T cell deficiency precipitates antibody evasion and emergence of neurovirulent polyomavirus
Figures
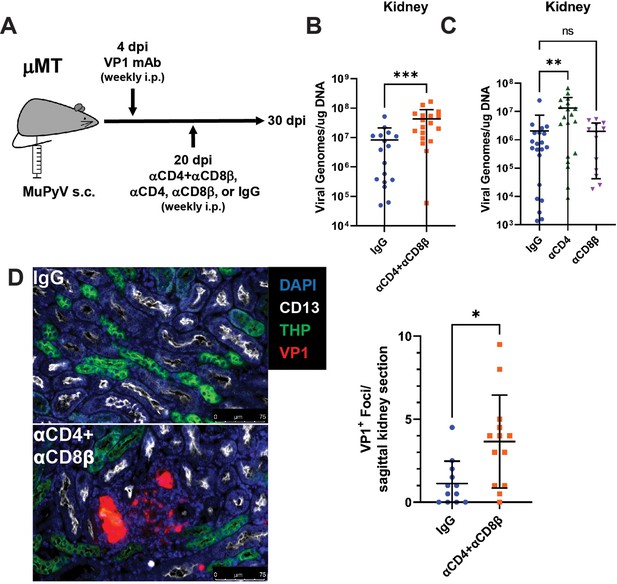
Increased MuPyV replication in the kidney following T cell depletion in VP1 mAb-treated μMT mice.
(A) Experimental approach for VP1 mAb treatment and T cell depletion. (B) Viral DNA levels in the kidney 10 days post T cell depletion with combined αCD4 and αCD8β. Viral DNA was quantified by qPCR and compared to a standard curve (n=16–18). (C) Viral DNA levels in the kidney 10 days post depletion with αCD4 or αCD8β. Viral DNA was quantified by qPCR and compared to a standard curve (n=13–21). (D) (Left) Foci of virus replication the kidney cortex 10 days post T cell depletion. Kidneys were stained for CD13 (white), THP (green) and VP1 (red). (Right) Quantification of virus foci in the kidney. Data are the average of two kidney sections per mouse (n=12–13). Error bars are mean ± SD. Data are from at least two independent experiments. Data were analyzed by Mann-Whitney U test (B, D) or Kruskal-Wallis test with Dunn’s multiple comparisons test (C). *p<0.05, **p<0.01, ***p<0.001.
-
Figure 1—source data 1
Data for the graphs in the figure.
- https://cdn.elifesciences.org/articles/83030/elife-83030-fig1-data1-v2.xlsx
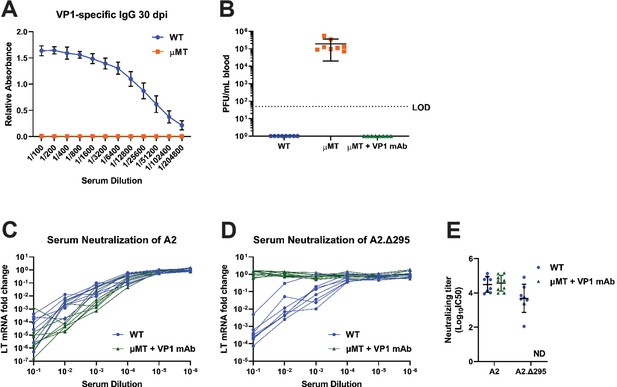
Passive immunization with a VP1 mAb in B cell-deficient mice neutralizes WT virus but not a VP1 mutant virus.
(A) VP1-specifc IgG 30 dpi from WT or μMT mice detected by ELISA with VP1 pentamers (n=10–12). (B) Viremia at 30 dpi in WT, μMT, or μMT mice treated with VP1 mAb starting 4 dpi. LOD: Limit of detection (n=8). (C–D) Neutralization of A2 (C) or A2.Δ295 (D) by sera from WT or VP1-mAb treated μMT mice 20 dpi. LT mRNA fold change is relative to infection by each virus in the absence of serum (n=8–9). (E) Neutralizing titers against A2 and A2.Δ295 from WT or VP1-mAb treated μMT mice 20 dpi. Values are the inverse log10 of the 50% neutralizing dilution of serum. ND: Not determined (n=8–9). Error bars are mean ± SD. Data are from at least two independent experiments.
-
Figure 1—figure supplement 1—source data 1
Data for the graphs in the figure.
