: p≤0.05 = *, p≤0.01 = **, p≤0.001 = ***, p≤0.0001 = ****. Error bars represent SEM. N = 2 individuals, 1 clone per individual, 2 neural inductions per clone and 10 dishes per condition per experiment.
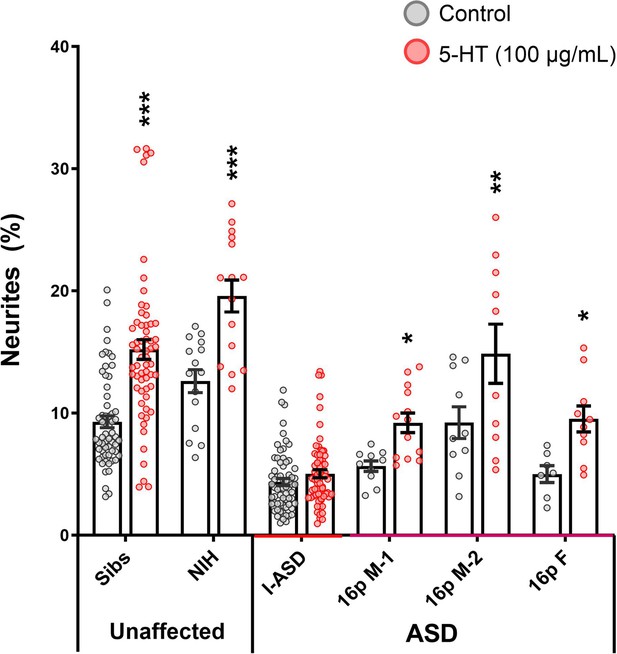
Effects of 5-HT on 16pDel neural precursor cells (NPCs).
5-HT stimulates neurite outgrowth in Sib, NIH, and all 16pDel neural precursor cells (NPCs) but not in I-ASD NPCs (two-way ANOVA). Asterisks: p≤0.05 = *, p≤0.01 = **, p≤0.001 = ***, p≤0.0001 = ****. Error bars represent SEM. N Values: 2 clones per individual (except NIH), 2 neural inductions per clone, 2-3 dishes per neural induction.
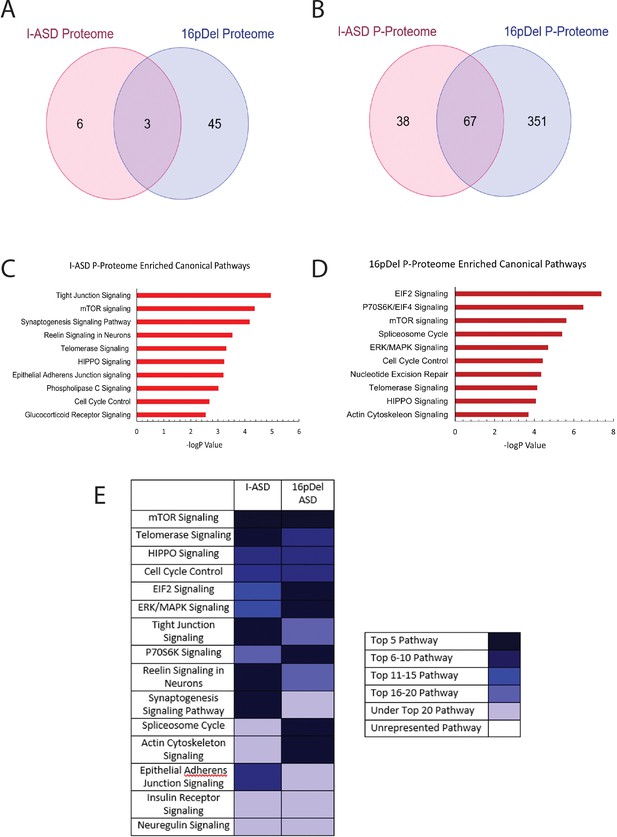
Phospho-proteome analysis of I-ASD and 16pDel data.
(A and B) Venn diagram illustrates proteomic (A) and p-proteomic (B) changes in both I-ASD and 16pDel NPCs and their overlap. (C and D) Ingenuity Pathway Analysis (IPA) canonical pathway analysis of I-ASD (C) and 16pDel (D) p-proteome data identifies the mTOR pathway as likely disrupted. (E) Heatmap of top overlapping canonical pathways between I-ASD and 16pDel p-proteome showing that the strongest overlap between I-ASD and 16pDel p-proteomes is the mTOR pathway. All p-proteome and proteome analyses were done with pooled proteins from all 3 I-ASD, all 3 Sib, and the 2 male 16pDel. Protein was extracted from at least 2 clones and 2 neural inductions per individual.
-
Figure 3—source data 1
IPA p-proteome canonical pathway proteins.
(A) Table showing the protein members in each enriched canonical pathway in I-ASD. (B) Table showing the protein members in each enriched canonical pathway in 16pDel ASD.
