Proteome-wide systems genetics identifies UFMylation as a regulator of skeletal muscle function
Figures
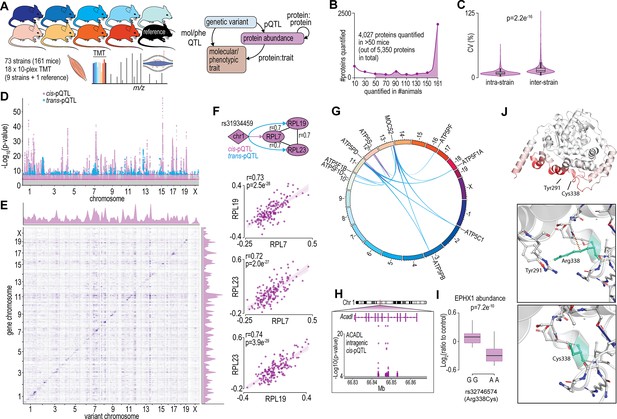
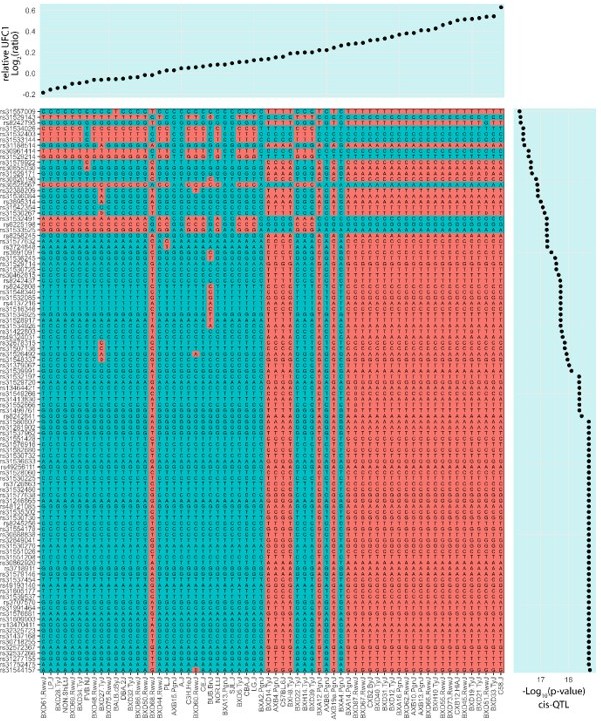
Proteome-wide systems genetics analysis of the mouse skeletal muscle proteome.
(A) Overview of the experimental design. (B) Number of proteins identified. (C) Intra- and inter-strain coefficient of variation. (D) Protein-quantitative trait loci (pQTL) Manhattan plot. (E) pQTL variant and gene location density. (F) Ribosomal proteins correlation and variant network (upper), and scatterplots expressed as Log2(ratio to control) showing correlation coefficient calculated using biweight midcorrelation (n=161) (lower). (G) Genetic associations of variant hotspot on chromosome 13 associated with mitochondrial complex V subunits in trans. (H) Intragenic variants associated to ACADL abundance. (I) Boxplot showing variant allele associated to EPHX1 abundance (Student’s t-test). (J) EPHX1 Arg338Cys mutation DynaMut protein flexibility analysis.
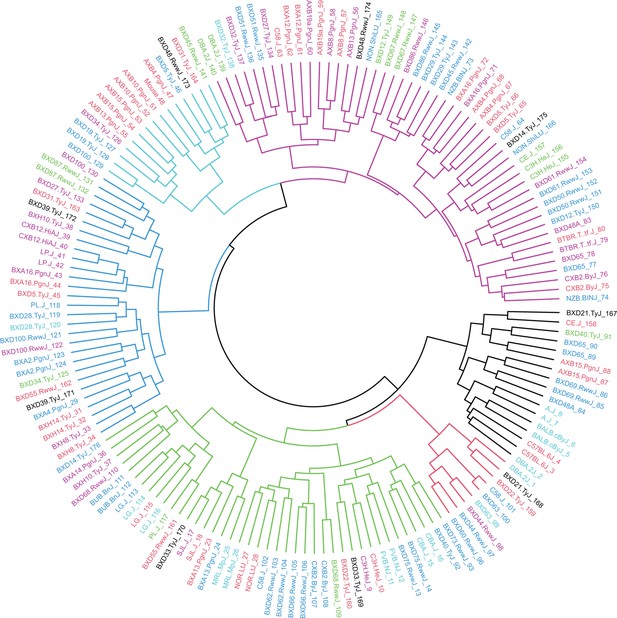
Hybrid Mouse Diversity Panel (HMDP) mouse sample dendrogram.