- https://cdn.elifesciences.org/articles/83030/elife-83030-fig1-figsupp1-data1-v2.xlsx
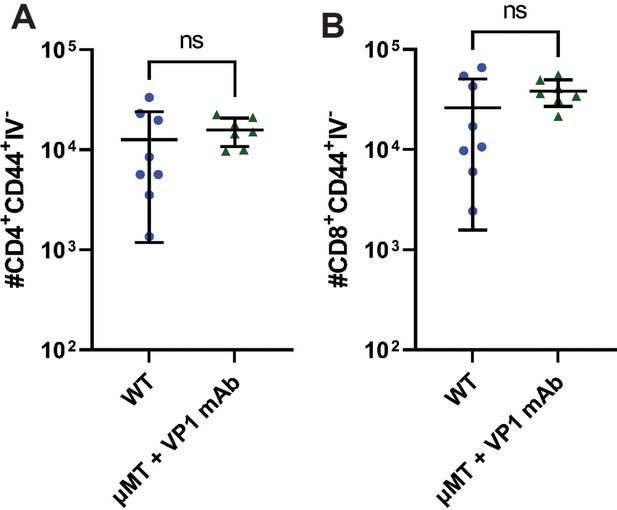
Kidney T cell infiltrates in WT and VP1-mAb treated μMT mice.
(A–B) Number of kidney-infiltrating CD4 T cells (A) and CD8 T cells (B) in the kidneys of WT and VP1-mAb-treated μMT mice 30 dpi. Kidney infiltrating cells were identified by i.v. injection of anti-CD45 prior to euthanasia (n=7–8). Error bars are mean ± SD. Data are from at least two independent experiments. Data were analyzed by Mann-Whitney U test (A–B).
-
Figure 1—figure supplement 2—source data 1
Data for the graphs in the figure.
- https://cdn.elifesciences.org/articles/83030/elife-83030-fig1-figsupp2-data1-v2.xlsx
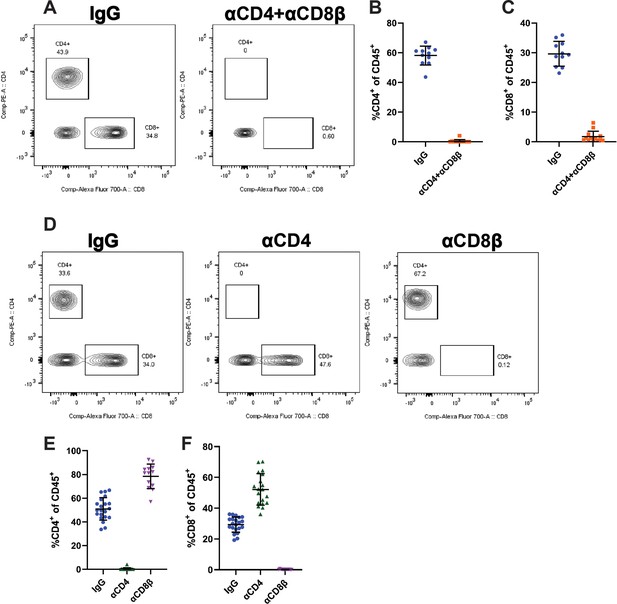
Confirmation of T cell depletion.
(A) Representative plot of CD4+ and CD8+ cells in the blood of mice treated with IgG or αCD4 and αCD8β at euthanasia 30 days post infection (10 days after the start of antibody treatment). Plots gated on CD45+ cells. (B–C) Frequency of CD4+ (B) and CD8+ (C) cells of total CD45+ cells in the blood of mice treated with IgG or αCD4 and αCD8β (n=11–13). (D) Representative plot of CD4+ and CD8+ cells in the blood of mice treated with IgG, αCD4, or αCD8β at euthanasia 30 days post infection (10 days after the start of antibody treatment). Plots gated on CD45+ cells. (E–F) Frequency of CD4+ (E) and CD8+ (F) cells of total CD45+ cells in the blood of mice treated with IgG, αCD4, or αCD8β (n=13–21). Error bars are mean ± SD. Data are from at least two independent experiments.
-
Figure 1—figure supplement 3—source data 1
Data for the graphs in the figure.
- https://cdn.elifesciences.org/articles/83030/elife-83030-fig1-figsupp3-data1-v2.xlsx
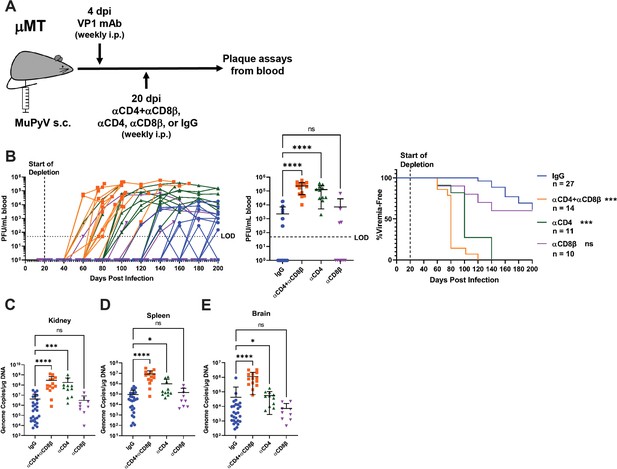
T cell loss enables viremia.