- https://cdn.elifesciences.org/articles/82809/elife-82809-fig3-data1-v2.docx
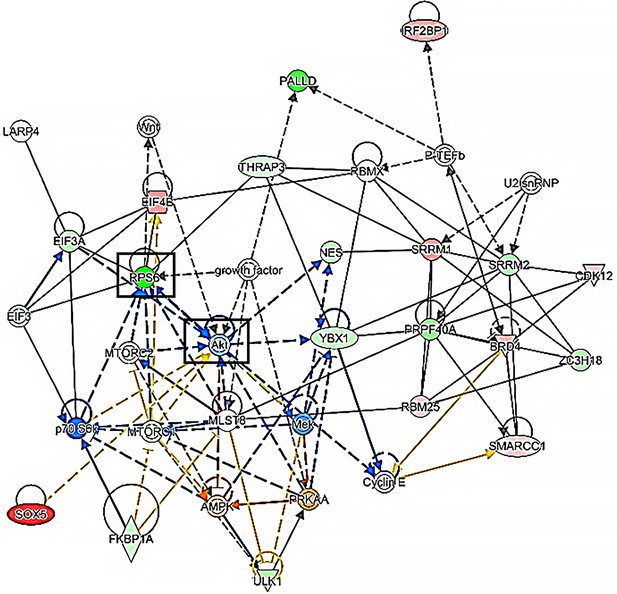
I-ASD p-proteome network analysis.
Network analysis of I-ASD p-proteome showing the most enriched/represented protein points of convergence. mTOR and associated pathway members (AKT, RPS6, EIFs, P70 S6K) are highly enriched particularly looking at the left side of the image.
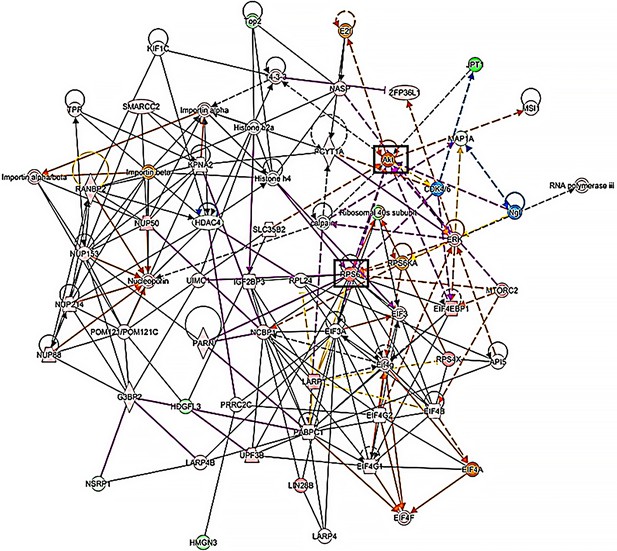
I6pDel p-proteome network analysis.
Network analysis of 16pDel p-proteome showing the most enriched/represented protein points of convergence. mTOR pathway members (RPS6, EIF, PS60K, AKT) are highly enriched, particularly looking at the right side of the network.
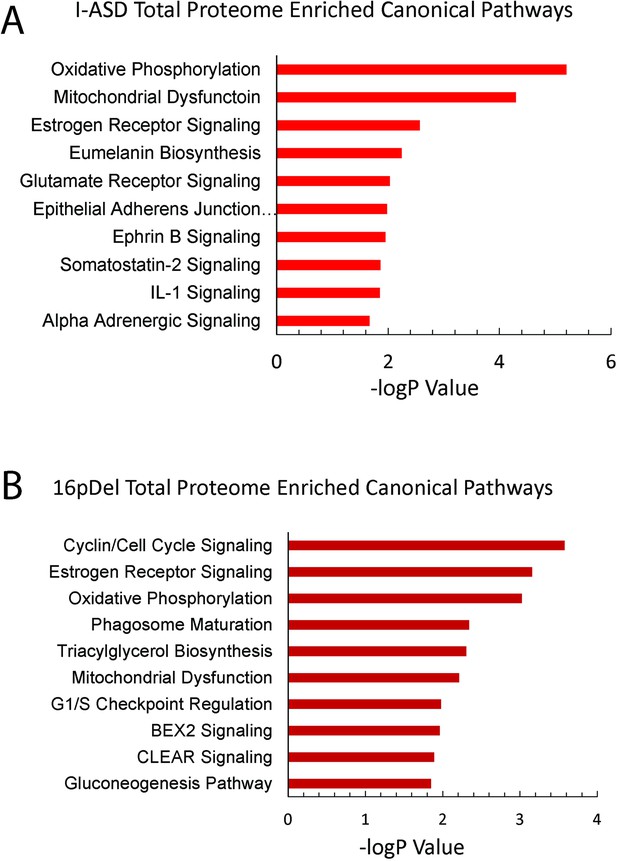
Ingenuity Pathway Analysis (IPA) total proteome canonical pathway analysis.
(A) IPA canonical pathway analysis of total proteome data from I-ASD showing top 10 enriched pathways. Total proteome does not show enrichment of mTOR pathway or mTOR pathway members. (B) IPA canonical pathway analysis of total proteome data from 16pDel showing top 10 enriched pathways. Total proteome does not show enrichment of mTOR pathway or mTOR pathway members.