Dendrogram of HMDP mouse samples based on Euclidean distance and Ward’s clustering criterion (n=161).
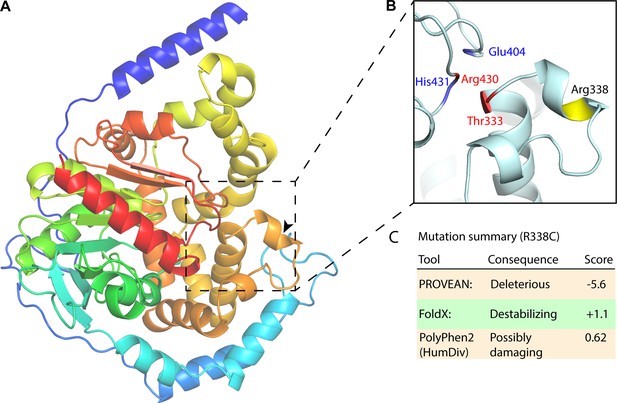
EPHX1 Arg338Cys mutation analysis.
(A) EPHX1 AlphaFold structure. Structure zoom-in highlighting Arg338Cys mutation and previously identified mutations with adverse health outcomes. (B) Arg338Cys is highlighted in yellow, the mutations and catalytic site key residues identified by Gautheron et al. in blue and red, respectively. (C) Mutation summary as determined by PROVEAN, FoldX, ELASPIC, and PolyPhen2.
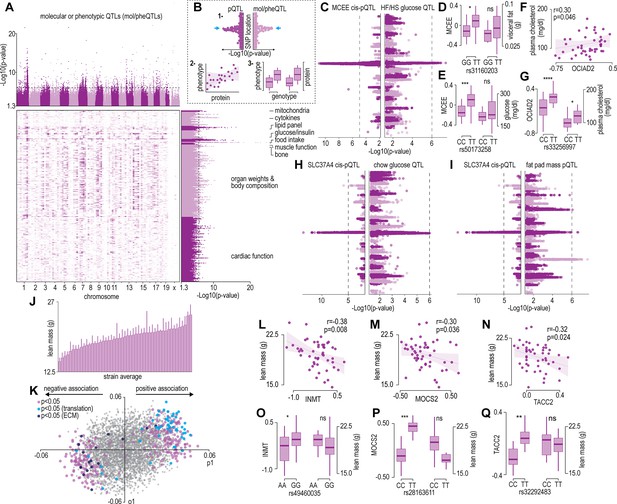
Protein and phenotype quantitative trait locus (QTL) analysis.
(A) Manhattan plot and genomic location distribution of mol/pheQTLs. (B) Overview of the three-step integrative analysis approach. (C) Mirrored Manhattan plots of MCEE and glucose QTLs. (D) Allelic variant boxplots of rs31160203 for MCEE and visceral fat. (E) Allelic variant boxplots of rs50173258 for MCEE and glucose. (F) Correlation scatterplot of OCIAD2 abundance expressed as Log2(ratio to control) and plasma cholesterol concentrations. (G) Allelic variant boxplots of rs33256997 for OCIAD2 and plasma cholesterol. (H) Mirrored Manhattan plots of SLC37A4 and glucose QTLs. (I) Mirrored Manhattan plots of SLC37A4 and fat pas mass QTLs. (J) Average distribution of lean mass per mouse strain. (K) Orthogonal partial least-squares (OPLS) loading plot of proteins explaining the variance related to strain lean mass. Separation on the x-axis shows variation related to the predictive component (p1), whilst the y-axis shows the orthogonal component (o1). Highlighted points reflect Student’s correlation p-values for multiple biweight midcorrelations of proteins correlated with lean mass (–0.3< r > 0.6, p<0.05). Correlation of lean mass and the protein abundance expressed as Log2(ratio to control) of INMT (L), MOCS2, (M) and TACC2 (N). Allelic variant boxplots of selected single nucleotide polymorphisms (SNPs) with lean mass and INMT (rs49460035) (O), MOCS2 (rs28163611) (P), and TACC2 (rs32292483) (Q). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 by Student’s t-test.