(A) Experimental scheme for VP1 mAb treatment, T cell depletion, and detection of viremia. (B) (Left) Viral titers in the blood of control and T cell depleted mice over time. Viremia was measured by plaque assay from whole blood. (Center) Peak levels of viremia detected in control and T cell depleted mice. (Right) Time to development of viremia in control and T-cell-depleted mice. Indicated significances are with comparison to the IgG group. LOD: Limit of detection (n=10–27). (C–E) Viral DNA levels in the kidney (C), spleen (D), and brain (E) at the time of euthanasia in control and T-cell-depleted mice. Viral DNA was quantified by qPCR and compared to a standard curve (n=9–27). Error bars are mean ± SD. Data are from at least two independent experiments. Data were analyzed by Kruskal-Wallis test with Dunn’s multiple comparisons test (B Center), (C–E) or Mantel-Cox test with Bonferroni’s correction for multiple comparisons (B Right). *p<0.05, ***p<0.001, ****p<0.0001.
-
Figure 2—source data 1
Data for the graphs in the figure.
- https://cdn.elifesciences.org/articles/83030/elife-83030-fig2-data1-v2.xlsx
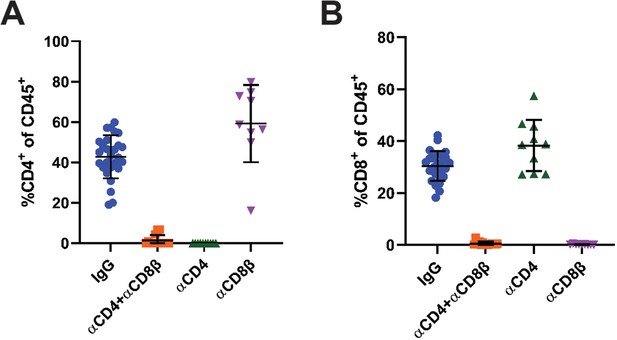
Confirmation of T cell depletion.
(A–B) Frequency of CD4+ (A) and CD8+ (B) cells of total CD45+ cells from the blood of mice treated with IgG, αCD4 and αCD8β, αCD4, or αCD8β. Blood was collected at euthanasia (n=9–27). Error bars are mean ± SD. Data are from at least two independent experiments.
-
Figure 2—figure supplement 1—source data 1
Data for the graphs in the figure.
- https://cdn.elifesciences.org/articles/83030/elife-83030-fig2-figsupp1-data1-v2.xlsx
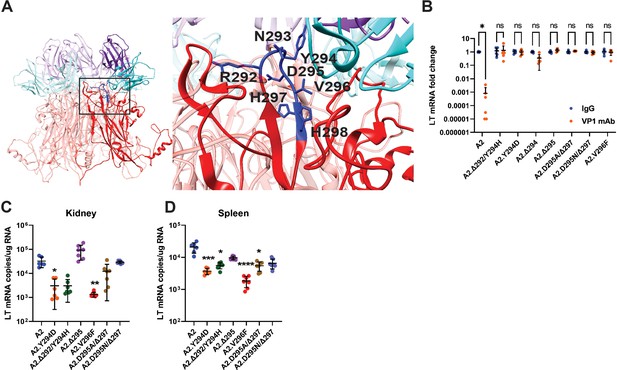
Viremia is mediated by VP1 Ab escape mutations.
(A) Location of VP1 mutations (blue) in the HI loop of one copy of VP1 relative to the location of VP1 mAb (cyan/purple) in the Cryo-EM structure of WT VP1 and the VP1 mAb. PDB ID: 7K22 (Lauver et al., 2020). (B) VP1 mAb neutralization of VP1 mutant viruses. Viruses were preincubated with 10 μg VP1 mAb or control IgG for 30 min prior to addition to 1x105 NMuMG epithelial cells. A2 was diluted to an MOI of 0.1 PFU/cell, mutant viruses were diluted to match A2 by genomic equivalents (g.e.). Viral LT mRNA levels were quantified 24 hpi and normalized for each virus to infection with control IgG (n=6). (C–D) Viral mRNA levels in the kidney (C) and spleen (D) 4 dpi with VP1 mutant viruses compared to WT. WT mice were infected i.v. with 1x106 PFU of A2 or mutant viruses matched by g.e. Viral LT mRNA levels were quantified by qPCR and compared to a standard curve (n=6–7). Error bars are mean ± SD. Data are from at least two independent experiments. Data were analyzed by Mann-Whitney U test with Holm-Šídák correction for multiple comparisons (B) or Kruskal-Wallis test with Dunn’s multiple comparisons test (C–D). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
-
Figure 3—source data 1
Data for the graphs in the figure.