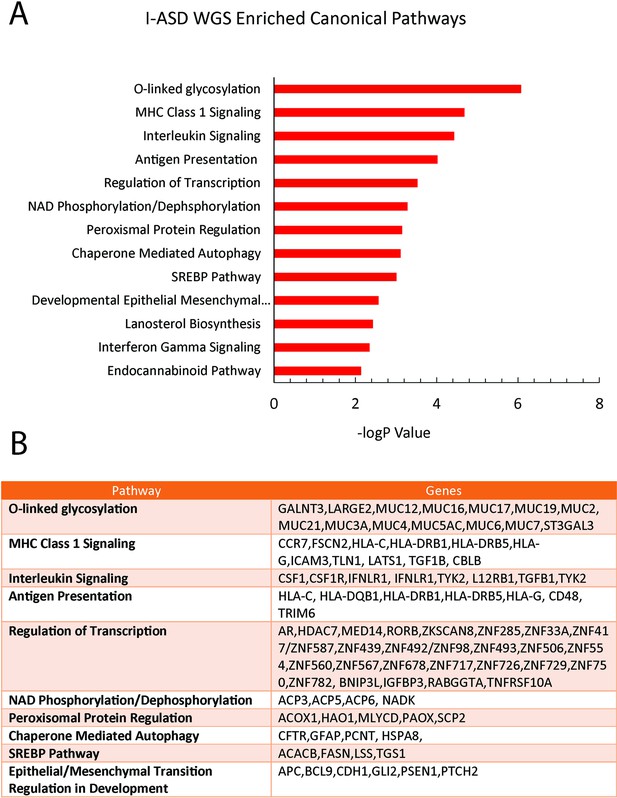
Ingenuity Pathway Analysis (IPA) of I-ASD whole genome sequencing (WGS).
(A) Top 15 enriched pathways in IPA of I-ASD-1 WGS. (B) Table showing the dysregulated genes in each enriched canonical pathway from I-ASD WGS analyses. WGS IPA does not show enrichment of mTOR pathway or mTOR pathway members.
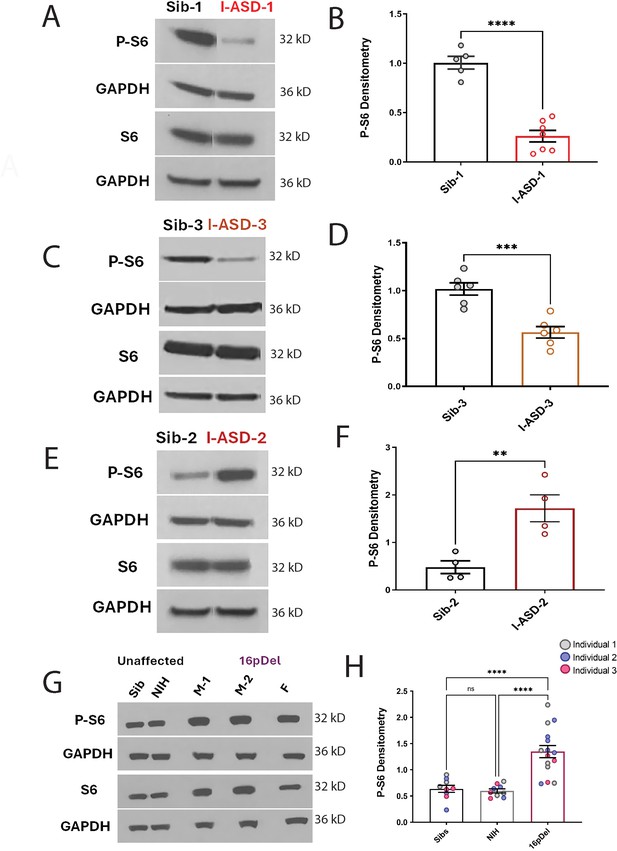
P-S6 and S6 western analysis for I-ASD and 16pDel neural precursor cells (NPCs).
All images show P-S6 and S6 blots with matched GAPDH loading control. Graphs show densitometry quantifications of normalized p-S6 (p-S6/GAPDH) divided by normalized S6 (S6/GAPDH). Student’s t-test utilized for all I-ASD vs Sib comparisons. (A–D) I-ASD-1 and -3 representative western blots showing reduced p-S6 but similar S6 and GAPDH in I-ASD-1 compared to Sib-1 (A) and in I-ASD-3 vs. Sib-3 (C). Graphs (B and D): reduced p-S6/S6 in I-ASD-1 vs. Sib-1 (B) and in I-ASD-3 vs Sib-3 (D). (E) I-ASD-2 representative western blot showing increased p-S6 but similar S6 in I-ASD-2 compared to Sib-2. (F) Graph: elevated p-S6/S6 in I-ASD-2 compared to Sib-2. (G) Western blot comparing a representative Sib, NIH control, and each 16pDel patient (M-1, M-2, F) showing increased p-S6 in all 16pDel NPCs compared to both NIH and Sib with similar total S6 and GAPDH. (H) Graph showing increased p-S6 in 16pDel compared to both Sibs and NIH (one-way ANOVA). For all graphs: p≤0.05 = *, p≤0.01 = **, p≤0.001 = ***, p≤0.0001 = ****. For detailed N values please see Supplementary file 1.