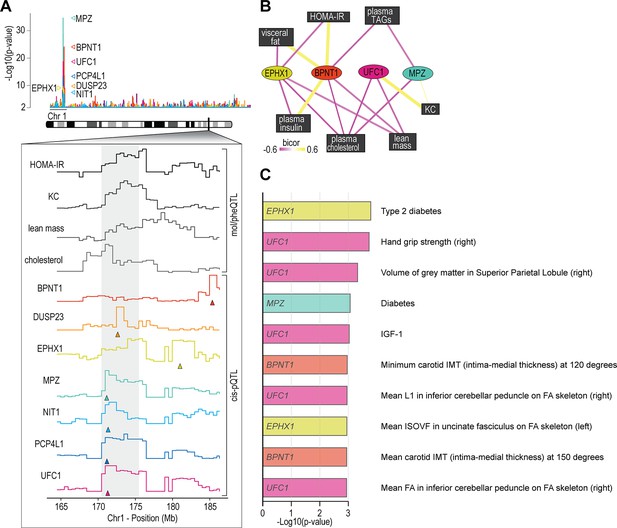
Proteome-phenotype associations of Qrr1 region on chromosome 1.
(A) Manhattan plot of selected genes located near the Qrr1 region, with corresponding traits. (B) Protein-trait correlation network. (C) Top 10 GeneBass associations from the ‘UK BioBank Assessment Centre’ and ‘Biological samples’ categories, excluding ‘Touchscreen’, ‘Medications’, and ‘Operations’ categories.
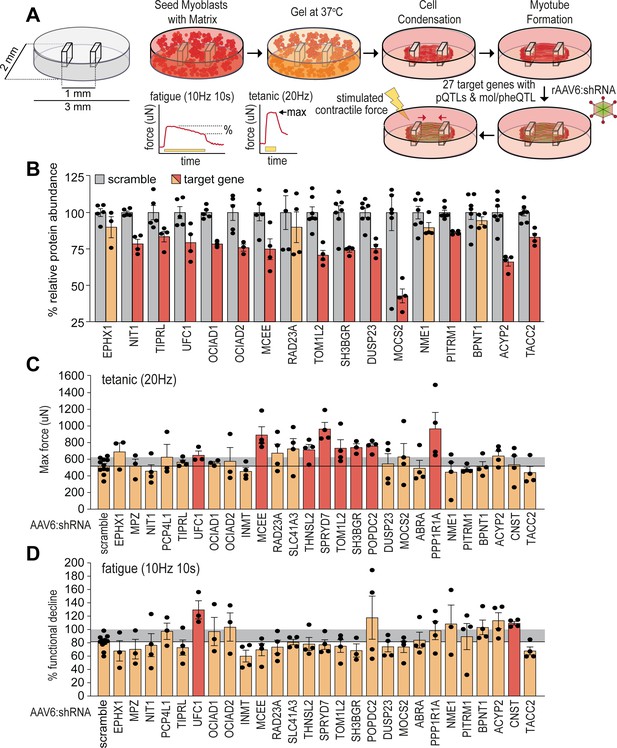
Functional screening of skeletal muscle function.
(A) Overview of experimental design. (B) Knockdown efficiency of target proteins (n=4–10). (C) Maximum tetanic force, and (D) % fatigue of rAAV6:shScramble and target proteins. Red: q<0.05; yellow: q>0.05 (Student’s t-test relative to scramble with Benjamini-Hochberg FDR).