- https://cdn.elifesciences.org/articles/83030/elife-83030-fig3-data1-v2.xlsx
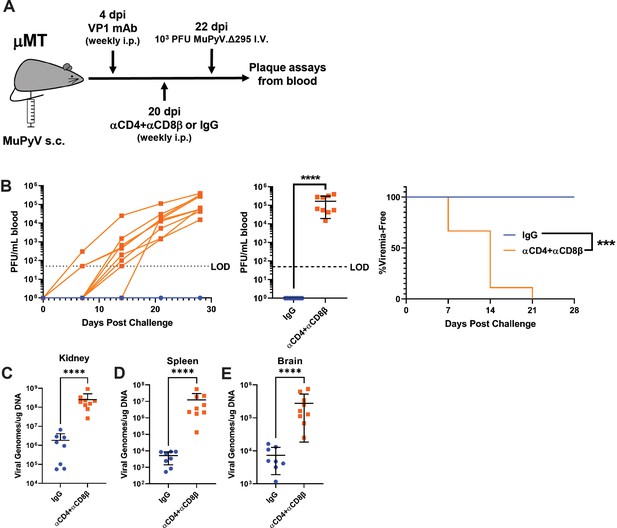
T cells prevent the outgrowth of Ab escape mutant virus.
(A) Experimental scheme for VP1 mAb treatment, T cell depletion, A2.Δ295 challenge, and detection of viremia in μMT mice. (B) (Left) Viral titers in the blood of control and T-cell-depleted mice over time. Viremia was measured by plaque assay from whole blood. (Center) Peak levels of viremia detected in control and T-cell-depleted mice. (Right) Time to development of viremia in control and T-cell-depleted mice in B. LOD: Limit of detection (n=8–9). (C–E) Viral DNA levels in the kidney (C), spleen (D), and brain (E) 28 days post challenge (n=8–9). Error bars are mean ± SD. Data are from two independent experiments. Data were analyzed by Mantel-Cox test (B Right) or Mann-Whitney U test (B center), (C–E). ***p<0.001, ****p<0.0001.
-
Figure 4—source data 1
Data for the graphs in the figure.
- https://cdn.elifesciences.org/articles/83030/elife-83030-fig4-data1-v2.xlsx
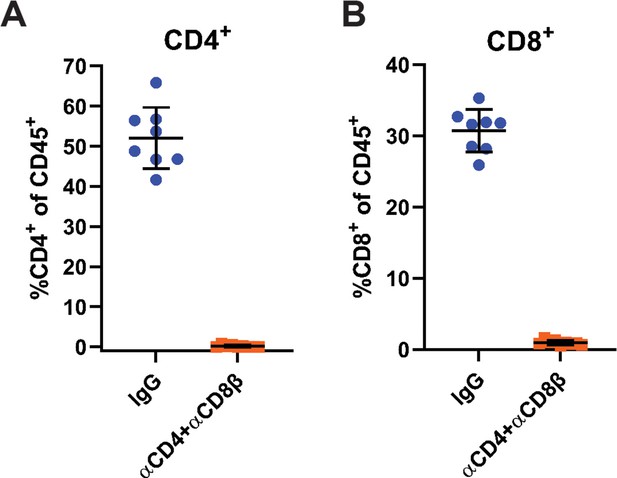
Confirmation of T cell depletion.
(A–B) Frequency of CD4+ (A) and CD8+ (B) cells out of total CD45+ cells the blood of mice treated with IgG or αCD4 and αCD8β. Blood was collected 2 weeks after the start of antibody treatment (n=8–9). Error bars are mean ± SD. Data are from at least two independent experiments.
-
Figure 4—figure supplement 1—source data 1
Data for the graphs in the figure.
- https://cdn.elifesciences.org/articles/83030/elife-83030-fig4-figsupp1-data1-v2.xlsx
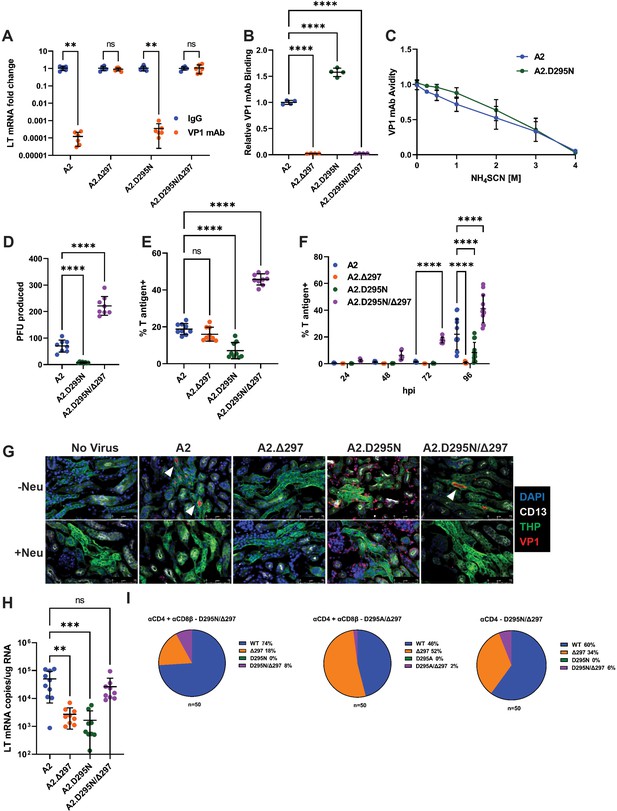
A compensatory mutation in VP1 arises to rescue defects in receptor binding caused by an Ab escape mutation.