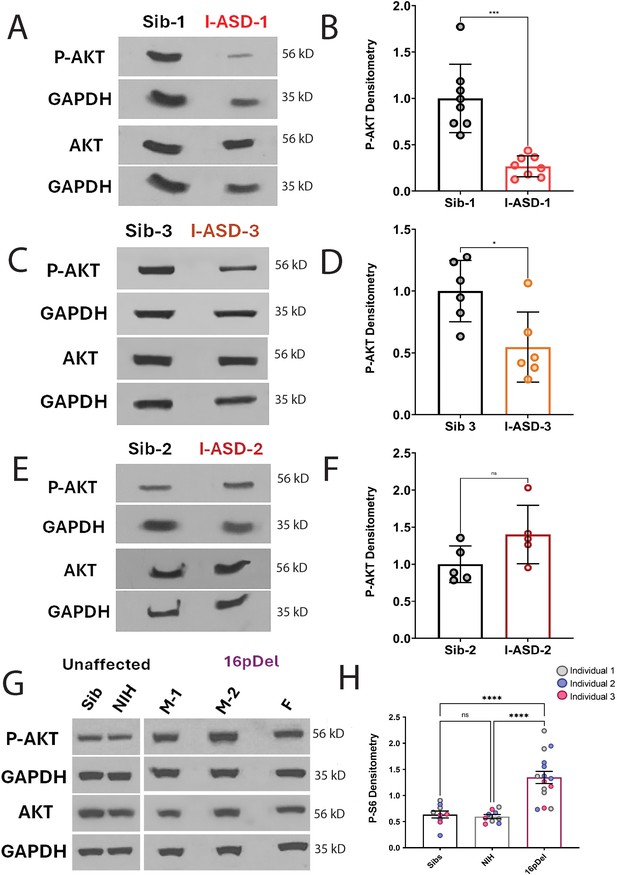
p-AKT in I-ASD and 16pdel neural precursor cells (NPCs).
All images show p-AKT and AKT western blots with matched GAPDH loading control. Graphs show densitometry quantifications of normalized p-AKT (p-AKT/GAPDH) divided by normalized AKT (AKT/GAPDH). Student’s t-test was used for all Sib vs I-ASD comparisons. (A) I-ASD-1 representative western blot showing reduced p-AKT but similar AKT and GAPDH in I-ASD-1 compared to Sib-1. (B) Graph: reduced p-AKT/AKT in I-ASD-1 vs. Sib-1. (C) I-ASD-3 representative western blot showing reduced p-AKT, but similar AKT and GAPDH in I-ASD-3 vs. Sib-3. (D) Graph: reduced p-AKT/AKT levels in I-ASD-3 compared to Sib-3. (E) Representative western blot showing slightly increased p-AKT but similar AKT and GAPDH in I-ASD-2 compared to Sib-2. (F) Graph: P-AKT levels in I-ASD-2 demonstrate a trend toward an increase compared to Sib-2. (G) Western blot showing increased p-AKT in all 16pDel NPCs compared to NIH and Sib with approximately equal total AKT and GAPDH. (H) Graph: increased p-AKT/AKT in all 16pdel NPCs compared to NIH NPCs (one-way ANOVA, p < 0.001) and but not compared to Sib NPCs (one-way ANOVA, p = 0.11). For all graphs: p≤0.05 = *, p≤0.01 = **, p≤0.001 = ***, p≤0.0001 = ****. For detailed N values please see Supplementary file 1.
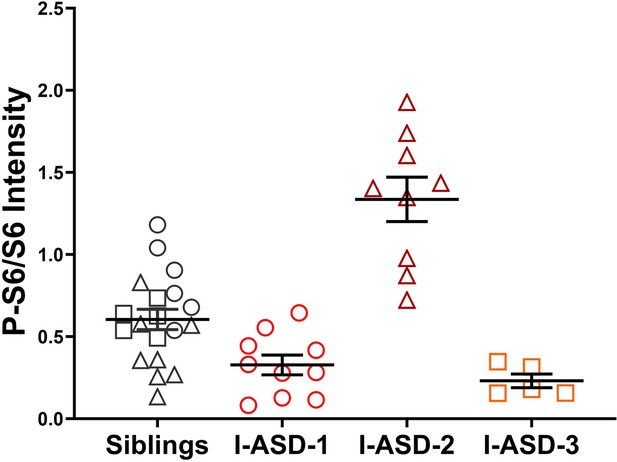
p-S6 levels compared across families.
Comparison of p-S6 levels between all I-ASD Sib controls (all Sibs) and each I-ASD-affected individual demonstrates that I-ASD-1 and -3 have reduced p-S6 while I-ASD-2 has elevated p-S6, indicating that phenotypes are relevant even when compared to unrelated Sib controls. For Sibs, circles represent Sib-1, triangles Sib-2, and squares Sib-3. p≤0.05 = *, p≤0.01 = **, p≤0.001 = ***, p≤0.0001 = ****. For detailed N values please see Supplementary file 1.