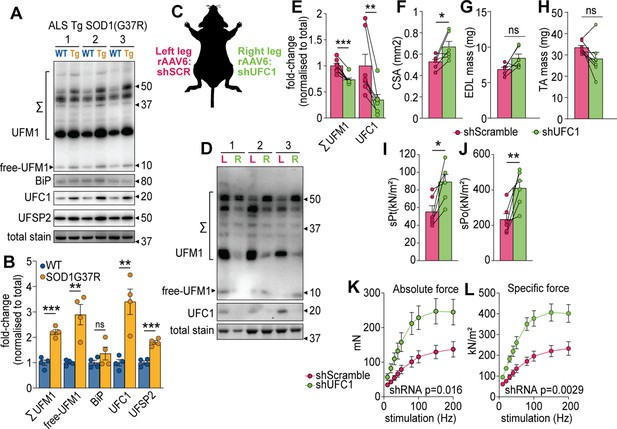
UFMylation is regulated in atrophy and influences skeletal muscle function.
(A) Western blot and (B) densitometry of UFMylation and BiP chaperone in a gastrocnemius muscle of a mouse model of amyotrophic lateral sclerosis (ALS). (C) Overview of the experimental design. (D) Western blot of extensor digitorum long (EDL) muscles treated with rAAV6:shScramble (red, left leg (L)) and rAAV:shUFC1 (green, right leg (R)). (E) Densitometry of western blot (n=6). (F) Muscle cross-sectional area (CSA) (n=6). (G) EDL mass and (H) tibialis anterior (TA) mass (n=6). Ex vivo analysis of contraction force in EDL muscles showing (I) single twitch contraction force normalized to CSA (sPt), (J) tetanic contraction force normalized to CSA (sPt), and (K) absolute, and (L) specific force normalized to CSA following shUFC1 or scrambled control. *p/q-value<0.05; **p/q-value<0.01; ***p/q-value<0.005; (B–C) paired Student’s t-test; (E–J) paired Student’s t-test; (K–L) two-way ANOVA.
-
Figure 5—source data 1
Zip file containing uncropped western blot image files as Image Lab Documents, tiff files, and a summarized.pdf highlighting the lane identifications, highlighted bands used to create Figure 5A, antibody information, and all densitometry results for each individual sample.
The top corner of each membrane is cut above lane 1.
- https://cdn.elifesciences.org/articles/82951/elife-82951-fig5-data1-v2.zip
-
Figure 5—source data 2
Zip file containing uncropped western blot image files as Image Lab Documents, tiff files, and a summarized.pdf highlighting the lane identifications, highlighted bands used to create Figure 5D, antibody information, and all densitometry results for each individual sample.
The top corner of each membrane is cut above lane 1.
- https://cdn.elifesciences.org/articles/82951/elife-82951-fig5-data2-v2.zip
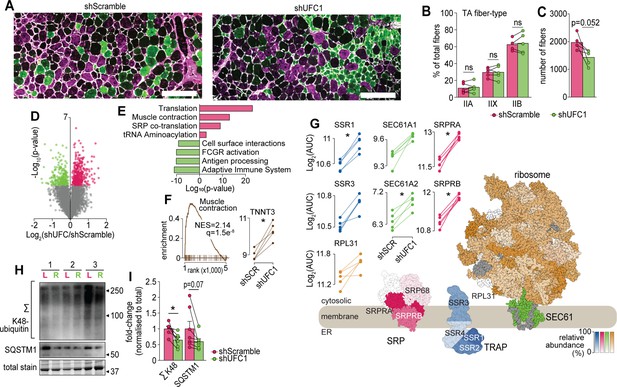
Characterization of skeletal muscles following UFC1 knockdown.