(A) VP1 mAb neutralization assay with D295N and Δ297 VP1 mutant viruses. Viruses were preincubated with 10 μg VP1 mAb or control IgG for 30 min prior to addition to 1x105 NMuMG epithelial cells. A2 was diluted to an MOI of 0.1 PFU/cell, mutant viruses were diluted to match A2 by g.e. Viral LT mRNA levels were quantified 24 hpi and normalized for each virus to infection with control IgG (n=6). (B) Binding of VP1 mAb to WT or VP1 mutant viruses. Wells were coated with 1x109 g.e. of WT or VP1 mutant virus and incubated with VP1 mAb. VP1 mAb binding was quantified using an anti-rat secondary antibody and values were normalized to binding to WT virus (n=4). (C) Binding avidity of VP1 mAb for A2 and A2.D295N. VP1 mAb binding to A2 and A2.D295N was performed as in B. Prior to detection of mAb binding, virus/mAb complexes were treated with varying concentrations of NH4SCN for 15 min. Binding at each concentration was normalized to binding at 0 M NH4SCN for each virus (n=4). (D) Quantification of virus production in a single round of replication by plaque assay. Virus was added to 1x105 A31 cells at an MOI of 0.1 PFU/cell. Cells were lysed at 60 hpi and infectious virus was quantified by plaque assay and divided by the input virus quantity. A2.Δ297 was not included due to inability of this mutant to form plaques (n=8). (E) Frequency of T-antigen-positive cells 24 hpi with WT or mutant viruses. 1x105 A31 cells were infected with A2 at an MOI of 1 PFU/cell or mutant viruses matched by g.e. Cells were collected at 24 hpi, permeabilized, stained for T ag protein, and quantified by flow cytometry. (F) Frequency of T-antigen-positive cells at 24, 48, 72, and 96 hpi with WT or mutant viruses. 1x105 A31 cells were infected with A2 at an MOI of 0.1 PFU/cell or mutant viruses matched by g.e. Cells were collected at each time point, permeabilized, stained for T ag protein, and quantified by flow cytometry (n=6–13). (G) Detection of virus binding in kidney sections. PFA-fixed frozen kidney sections were treated with neuraminidase or buffer alone prior to incubation with WT or VP1 mutant virus. Sections were then stained for VP1 (red) and kidney markers [CD13 (white), THP (green)]. Neu: Neuraminidase. Representative of three independent experiments. (H) LT mRNA levels in the kidney 4 dpi with WT or mutant viruses. WT mice were infected i.v. with 1x106 PFU of A2 or mutant viruses matched by g.e. Viral LT mRNA levels were quantified by qPCR and compared to a standard curve (n=9–10). (I) Detection of D295 and H297 mutations in the kidney. VP1 sequences were PCR amplified from kidney DNA samples of the mice that developed the D295N/Δ297 and D295A/Δ297 double mutant viruses. VP1 clones were sequenced and screened for the presence of mutations at D295 and H297; the frequency of each mutation in 50 clones is shown. Error bars are mean ± SD. Data are from at least two independent experiments. Data were analyzed by Mann-Whitney U test with Holm-Šídák correction for multiple comparisons (A), one-way ANOVA with Dunnett’s multiple comparisons test (B and D–E), two-way ANOVA with Dunnett’s multiple comparisons test (F), or Kruskal-Wallis test with Dunn’s multiple comparisons test (H). **p<0.01, ***p<0.001, ****p<0.0001.
-
Figure 5—source data 1
Data for the graphs in the figure.
- https://cdn.elifesciences.org/articles/83030/elife-83030-fig5-data1-v2.xlsx
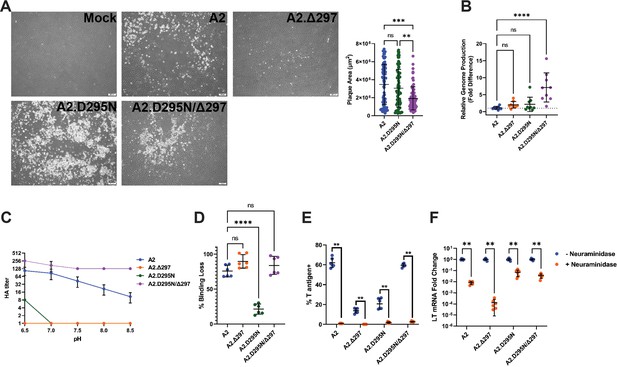
Defects in mutant virus spread and receptor binding.