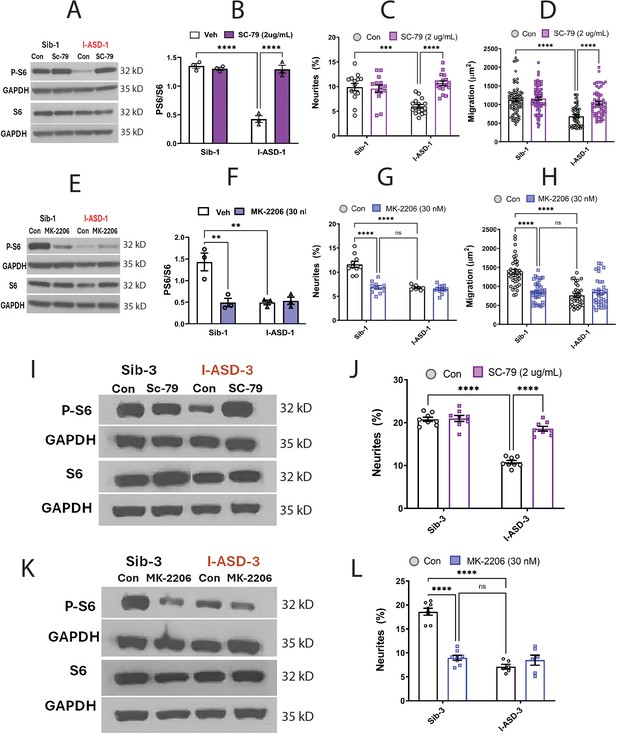
Effects of mTOR pathway manipulation in low mTOR cohort.
(A) Representative western blot: treatment of Family-1 neural precursor cells (NPCs) with SC-79 increases p-S6 levels in I-ASD-1 but not Sib-1 with no changes in total S6. (B) Graph: SC-79 vs Veh: increases p-S6/S6 levels in I-ASD-1 but not in Sib-1. (C) SC-79 treatment rescues neurite outgrowth in low mTOR I-ASD-1 NPCs without affecting Sib-1 NPC neurites. (D) SC-79 treatment rescues migration in low mTOR I-ASD-1 NPCs without affecting Sib-1 NPC neurites. (E) Representative western blot: MK-2206 treatment of Family-1 NPCs decreases p-S6 in Sib-1 but not I-ASD-1 with no changes in total S6. (F) Graph: MK-2206 vs Veh: decreases PS6 in Sib-1 but not I-ASD-1. (G) MK-2206 treatment reduces neurite outgrowth in Sib-1 to the level of I-ASD-1 without affecting I-ASD-1 neurite outgrowth. (H) MK-2206 treatment reduces migration in Sib-1 to the level of I-ASD-1 without affecting I-ASD-1. (I) Proof of principle western blot showing that SC-79 increases p-S6 levels in I-ASD-3 but not in Sib-3. (J) SC-79 treatment of low mTOR I-ASD-3 increases neurite outgrowth to the level of Sib-3. (K) Proof of principle western blot showing that MK-2206 decreases p-S6 in Sib-3 without affecting I-ASD-3 p-S6 levels. (L) Treatment of Sib-3 NPCs with MK-2206 reduces neurite outgrowth to the level of I-ASD-3. For all graphs: p≤0.05 = *, p≤ 0.01 = **, p≤0.001 = ***, p≤0.0001 = ****, two-way ANOVA. For detailed N values please see Supplementary file 1.
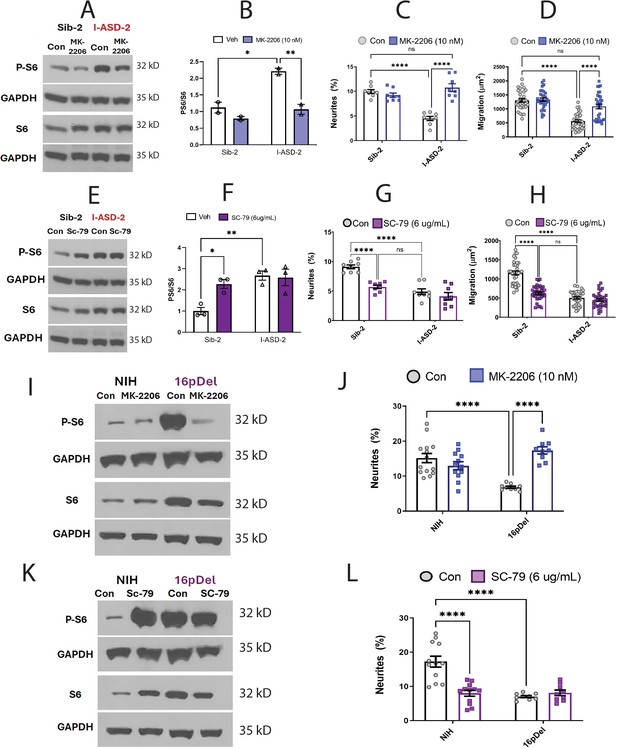
Effects of mTOR pathway manipulation on high mTOR cohort.