(A) Representative immunofluorescence microscopy of fiber-type composition in tibialis anterior (TA). Myosin heavy chain isoforms (MYH2, green, type IIa; MYH4, purple, type IIb; MYH1, unstained, type IIx) while laminin is white. Scale bar = 200 µm. (B) TA fiber-type distribution, and (C) TA total fiber number (n=5). (D) Volcano plot and (E) gene set enrichment analysis of proteins affected by UFC1 knockdown. (F) Enrichment plot of the muscle contraction gene set (REACTOME_MUSCLE_CONTRACTION, MSigDB C2 collection) and paired analysis of TNNT3. (G) Knockdown of UFC1 up-regulates the ribosome-SEC61 complex, signal recognition particle, and translocon-associated protein. Protein constituents of each structure were coloured based on the relative increased abundance following shUFC1, where the colours are scaled based on the relative fold change per complex (signal recognition particle [SRP] – red; translocon-associated protein (TRAP) – blue; SEC61 – green; ribosome – yellow/orange, grey – not measured). (H) Western blot of extensor digitorum longus (EDL) muscles treated with rAAV6:shScramble (red, left leg (L)) and rAAV:shUFC1 (green, right leg (R)). (I) Densitometry of western-blot (n=6). *p/q-value<0.05; (B, C, I): paired Student’s t-test; (F–G): paired Student’s t-test with Benjamini-Hochberg FDR.
-
Figure 6—source data 1
Zip file containing uncropped western blot image files as Image Lab Documents, tiff files, and a summarized.pdf highlighting the lane identifications, highlighted bands used to create Figure 6H, antibody information and all densitometry results for each individual sample.
The top corner of each membrane is cut above lane 1.
- https://cdn.elifesciences.org/articles/82951/elife-82951-fig6-data1-v2.zip
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Rabbit monoclonal anti-UFC1 | Abcam | EPR15014-102 (ab189252) | (1:1000) |
| Antibody | Rabbit monoclonal anti-UFSP2 | Abcam | EP13424-49 (ab192597) | (1:1000) |
| Antibody | Rabbit monoclonal anti-UFM1 | Abcam | EPR4264(2) (ab109305) | (1:1000) |
| Antibody | Rabbit monoclonal antiBiP | Cell Signaling Technologies | 3177 | (1:1000) |
| Antibody | Rabbit monoclonal SQSTM1/p62 (D1Q5S) | Cell Signaling Technologies | 39749 | (1:1000) |
| Antibody | Rabbit monoclonal K48-linkage Specific Polyubiquitin (D9D5) | Cell Signaling Technologies | 8081 | (1:1000) |
| Antibody | Donkey polyclonal Anti-Rabbit-HRP | Jackson ImmunoResearch | 711-035-152 (RRID:AB_10015282) | (1:10000) |
| Genetic reagent (Homo sapiens) | Human Skeletal Myoblasts | Lonza | CC-2580 (lot #18TL269121) | |
| Genetic reagent (Homo sapiens) | Human embryonic kidney 293 cells expressing SV40 large T antigen | ATCC | CRL-1573 | |
| Strain, strain background (Mus musculus) | A/J | JAX | RRID:IMSR_JAX:000646 | |
| Strain, strain background (Mus musculus) | AXB10/PgnJ | JAX | RRID:IMSR_JAX:001681 | |
| Strain, strain background (Mus musculus) | AXB13/PgnJ | JAX | RRID:IMSR_JAX:001684 | |
| Strain, strain background (Mus musculus) | AXB15/PgnJ | JAX | RRID:IMSR_JAX:001685 | |
| Strain, strain background (Mus musculus) | AXB19a/PgnJ | JAX | RRID:IMSR_JAX:001686 | |
| Strain, strain background (Mus musculus) | AXB4/PgnJ | JAX | RRID:IMSR_JAX:001676 | |
| Strain, strain background (Mus musculus) | AXB8/PgnJ | JAX | RRID:IMSR_JAX:001679 | |
| Strain, strain background (Mus musculus) | B6.Cg-Tg(SOD1*G37R)42Dpr/J | JAX | RRID:IMSR_JAX:008342 | |