(A) (Left) Plaque formation by WT and mutant viruses in A31 fibroblasts 6 dpi. (Right) Area of individual plaques formed by A2, A2.D295N, and A2.D295N/Δ297. Plaque area was measured using ImageJ (n=80). (B) Encapsidated genomes produced by WT and mutant viral DNA transfection. Data is relative to WT (n=9). (C) pH-dependent hemagglutination curves for WT and mutant viruses. The reported HA titer was the inverse of the highest dilution showing hemagglutination (n=3). (D) Percent of virus binding lost with neuraminidase pre-treatment. Bound virus was detected with polyclonal VP1 Ab and quantified by flow cytometry. For each virus, the percent of binding lost to neuraminidase treatment was calculated by comparing virus binding with and without neuraminidase treatment (n=6). (E) Quantification of T antigen positive cells 24 hpi with WT or mutant viruses with neuraminidase pretreatment. Cells were treated and infected as in (E). At 24 hpi, cells were collected, permeabilized, stained for T ag protein, and quantified by flow cytometry (n=6). (F) Quantification of LT mRNA levels with neuraminidase pretreatment. A31 cells were treated with neuraminidase and then infected with A2 at an MOI of 0.1 PFU/cell or mutant viruses matched by g.e. Viral LT mRNA levels were quantified by qPCR and compared to a standard curve (n=6). Error bars are mean ± SD. Data are from at least two independent experiments. Data were analyzed by Kruskal-Wallis test with Dunn’s multiple comparisons test (A), one-way ANOVA with Dunnett’s multiple comparisons test (B and D), or Mann-Whitney U test with Holm-Šídák correction for multiple comparisons (E–F). **p<0.01, ***p<0.001, ****p<0.0001.
-
Figure 5—figure supplement 1—source data 1
Data for the graphs in the figure.
- https://cdn.elifesciences.org/articles/83030/elife-83030-fig5-figsupp1-data1-v2.xlsx
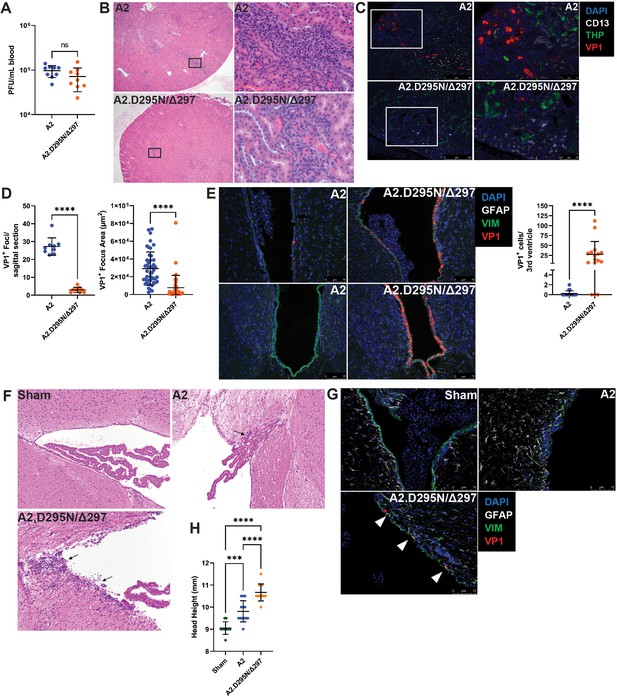
Reduced kidney tropism and heightened neurovirulence by a VP1 double mutant virus.
(A) Viremia in μMT mice 30 dpi with A2 or A2.D295N/Δ297 i.v. Viremia was quantified by plaque assay from whole blood (n=9). (B) H&E stained sagittal sections of kidneys from μMT mice 30 dpi after i.v. inoculation with A2 or A2.D295N/Δ297. Left: ×50 magnification. Right: ×500 magnification. (C) Foci of VP1+ cells in kidneys from μMT mice 30 dpi with A2 or A2.D295N/Δ297 i.v. Kidneys were stained for CD13 (white), THP (green) and VP1 (red). (D) Quantification of the number (Left, n=9) and size (Right, n=42–45) of VP1+ foci in kidneys from μMT mice 30 dpi with A2 or A2.D295N/Δ297 i.v. Foci number is the average of two sagittal kidney sections per mouse. For quantifying foci area, five random foci per mouse or the maximum number of foci found were imaged and the area of each VP1+ focus was calculated using ImageJ. (E) VP1+ cells in the lateral (top) and third (bottom) ventricles of WT mice 4 dpi with A2 or A2.D295N/Δ297 inoculated i.c. and quantification of VP1+ cells in the third ventricle (n=16–18). (F) H&E stained coronal sections from brains of sham (top left), A2 (top right), or A2.D295N/Δ297 (bottom left) i.c.-inoculated mice 30 dpi (×200 magnification). Arrows indicate sites of ependymal inflammation. (G) VP1 staining in the ventricles of WT mice 30 dpi with A2 or A2.D295N/Δ297 i.c. VP1+ cells are indicated with white markers. (H) Quantification of hydrocephalus 30 dpi after i.c. inoculation with vehicle, A2, or A2.D295N/Δ297. Coronal head height was measured with a Vernier caliper in line with the ear canal to the nearest 0.5 mm (n=10–13). Error bars are mean ± SD. Data are from at least two independent experiments. Data were analyzed by Mann-Whitney U test (A, D, and E) or one-way ANOVA with Dunnett’s multiple comparisons test (H). ***p<0.001, ****p<0.0001.