(A) Representative western: MK-2206 treatment of I-ASD-2 neural precursor cells (NPCs) leads to reduction of p-S6 in I-ASD-2 to Sib-2 levels without changing total S6. (B) Quantification of PS6/S6 western blots showing decreased PS6 in I-ASD-2 but no change in Sib-2 with MK-2206 treatment. (C) MK-2206 rescues neurite outgrowth in high mTOR I-ASD-2 without changing Sib-2 neurites. (D) MK-2206 rescues migration in high mTOR I-ASD-2 without changing Sib-2 migration. (E) Representative western: SC-79 treatment of Family-2 NPCs increases p-S6 in Sib-2 to the level of I-ASD-2 without changing total S6. (F) Quantification of Veh vs.SC-79 westerns showing increased PS6 in Sib-2 but no change in I-ASD-2. (G) Treatment of Sib-2 with SC-79 diminished neurite to the level of I-ASD-2. (H) Treatment of Sib-2 with SC-79 diminished migration to the level of I-ASD-2. (I) Proof of principal western: MK-2206 treatment of 16pDel NPCs leads to reduction of p-S6 in 16pDel NPCs to NIH levels without changing total S6. (J) In high mTOR 16pDel NPCs, MK-2206 treatment rescues neurites without significantly affecting NIH NPCs. (K) Proof of principle western: SC-79 treatment of NIH NPCs leads to increase in p-S6 to the level of 16pDel NPCs without changing total S6. (L) Treatment of NIH NPCs with SC-79 diminishes neurite outgrowth to 16pDel levels without affecting 16pDel NPCs. For all graphs: p≤0.05 = *, p≤0.01 = **, p≤0.001 = ***, p≤0.0001 = ****, two-way ANOVA. For detailed N values please see Supplementary file 1.
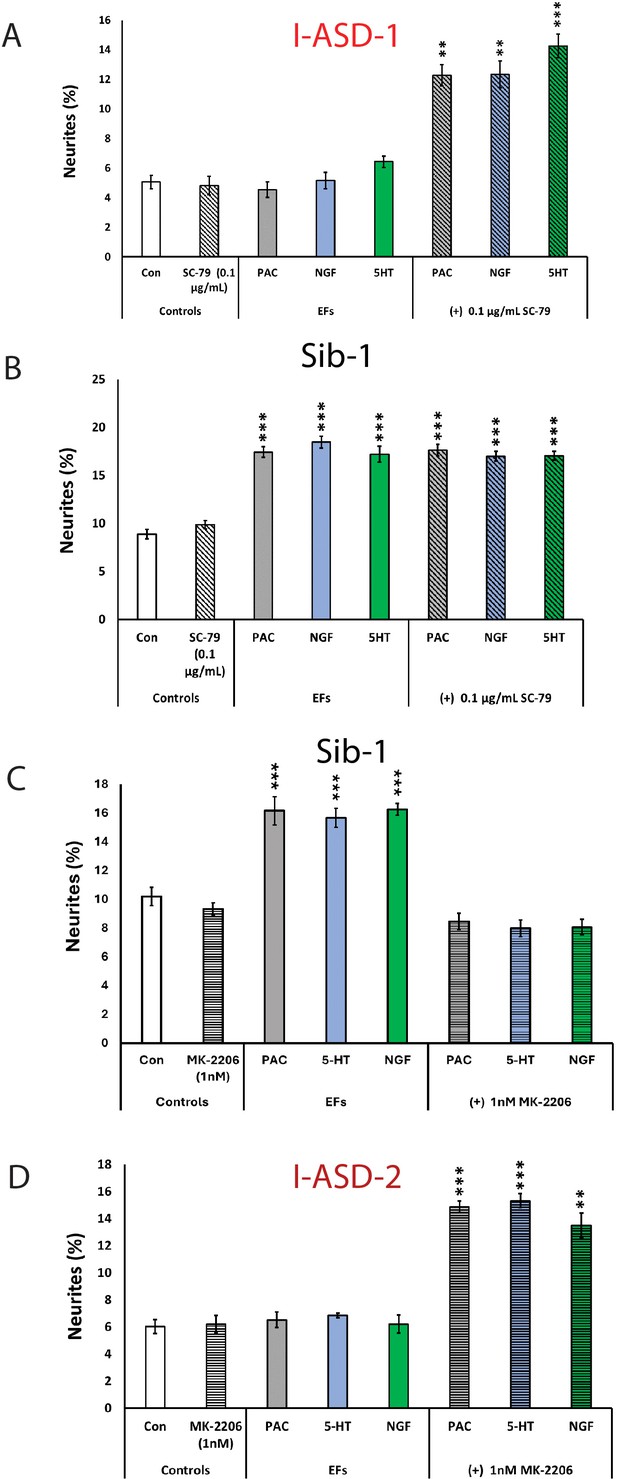
Modulation of mTOR can facilitate or abolish extracellular factor (EF) responses in I-ASD.