| Strain, strain background (Mus musculus) | BALB/cByJ | JAX | RRID:IMSR_JAX:001026 | |
| Strain, strain background (Mus musculus) | BTBR T+tf/J | NA | NA | |
| Strain, strain background (Mus musculus) | BUB/BnJ | JAX | RRID:IMSR_JAX:000653 | |
| Strain, strain background (Mus musculus) | BXA12/PgnJ | JAX | RRID:IMSR_JAX:001700 | |
| Strain, strain background (Mus musculus) | BXA13/PgnJ | JAX | RRID:IMSR_JAX:001826 | |
| Strain, strain background (Mus musculus) | BXA14/PgnJ | JAX | RRID:IMSR_JAX:001702 | |
| Strain, strain background (Mus musculus) | BXA16/PgnJ | JAX | RRID:IMSR_JAX:001703 | |
| Strain, strain background (Mus musculus) | BXA2/PgnJ | JAX | RRID:IMSR_JAX:001693 | |
| Strain, strain background (Mus musculus) | BXA4/PgnJ | JAX | RRID:IMSR_JAX:001694 | |
| Strain, strain background (Mus musculus) | BXD100 | NA | NA | |
| Strain, strain background (Mus musculus) | BXD100/RwwJ | JAX | RRID:IMSR_JAX:007143 | |
| Strain, strain background (Mus musculus) | BXD12/TyJ | JAX | RRID:IMSR_JAX:000045 | |
| Strain, strain background (Mus musculus) | BXD14/TyJ | JAX | RRID:IMSR_JAX:000329 | |
| Strain, strain background (Mus musculus) | BXD19/TyJ | JAX | RRID:IMSR_JAX:000010 | |
| Strain, strain background (Mus musculus) | BXD21/TyJ | JAX | RRID:IMSR_JAX:000077 | |
| Strain, strain background (Mus musculus) | BXD22/TyJ | JAX | RRID:IMSR_JAX:000043 | |
| Strain, strain background (Mus musculus) | BXD27/TyJ | JAX | RRID:IMSR_JAX:000041 | |
| Strain, strain background (Mus musculus) | BXD28/TyJ | JAX | RRID:IMSR_JAX:000047 | |
| Strain, strain background (Mus musculus) | BXD29/TyJ | NA | NA | |
| Strain, strain background (Mus musculus) | BXD31/TyJ | JAX | RRID:IMSR_JAX:000083 | |
| Strain, strain background (Mus musculus) | BXD32/TyJ | JAX | RRID:IMSR_JAX:000078 | |
| Strain, strain background (Mus musculus) | BXD33/TyJ | JAX | RRID:IMSR_JAX:003222 | |
| Strain, strain background (Mus musculus) | BXD34/TyJ | JAX | RRID:IMSR_JAX:003223 | |
| Strain, strain background (Mus musculus) | BXD39/TyJ | JAX | RRID:IMSR_JAX:003228 | |
| Strain, strain background (Mus musculus) | BXD40/TyJ | JAX | RRID:IMSR_JAX:003229 | |
| Strain, strain background (Mus musculus) | BXD44/RwwJ | JAX | RRID:IMSR_JAX:007094 | |
| Strain, strain background (Mus musculus) | BXD45/RwwJ | JAX | RRID:IMSR_JAX:007096 | |
| Strain, strain background (Mus musculus) | BXD48/RwwJ | JAX | RRID:IMSR_JAX:007097 | |
| Strain, strain background (Mus musculus) | BXD48A | NA | NA | |
| Strain, strain background (Mus musculus) | BXD5/TyJ | JAX | RRID:IMSR_JAX:000037 | |
| Strain, strain background (Mus musculus) | BXD50/RwwJ | JAX | RRID:IMSR_JAX:007099 | |
| Strain, strain background (Mus musculus) | BXD51/RwwJ | JAX | RRID:IMSR_JAX:007100 | |
| Strain, strain background (Mus musculus) | BXD55/RwwJ | JAX | RRID:IMSR_JAX:007103 | |
| Strain, strain background (Mus musculus) | BXD60/RwwJ | JAX | RRID:IMSR_JAX:007105 | |
| Strain, strain background (Mus musculus) | BXD61/RwwJ | JAX | RRID:IMSR_JAX:007106 | |
| Strain, strain background (Mus musculus) | BXD62/RwwJ | JAX | RRID:IMSR_JAX:007107 | |
| Strain, strain background (Mus musculus) | BXD63 | NA | NA | |
| Strain, strain background (Mus musculus) | BXD65 | NA | NA | |
| Strain, strain background (Mus musculus) | BXD66/RwwJ | JAX | RRID:IMSR_JAX:007111 | |
| Strain, strain background (Mus musculus) | BXD67/RwwJ | JAX | RRID:IMSR_JAX:007112 | |
| Strain, strain background (Mus musculus) | BXD68/RwwJ | JAX | RRID:IMSR_JAX:007113 | |