-
Figure 6—source data 1
Data for the graphs in the figure.
- https://cdn.elifesciences.org/articles/83030/elife-83030-fig6-data1-v2.xlsx
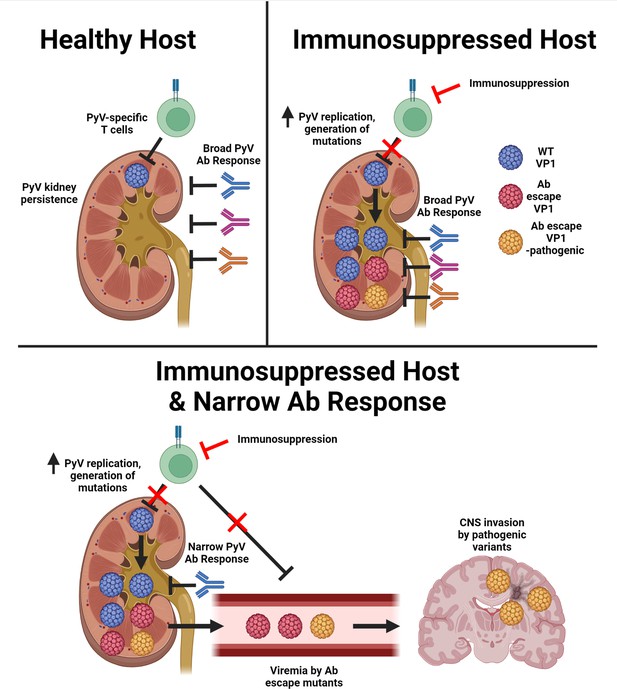
T cell deficiency and a narrow antiviral Ab response enable emergence of Ab-escape, neurovirulent PyV variants.
Model for the evolution of Ab escape, neurovirulent PyV VP1 variants in the setting of T cell deficiency and a neutralizing Ab response with limited coverage of VP1 epitopes on the virus capsid. Figure was created with Biorender.com.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-VP1 (Rat Clone 8A7H5) | Swimm et al., 2010 | Clone 8A7H5 | 250 µg/week |
| Antibody | ChromPure Rat IgG | Jackson ImmunoResearch | Cat#012-000-003 RRID: AB_2337136 | 250 µg/week |
| Antibody | Anti-CD8β (H35-17.2) | Golstein et al., 1982 | N/A | 250 µg/week |
| Antibody | Anti-CD4 (GK1.5) | Dialynas et al., 1983 | N/A | 250 µg/week |
| Antibody | Anti-VP1 (Rabbit polyclonal) | Provided by Robert Garcea | N/A | IF (1:1,000) FC (1:10,000) |
| Antibody | Anti-Vimentin (Rat Clone 280618) | R&D Systems | Cat#MAB2105 RRID: AB_2241653 | IF (1:500) |
| Antibody | Anti-GFAP (Goat polyclonal) | Abcam | Cat#ab53554 RRID: AB_880202 | IF (1:1,000) |
| Antibody | Anti-THP (Rat monoclonal) | R&D Systems | Cat#MAB5175 RRID: AB_2890000 | IF (1:1,000) |
| Antibody | Anti-CD13 (Goat polyclonal) | R&D Systems | Cat#AF2335 RRID: AB_2227288 | IF (1:500) |
| Antibody | Anti-Goat IgG AF488 (Bovine polyclonal) | Jackson ImmunoResearch | Cat#805-545-180 RRID: AB_2340883 | IF (1:500) |
| Antibody | Anti-Rat IgG AF555 (Donkey polyclonal) | Abcam | Cat#ab150154 RRID: AB_2813834 | IF (1:500) |
| Antibody | Anti-Rabbit IgG AF647 (Donkey polyclonal) | Jackson ImmunoResearch | Cat#711-605-152 RRID: AB_2492288 | IF (1:500) |
| Antibody | Anti-CD8α-AF700 (Clone 53–6.7) | Biolegend | Cat#100730 RRID:AB_493703 | FC (1:200) |
| Antibody | Anti-CD45-PerCP/Cy5.5 (Clone 30-F11) | Biolegend | Cat#103132 RRID:AB_893340 | FC (1:200) |
| Antibody | Anti-CD45-FITC (Clone 30-F11) | BD | Cat#553080 RRID:AB_394610 | FC (3 µg/mouse) |
| Antibody | Anti-CD4-PE (Clone RM4-5) | Biolegend | Cat#100512 RRID:AB_312715 | FC (1:200) |
| Antibody | Anti-CD4-BV421 (clone GK1.5) | BD | Cat#562891 RRID: AB_2737870 | FC (1:200) |
| Antibody | Anti-Rat IgG-APC (Goat polyclonal) | BD | Cat#551019 RRID:AB_398484 | FC (1:200) |
| Antibody | Anti-Mouse IgG-HRP (Goat polyclonal) | Bethyl Laboratories INC | Cat#A90-116P RRID:AB_67183 | ELISA (1:7,000) |
| Other | Fixable Viability Dye eFluor 780 | ThermoFisher | Ref#65-0865-14 | FC (1:1,000) |
| Other | Flow Cytometry Absolute Count Standard | Bangs Laboratories | Cat#580 | |
| Other | MuPyV (Strain A2) | N/A | N/A | |
| Other | Sheep Red Blood Cells | Innovative Research | Cat#ISHRBC10P15ML | |
| Peptide, recombinant protein | VP1 pentamers | Provided by Robert Garcea | N/A | |
| Peptide, recombinant protein | Benzonase Nuclease | Sigma | Cat#E1014 | |
| Peptide, recombinant protein | Neuraminidase from Vibrio cholerae | Sigma | Cat#N6514 | IF (1:100) FC (1:200) |
| Peptide, recombinant protein | Collagenase (Type I) | Worthington | Cat#LS004197 | |