(A) Treatment of low mTOR I-ASD-1 neural precursor cells (NPCs) with subthreshold dose of SC-79 (0.1 µg/mL) results in NPCs responding to EFs. (B) Treatment of Sib-1 NPCs with a subthreshold dose of SC-79 does not alter EF response. (C) Treatment of Sib-1 NPCs with subthreshold MK-2206 (1 nM) abolishes NPC response to EFs. (D) Treatment of high mTOR I-ASD-2 with subthreshold MK-2206 (1 nM) establishes EF responses. p < 0.001 for all comparisons, one-way ANOVA. For all graphs: p≤0.05 = *, p≤0.01 = **, p≤0.001 = ***, p≤0.0001 = ****. For detailed N values please see Supplementary file 1.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Phospho-S6 Ribosomal Protein (Ser235/236) Antibody Rabbit polyclonal | Cell Signaling Technology | Catalog #: 2211 | ‘p-S6’ WB: (1:2000) |
| Antibody | S6 Ribosomal Protein (54D2) Mouse monoclonal | Cell Signaling Technology | Catalog #: 2317 | ‘Total S6 or S6’ WB: (1:2000) |
| Antibody | Phospho-Akt (Ser473) Antibody Rabbit polyclonal | Cell Signaling Technology | Catalog #: 9271 | ‘p-AKT’ WB: (1:1000) |
| Antibody | Akt Antibody Rabbit polyclonal | Cell Signaling Technology | Catalog #: 9272 | ‘AKT’ WB: (1:2000) |
| Antibody | Glyceraldehyde-3-phosphate dehydrogenase Mouse Monoclonal Antibody, Unconjugated, Clone 4G5 | Meridian Life Science | Catalog #: H86045M | ‘GAPDH’ WB: (1:10,000) |
| Antibody | Goat anti-Rabbit IgG (H+L) Secondary Antibody, HRP | Thermo Fisher Scientific | Catalog #: 31460 | WB: (1:2000) WB GAPDH: (1:5000) |
| Antibody | Goat anti-Mouse IgG (H+L) Secondary Antibody, HRP | Thermo Fisher Scientific | Catalog #: 31430 | WB: (1:2000) WB GAPDH: (1:5000) |
| Antibody | Recombinant Anti-SOX-2 antibody Rabbit monoclonal | Abcam | Catalog #: Ab92494 | ‘SOX2 antibody’ WB: (1:4000) ICC: (1:1000) |
| Antibody | Human Nestin Antibody Mouse Monoclonal | R&D Systems | Catalog #: MAB1259 | ‘Nestin’ WB: (1:5000) ICC: (1:500) |
| Antibody | Purified anti-PAX-6 antibody Rabbit Polyclonal | BioLegend (Previously Covance) | Covance Catalog #: PRB-278P | ‘Pax-6’ WB: (1:2000) ICC: (1:500) |
| Antibody | Purified anti-tubulin β 3 (TUBB3) Antibody Mouse Monoclonal | BioLegend (Previously Covance) | Covance Catalog #: MMS-435P | ‘TUJ1’ WB: (1:1000) ICC: (1:100) |
| Antibody | Goat anti-mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Catalog #: A-11001 | ICC: (1:2000) |
| Antibody | F(ab’)2-Goat anti-rabbit IgG (H+L) Cross-adsorbed secondary antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Catalog #: A-11070 | ICC: (1:2000) |
| Antibody | Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 | Thermo Fisher Scientific | Catalog #: A-11032 | ICC: (1:2000) |
| Antibody | Goat anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 | Thermo Fisher Scientific | Catalog #: A-11037 | ICC: (1:2000) |
| Cell Line (Human) | NCRM-1 | NIH | Research Grade: RMP Generated iPSC Line, control reference line, CD34+ Cord blood, episomal plasmid reprogramming | |
| Cell Line (Human) | NCRM-3 | NIH | Research Grade: RMP Generated iPSC Line, control reference line, CD34+ Cord blood, episomal plasmid reprogramming | |
| Cell Line (Human) | NCRM-6 | NIH | Research Grade: RMP Generated iPSC Line, control reference line, CD34+ Cord blood, episomal plasmid reprogramming | |
| Cell Line (Human) | iPSC Lines (16p11.2 deletion) | This paper | 2 male and 1 female line. One clone each. Maintained by Millonig lab | |
| Cell Line (Human) | iPSC Lines (I-ASD) | This paper | 3 male lines, 3 clones each Maintained by Millonig lab | |
| Cell Line (Human) | IPSC Lines (Sib) | This paper | 3 male lines, 3 clones each. Maintained by Millonig lab | |
| Chemical Compound, drug | Primocin | Invivogen | Catalog #: ant-pm-1 | Antimicrobial for culture Concentration: 100 µg/mL |
| Chemical Compound, drug | Serotonin Hydrochloride | Sigma-Aldrich | Catalog #: H9523 | ‘Serotonin or 5-HT’ |
| Chemical Compound, drug | Y-27632 | STEMCELL Technologies | Catalog #: 72302 | Noted as ‘Y- compound’ in manuscript, ROCK inhibitor |
| Chemical Compound, drug | SC79 | Selleckchem | Catalog #: S7863 | Small molecule drug, AKT activator |
| Chemical Compound, drug | MK-2206 2HCl | Selleckchem | Catalog #: S107 | Small molecule drug, AKT inhibitor |