| Strain, strain background (Mus musculus) | BXD69/RwwJ | JAX | RRID:IMSR_JAX:007114 | |
| Strain, strain background (Mus musculus) | BXD73/RwwJ | JAX | RRID:IMSR_JAX:007117 | |
| Strain, strain background (Mus musculus) | BXD75/RwwJ | JAX | RRID:IMSR_JAX:007119 | |
| Strain, strain background (Mus musculus) | BXD86/RwwJ | JAX | RRID:IMSR_JAX:007129 | |
| Strain, strain background (Mus musculus) | BXD87/RwwJ | JAX | RRID:IMSR_JAX:007130 | |
| Strain, strain background (Mus musculus) | BXH10/TyJ | JAX | RRID:IMSR_JAX:000032 | |
| Strain, strain background (Mus musculus) | BXH14/TyJ | JAX | RRID:IMSR_JAX:000009 | |
| Strain, strain background (Mus musculus) | BXH8/TyJ | JAX | RRID:IMSR_JAX:000076 | |
| Strain, strain background (Mus musculus) | C3H/HeJ | JAX | RRID:IMSR_JAX:000659 | |
| Strain, strain background (Mus musculus) | C57BL/6J | JAX | RRID:IMSR_JAX:000664 | |
| Strain, strain background (Mus musculus) | C58/J | JAX | RRID:IMSR_JAX:000669 | |
| Strain, strain background (Mus musculus) | CBA/J | JAX | RRID:IMSR_JAX:000656 | |
| Strain, strain background (Mus musculus) | CE/J | JAX | RRID:IMSR_JAX:000657 | |
| Strain, strain background (Mus musculus) | CXB12/HiAJ | JAX | RRID:IMSR_JAX:001633 | |
| Strain, strain background (Mus musculus) | CXB2/ByJ | JAX | RRID:IMSR_JAX:000352 | |
| Strain, strain background (Mus musculus) | DBA/2J | JAX | RRID:IMSR_JAX:000671 | |
| Strain, strain background (Mus musculus) | FVB/NJ | JAX | RRID:IMSR_JAX:001800 | |
| Strain, strain background (Mus musculus) | LG/J | JAX | RRID:IMSR_JAX:000675 | |
| Strain, strain background (Mus musculus) | LP/J | JAX | RRID:IMSR_JAX:000676 | |
| Strain, strain background (Mus musculus) | MRL/MpJ | JAX | RRID:IMSR_JAX:000486 | |
| Strain, strain background (Mus musculus) | NON/ShiLtJ | JAX | RRID:IMSR_JAX:002423 | |
| Strain, strain background (Mus musculus) | NOR/LtJ | JAX | RRID:IMSR_JAX:002050 | |
| Strain, strain background (Mus musculus) | NZB/BINJ | JAX | RRID:IMSR_JAX:000684 | |
| Strain, strain background (Mus musculus) | PL/J | JAX | RRID:IMSR_JAX:000680 | |
| Strain, strain background (Mus musculus) | SJL/J | JAX | RRID:IMSR_JAX:000686 | |
| Software, algorithm | R version 4.1.1 | R Development Core Team, 2016 | https://www.R-project.org/ | |
| Software, algorithm | Limma 3.32.2 | Ritchie et al., 2015 | https://bioconductor.org/packages/release/bioc/html/limma.html | |
| Software, algorithm | CoffeeProt | Molendijk and Parker, 2021a | https://www.coffeeprot.com | |
| Software, algorithm | TeaProt | Molendijk et al., 2022 | https://tea.coffeeprot.com | |
| Software, algorithm | Mol* (Molstar) | Sehnal et al., 2021 | https://molstar.org/ | |
| Software, algorithm | WGCNA | Langfelder and Horvath, 2008 | https://cran.r-project.org/web/packages/WGCNA/ | |
| Software, algorithm | ColabFold (Alphafold2) | Mirdita et al., 2022 | https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb | |
| Software, algorithm | Genebass | Karczewski et al., 2022 | https://app.genebass.org/ | |
| Software, algorithm | FoldX | Delgado et al., 2019 | http://foldxsuite.crg.eu/ | |
| Software, algorithm | PROVEAN | Choi and Chan, 2015 | http://provean.jcvi.org/index.php | |
| Software, algorithm | DynaMut | Rodrigues et al., 2018 | http://biosig.unimelb.edu.au/dynamut/ |
Additional files
-
Supplementary file 1
Hybrid Mouse Diversity Panel (HMDP) skeletal muscle proteomics.