| Chemical compound, drug | TRIzol Reagant | ThermoFisher | Ref#15596018 | |
| Chemical compound, drug | RevertAid H Minus Reverse Transcriptase | ThermoFisher | Cat#EP0451 | |
| Chemical compound, drug | Lipofectamine 2000 Transfection Reagent | ThermoFisher | Cat#11668030 | |
| Chemical compound, drug | Lipofectamine 3000 Transfection Reagent | ThermoFisher | Cat# L3000008 | |
| Commercial assay, kit | PFHM-II Protein-Free Hybridoma Medium | ThermoFisher | Ref#12040–077 | |
| Commercial assay, kit | TOPO TA Cloning Kit | ThermoFisher | Ref#45–0641 | |
| Commercial assay, kit | TBP PrimeTime XL qPCR Assay | IDT | Mm.PT.39a.22214839 | |
| Commercial assay, kit | 1-Step Ultra TMB-ELISA | ThermoFisher | Ref#34028 | |
| Commercial assay, kit | 96 Well EIA/RIA Polystyrene High Bind Microplate | Fisher Scientific | Cat#3590 | |
| Commercial assay, kit | PerfectCTa FastMix II ROX | Quantabio | P/N 84210 | |
| Commercial assay, kit | PureLink Viral RNA/DNA mini Kit | ThermoFisher | Ref#12280–050 | |
| Commercial assay, kit | Wizard Genomic DNA Purification Kit | Promega | Ref#A1120 | |
| Commercial assay, kit | QuikChange II Site-Directed Mutagenesis Kit | Agilent | Cat#200523 | |
| Commercial assay, kit | CELLine Disposable Bioreactor | Fisher Scientific | Cat#353137 | |
| Cell Line (M. musculus) | BALB/3T3 Clone A31 | ATCC | CCL-163; RRID:CVCL_0184 | |
| Cell Line (M. musculus) | NMuMG | ATCC | CRL-1636; RRID:CVCL_0075 | |
| Strain, strain background (M. musculus) | C57BL/6 Mice | National Cancer Institute | Cat#OIC55 | |
| Genetic reagent (M. musculus) | μMT Mice | Jackson Laboratory | Cat#002288; RRID:IMSR_JAX:002288 | |
| Software, algorithm | Prism | Graphpad | v 9.3.1; RRID:SCR_002798 | |
| Software, algorithm | FlowJo | BD | v 10.6.1; RRID:SCR_008520 | |
| Software, algorithm | ImageJ | NIH | v 1.8.0; RRID:SCR_003070 | |
| Software, algorithm | Leica LAS X | Leica | v 3.7.2; RRID:SCR_013673 |
Additional files
-
Supplementary file 1
VP1 mutations.
Identity, location, and frequency of detected VP1 mutations. Superscripted numbers indicate the VP1 loop in which the mutations are located (1: BC, 2: DE, 3: EF, 4: HI). Deletions are indicated by a “Δ” followed by the deleted residues; the identity of the deleted amino acids is indicated in “()”. The duplication of a residue is indicated with “dup.” The presence of two mutations in a virus is indicated with “+”. Sets of mutations separated by “and” indicate that both of the listed mutant viruses were isolated from the same mouse.
- https://cdn.elifesciences.org/articles/83030/elife-83030-supp1-v2.docx
-
Supplementary file 2
Oligonucleotide sequences.
Sequences of oligonucleotides used for site-directed mutagenesis, cloning, qPCR, and sequencing.
- https://cdn.elifesciences.org/articles/83030/elife-83030-supp2-v2.docx
-
Supplementary file 3
Statistical information.
Statistical tests used and exact p values for the comparisons of data presented in each figure as indicated.
- https://cdn.elifesciences.org/articles/83030/elife-83030-supp3-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/83030/elife-83030-mdarchecklist1-v2.docx