| Commercial assay, kit | MycoAlert Mycoplasma Detection Kit | Lonza | Catalog #: LT07-318 | |
| Commercial assay, kit | mTeSR1 Complete Kit | STEMCELL Technologies | Catalog #: 5850 | Includes - mTeSR Basal medium (400 mL) - mTeSR 5× supplement 100 mL |
| Commercial assay, kit | Gibco PSC Neural Induction Medium | Thermo Fisher Scientific | Catalog #: A1647801 | Includes: - Basal medium (500 mL) - Supplement (10 mL) |
| Other | Gibco DMEM/F12 | Thermo Fisher Scientific | Catalog #: 11320033 | Cell Culture Media |
| Other | Gibco Neurobasal Medium | Thermo Fisher Scientific | Catalog #: 21103049 | Cell Culture Media |
| Other | Costar 6-well clear TC treated multiple well plates | Corning LifeSciences | Product #: 3506 | Cultureware; six-well plates |
| Other | 35 mm TC-treated culture dish | Corning | Product #: 430165 | Cultureware |
| Other | Accutase | Thermo Fisher Scientific | Catalog #: A111050 | Cell detachment solution |
| Other | Matrigel Matrix | Corning LifeSciences | Product #: 354277 | Extracellular Matrices |
| Other | Fibronectin bovine plasma | Sigma-Aldrich | Catalog # F1141 | Sterile filtered Extracellular Matrices |
| Other | NuPAGE LDS Sample Buffer (4×) | Thermo Fisher Scientific | Catalog #: NP0007 | Gel Electrophoresis Equipment and Supplies |
| Other | NuPAGE Sample Reducing Agent (10×) | Thermo Fisher Scientific | Catalog #: NP0004 | Gel Electrophoresis Equipment and Supplies |
| Other | NuPAGE 12%, Bis-Tris, 1.0 mm, Mini Protein Gels | Thermo Fisher Scientific | Catalog #: NP0342PK2 | Gel Electrophoresis Equipment and Supplies |
| Other | NuPAGE MOPS SDS Running Buffer (20×) | Thermo Fisher Scientific | Catalog #: NP000102 | Gel Electrophoresis Equipment and Supplies |
| Other | PVDF Transfer Membranes, 0.45 μm | Thermo Fisher Scientific | Catalog #: 88585 | Gel Electrophoresis Equipment and Supplies |
| Other | NuPAGE Transfer Buffer (20×) | Thermo Fisher Scientific | Catalog #: NP00061 | Gel Electrophoresis Equipment and Supplies |
| Other | Pierce ECL Western Blotting Substrate | Thermo Fisher Scientific | Catalog #: 32106 | Gel Electrophoresis Equipment and Supplies |
| Other | Pierce ECL Western Blotting Substrate | Thermo Fisher Scientific | Catalog #: 32106 | Gel Electrophoresis Equipment and Supplies |
| Other | LucentBlue X-Ray Film | Advantsa | Catalog #: 1190V51 or EK-5150 | Gel Electrophoresis Equipment and Supplies |
| Peptide, recombinant protein | Recombinant human β-NGF | Peprotech | Catalog #: 450-01 | ‘Nerve Growth Factor or NGF’ |
| Peptide, recombinant protein | PACAP-38 (human, mouse, ovine, porcine, rate) | BACHEM | Product #: 4031157 Previous Product #: H-8430 | ‘Pituitary adenylate cyclase activating polypeptide or PACAP’ |
| Software | Ingenuity Pathway Analysis | QIAGEN | p-Proteome, proteome, and WGS pathway analysis | |
| Software | Photoshop 2023 | Adobe | Image Editing | |
| Software | ImageJ | NIH | Western blot quantification | |
| Software | Prism | GraphPad by Dotmatics | Statistical analysis, graph generation | |
| Software | RStudio | GNU Project | Modeling, statistical analysis |
Additional files
-
Supplementary file 1
Tabulation of neural precursor cell (NPC) N-values for Figures 1—7.
In each cell, multiple different kinds of N-values are represented: # of clones (C)/Total # of neural inductions (NI)/# of experiments (E) and for neurite experiments, # of dishes (D) whereas for neurospheres experiment, # of neurospheres (NS).
- https://cdn.elifesciences.org/articles/82809/elife-82809-supp1-v2.docx
-
Supplementary file 2
Alternative allele genotypes in the chr16.p11.2 deletion region in three I-ASD families.
The count of the heterozygous and homozygous alternative allele genotypes and their ratio in the chr16.p11.2 deletion region (chr16: 28,500,001–35,300,000). In the event of a chr16.p11.2 deletion, we expect no heterozygous genotypes in the region. The large number of heterozygous genotypes across all individuals in this region indicates that the deletion does not appear in any of the individuals in the three I-ASD families.
- https://cdn.elifesciences.org/articles/82809/elife-82809-supp2-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/82809/elife-82809-mdarchecklist1-v2.docx