Proteomics data of gastrocnemius muscle displaying quantification ratios of each sample compared to its corresponding pooled tandem mass tag (TMT) control. PEP: posterior error probability. Related to Figures 1—3, Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/82951/elife-82951-supp1-v2.xlsx
-
Supplementary file 2
Hybrid Mouse Diversity Panel (HMDP) skeletal muscle protein-quantitative trait loci (pQTLs).
Protein quantitative trait loci from 161 HMDP cohort mice. Table contains cis-pQTLs (p < 1×10−4) and trans-pQTLs (p < 5 × 10−8), including genomic locations of the single nucleotide polymorphism (SNP) and associated gene. pQTLs are annotated with proxy (cis/trans), intragenic variants, known LD blocks, variant effect, variant impact, and target screen prioritization columns. Related to Figures 1—3, and Figure 1—figure supplement 2.
- https://cdn.elifesciences.org/articles/82951/elife-82951-supp2-v2.xlsx
-
Supplementary file 3
Hybrid Mouse Diversity Panel (HMDP) skeletal muscle pairwise protein-protein correlations.
Protein-protein correlation as determined using biweight midcorrelation. p-Values and q-values derived using the Benjamini-Hochberg procedure, and only positive correlations are shown (cor > 0.3 and q < 0.05). Correlated protein pairs are annotated with protein:protein interactions from the CORUM and BioPlex databases, and subcellar. Related to Figures 1 and 2.
- https://cdn.elifesciences.org/articles/82951/elife-82951-supp3-v2.xlsx
-
Supplementary file 4
Hybrid Mouse Diversity Panel (HMDP) molecular or phenotypic traits.
- https://cdn.elifesciences.org/articles/82951/elife-82951-supp4-v2.xlsx
-
Supplementary file 5
Hybrid Mouse Diversity Panel (HMDP) molecular or phenotypic quantitative trait loci (QTLs).
- https://cdn.elifesciences.org/articles/82951/elife-82951-supp5-v2.xlsx
-
Supplementary file 6
Proteomics of skeletal muscle treated with either rAAV6:shScramble or AAV6:shUFC1.
Proteomics of extensor digitorum long (EDL) muscles displaying tandem mass tag (TMT) quantification expressed as Log2(area under the curve). Significance was calculated using paired Student’s t-test with Benjamini-Hochberg FDR. PEP: posterior error probability. Related to Figure 4.
- https://cdn.elifesciences.org/articles/82951/elife-82951-supp6-v2.xlsx
-
Supplementary file 7
shRNA sequences used in the human micro-muscle screen and mouse shUFC1 experiments.
Related to Figure 4.
- https://cdn.elifesciences.org/articles/82951/elife-82951-supp7-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/82951/elife-82951-mdarchecklist1-v2.docx
-
Source data 1
Genebass datasets for UFC1, EPHX1, DUSP23, NIT1, MPZ, BPNT1 and PCP4L1.
- https://cdn.elifesciences.org/articles/82951/elife-82951-data1-v2.zip